NRMT1 is an N-terminal methyltransferase that methylates histone CENP-A as well as nonhistone substrates. Wu et al. report the crystal structure of human NRMT1 bound to CENP-A peptide at 1.3 Å. NRMT1 adopts a core methyltransferase fold that resembles DOT1L and PRMT but not SET domain family histone methyltransferases.
Keywords: histone modification, N-terminal methylation, NRMT1, CENP-A, crystal structure, epigenetic regulation
Abstract
NRMT1 is an N-terminal methyltransferase that methylates histone CENP-A as well as nonhistone substrates. Here, we report the crystal structure of human NRMT1 bound to CENP-A peptide at 1.3 Å. NRMT1 adopts a core methyltransferase fold that resembles DOT1L and PRMT but not SET domain family histone methyltransferases. Key substrate recognition and catalytic residues were identified by mutagenesis studies. Histone peptide profiling revealed that human NRMT1 is highly selective to human CENP-A and fruit fly H2B, which share a common “Xaa-Pro–Lys/Arg” motif. These results, along with a 1.5 Å costructure of human NRMT1 bound to the fruit fly H2B peptide, underscore the importance of the NRMT1 recognition motif.
Histone post-translational modifications (PTMs) carry critical epigenetic information that governs the decoding of the genetic message stored in the DNA sequence at the chromosomal level (Jenuwein and Allis 2001). Histone methylation, which usually occurs on side chains of histone lysine or arginine residues, is catalyzed by groups of histone lysine methyltransferases (KMTs) or protein arginine methyltransferases (PRMTs) (Campagna-Slater et al. 2011). It has been well documented that histone lysine and arginine methylations are important in regulating diverse cellular processes ranging from transcriptional regulation to high-order chromatin organization and repair (Bedford and Clarke 2009; Black et al. 2012). In addition to histone lysine/arginine side chain methylation, methylation can also occur at the histone α-N terminus. For instance, methylations on the N terminus of histone H2B have been reported in Tetrahymena (Nomoto et al. 1982), Arabidopsis thaliana (Bergmueller et al. 2007), Drosophila melanogaster (Desrosiers and Tanguay 1988), and other invertebrates (Webb et al. 2010). Recently, the N terminus of CENP-A, a centromere-specific histone H3 variant, was shown to be trimethylated by NRMT1, an α-N-methyltransferase. This type of methylation has been proposed to prompt the phasing of CENP-A nucleosomes on centromeric α-satellite DNA (Bailey et al. 2013).
α-N-methylations have been shown to occur in a wide range of proteins in both prokaryotes and eukaryotes. Human NRMT1 is the first α-N-methyltransferase identified in eukaryotes and methylates histone as well as nonhistone proteins. For example, in addition to CENP-A, the regulator of chromosome condensation 1 (RCC1), retinoblastoma protein, CENP-B, kelch-like protein 31, ribosomal protein L23a, myosin light chain 2, and myosin light chain 3 are all reported NRMT1 substrates (Okada et al. 2007; Tooley et al. 2010; Bailey et al. 2013). A sequence motif of “Xaa–Pro–Lys/Arg” (Xaa denotes small side chain amino acids) has been identified to be the recognition signature of NRMT1 (Petkowski et al. 2012). Although a complex structure of NRMT1 bound to S-adenosyl-L-homocysteine (SAH) modeled with RCC1 peptide has been reported (Tooley et al. 2010), an experimental ternary complex structure is lacking. Detailed molecular mechanisms regarding how NRMT1 recognizes and methylates its substrates, especially histone substrates, remain largely unexplored.
Here, we report ternary structures of human NRMT1 bound to α-N-methylated peptides of human histone CENP-A or fruit fly histone H2B in the presence of SAH. Our work provides the first structural insights into histone α-N-methylation by NRMT1 and sheds light on the catalytic properties of NRMT1 and other NRMT family members.
Results and Discussion
Overall structure of NRMT1
To decipher the substrate recognition and catalytic mechanisms of NRMT1, we solved the crystal structure of full-length human NRMT1 in a ternary complex with the α-N-dimethylated CENP-A peptide (1Gme2PRRRSRKP9) and SAH at 1.3 Å (Supplemental Table 1).
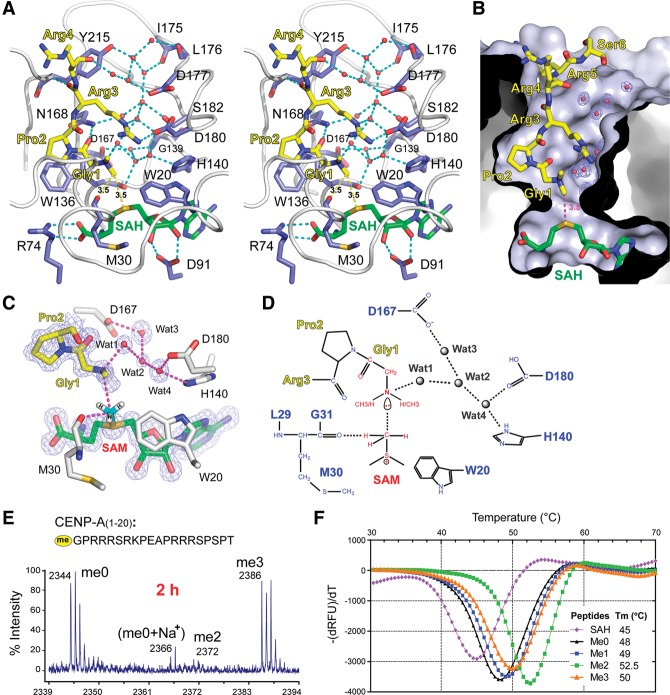
NRMT1 harbors a canonical SAM-dependent methyltransferase (SAM-MTase) core fold consisting of a seven-stranded β sheet (β1–β7) sandwiched by five flanking α helices (Fig. 1A,B). The enzyme also has several additional structural elements, including three helices (ηA, αB, and αC) from the N terminus and a pair of β hairpins (βAB) inserted between β5 and α6 of the core domain. These elements cluster on top of the core domain to form the CENP-A-binding platform and cover SAH as a lid (Fig. 1A,B). Our complex structure suggested that the methyl transfer takes place in a cleft formed by the CENP-A platform and the core domain base.
Figure 1.
Overall structure of the NRMT1–SAH–CENP-A ternary complex. (A) Topology of NRMT1. (Slate blue) Helices; (pink) β strands. CENP-A- and SAH-binding motifs are shaded yellow and green, respectively. CENP-A-interacting loops (LC, L4, LAB, and L67) are highlighted in cyan. (B) NRMT1 in complex with SAH and dimethylated CENP-A peptide. The CENP-A peptide and SAH are depicted as sticks. Color coding is the same as in A. (C) Composition of the CENP-A platform. Several segments of NRMT1 constitute the CENP-A peptide-binding pocket, including N-terminal helices (slate blue), loop L4 (green), βAB hairpin (wheat), and loop L67 (cyan). (D) Interaction details that enable an intimate integration of the CENP-A platform. Key residues are shown as sticks and are color-coded in the background of cartoon.
The CENP-A platform is stabilized by structural integration of the extended N-terminal segments (ηA, αB, αC, LC, and η1′), the βAB hairpin, and the L67 and L4 loops (Fig. 1C). Specifically, the N-terminal ηA and αB segments form extensive interactions with the βAB hairpin involving hydrophobic residues V5, I6, F12, V174, L176, and V183 and hydrogen-bonding pairs F12–S181 and K14–D178 (Fig. 1C,D). In parallel, the αC, LC, and η1′ segments cluster with the L67 and L4 loops to create the peptide-binding channel, integrated by residues L31, Y34, I37, W136, L210, P211, I214, V217, Y215, and E213 (Fig. 1C,D). Additionally, βAB and L67 are connected by a C184–Y215 stacking pair, which zips up the active pocket (Fig. 1D).
NRMT1 is conserved from yeast to humans. Structure-based sequence alignment and LigPlot (Laskowski and Swindells 2011) analyses (Supplemental Fig. 1) revealed that key residues responsible for CENP-A peptide and SAM/SAH recognition are highly conserved. Superimposition of NRMT1 with other histone methyltransferases revealed that NRMT1 adopts a core SAM-MTase fold similar to DOT1L and PRMT7 but not the SET domain methyltransferases (Supplemental Fig. 2). Interestingly, the SAM-MTase core domains superimpose well among NRMT1, PRMT7, and DOT1L. In contrast, the inserted motifs either at the N-terminal or within the core domain are highly divergent, which defines the peptide substrate specificity of NRMT1, PRMT7, and DOT1L (Supplemental Fig. 2C,D).
Molecular details for CENP-A recognition by NRMT1
In the ternary structure, the CENP-A peptide is inserted into an acidic pocket of NRMT1 (Fig. 2A). The pocket is relatively wide and contains two candidate peptide-binding channels (βAB and L67) (Fig. 2A). Our experimental structure revealed that the CENP-A peptide occupies the L67 channel, and a string of waters occupies the βAB channel, different from previous modeling studies (Tooley et al. 2010). The CENP-A pocket is largely preformed, and superimposition of the peptide-bound and free NRMT1 structures revealed little conformational change of the pocket residues (Supplemental Fig. 3).
Figure 2.
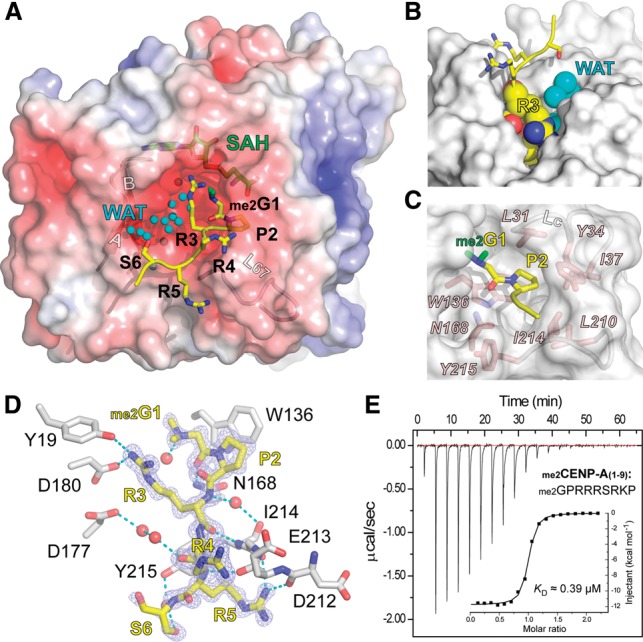
CENP-A peptide recognition by NRMT1. (A) Structure of NRMT1 bound to the dimethylated CENP-A peptide. NRMT1 is in surface view and is colored according to its electrostatic potential as a spectrum ranging from −6 kT/e (red) to +6 kT/e (blue). The CENP-A peptide is depicted as yellow sticks, with the two α-N-methyl groups highlighted in green. Note the two channels that are close to the βAB hairpin and the L67 loop. Waters (WAT) are depicted as smal cyan balls. (B) Burial of the CENP-A “G1–P2–R3” segment in the NRMT1 L67 channel. NRMT1 is represented as a gray surface. Residues G1, P2, and R3 and waters are shown as spheres. Note that the βAB channel is fully occupied with waters (cyan). (C) Pocket composition for “G1–P2” recognition. Key residues of NRMT1 are depicted as pink sticks. (D) Hydrogen-bonding network involved in CENP-A peptide recognition. The Fo–Fc omit map of CENP-A is contoured at the 2.5 σ level. Waters are depicted as red balls, and hydrogen bonds are shown as cyan dashes. (E) Calorimetric titration and fitting curves of N-dimethylated CENP-A titrated into NRMT1.
The first three residues of CENP-A (G1–P2–R3) are deeply buried in the active pocket and, together with the water molecules, fill up the pocket (Fig. 2A,B). Notably, the long side chain of R3 caps the α-N-dimethylated G1 from the top, seals the “G1–P2” step of CENP-A, and coordinates the active center formation (Fig. 2B). A motif of “Xaa–Pro–Lys/Arg” has been identified as a recognition signature for NRMT1. In our complex structure, G1 is positioned in a narrow pocket whose dimension is constrained by the LC loop (L31–Y34) (Fig. 2C), accounting for the requirement of small residues (Xaa) at position 1. Proline 2 has been biochemically demonstrated to be a key determinant for NRMT1 substrates. In the complex structure, P2 of CENP-A is snugly placed into a conserved hydrophobic pocket formed by residues L31, Y34, I37, W136, and I214 (Fig. 2C). The P2 pocket is relatively flat and shallow, best for accommodating a proline ring.
Besides hydrophobic contacts introduced by P2, multiple sets of polar interactions contribute to the specific recognition between NRMT1 and CENP-A. As illustrated in Figure 2D, G1 is stabilized by direct hydrogen bonding with N168 of NRMT1, and R3 forms hydrogen-bonding and electrostatic interactions with residues Y19, N168, D180, and Y215 of NRMT1. The relatively exposed CENP-A segment “R4–R5–S6” further contributes to enzyme-substrate recognition through direct or water-mediated hydrogen-bonding interactions involving residues D212, E213, I214, and Y215 from the L67 loop.
Using isothermal titration calorimetry (ITC), we measured a binding KD of 0.39 μM, ΔH of −11.8 kcal/mol, and ΔS of −10.2 cal/mol/deg between NRMT1 and the α-N-dimethylated CENP-A (1–9) peptide (Fig. 2E), underscoring the contributions of specific enzyme–substrate interactions revealed in our structure studies.
Mutagenesis studies
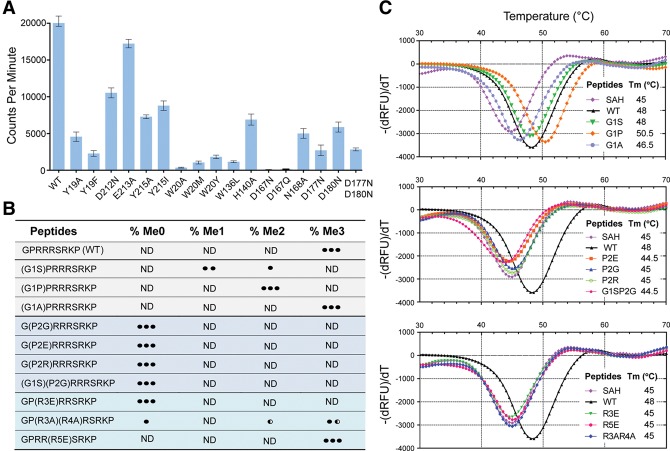
To determine the functional importance of the key residues involved in CENP-A binding, we next generated NRMT1 point mutants of Y19A, Y19F, W136L, N168A, D180N, D212N, E213A, Y215A, and Y215I and performed an in vitro tritium label methylation assay. As expected, these NRMT1 mutants displayed compromised enzymatic efficiency with retained activities ranging from 8% (W136L) to 85% (E213A) compared with wild-type enzyme (Fig. 3A). The fact that Y19, W136, N168, and D180 mutants were among the most affected mutations underscores the importance of these residues in the CENP-A “G1–P2–R3” motif recognition. Notably, an activity loss of ∼14-fold in the case of W136L suggests that, in addition to hydrophobicity, a planar shape at position 136 of NRMT1 is critical for P2 recognition through ring–ring stacking (Fig. 2C). Loop L67 mutants D212N, E213A, Y215A, and Y215I exhibited moderate activity reduction, suggesting an augmented role of these residues in peptide substrate recognition outside of the active center (Fig. 2D).
Figure 3.
Mutagenesis and biochemical analysis. (A) In vitro methyltransferase assay of wild-type and mutant NRMT1. Enzymatic activity was monitored by scintillation counts of [3H]-labeled CENP-A methylated products. (B) Summary of methylation products catalyzed by NRMT1 using native and mutant CENP-A peptides after 5 h of incubation. MALDI-TOF mass spectrometry (MS) was adopted to detect the product. (ND) Not detected. The number of black dots denotes the abundance of the methylated products. (Half-black dot) Half abundance. (C) Thermofluor shift assay (TSA) analysis of NRMT1 with wild-type and mutant CENP-A peptides (1–9). All assays were performed in the presence of saturated SAH. A peptide-free control curve is labeled as SAH.
To explore the importance of the CENP-A sequence motif in NRMT1 recognition, we further performed a mass spectrometry (MS)-based enzymatic assay using unmodified wild-type and different mutant CENP-A (1–9) peptides. As outlined in Figure 3B, mutation of G1 to small side chain residues, such as serine, proline, and alanine, retained the methylation capacity of these peptides, with methylation state preferences of G1P and G1A to full state (note that proline is an imino acid, and dimethylation represents its full methylation state) and G1S to α-N-monomethylation. In contrast, mutation of P2 to glycine, glutamate, or arginine completely abolished α-N-methylation, recapitulating the signature role of a proline at position 2. Substitution of R3 but not R5 with a glutamate residue abolished the methylation capacity of the CENP-A peptide, suggesting a critical role of the basic arginine residue at position 3 but not position 5. Alanine mutations of both R3 and R4 displayed compromised α-N-trimethylation, further highlighting the contribution of their positive charge.
We next performed a thermofluor shift assay (TSA) to evaluate the stabilization effect of wild-type and mutant CENP-A peptides on NRMT1. As summarized in Figure 3C, peptide-free yet SAH-saturated NRMT1 displayed a melting temperature (Tm) of 45°C, and the incubation with CENP-A (1–9) peptide further elevated the Tm by 3°C. Notably, all of the mutant peptides that lost methylation capacity displayed no or negligible stabilization effect of NRMT1, suggesting loss of peptide binding due to residue substitution. In contrast, G1A and G1P CENP-A mutants stabilized NRMT1 by 1.5°C and 5.5°C compared with that of the NRMT1–SAH binary complex, consistent with their retained methylation capacity. Despite α-N-monomethylation being the major product form for G1S (Fig. 3B), a 3°C stabilization effect was observed (Fig. 3C), which suggests that unmethylated G1S peptide is a good substrate for NRMT1. Failure to achieve trimethylation is likely due to the bulkier size of Ser (compared with Gly, Ala, and Pro), whose dimethylation conceivably precludes higher-state methylation due to the spatial constraint at the G1 pocket. R5E that displayed no stabilization effect could still undergo trimethylation (Fig. 3B); however, its methylation efficiency was reduced in our time-course MS-based methylation assay (data not shown).
Active site and catalysis
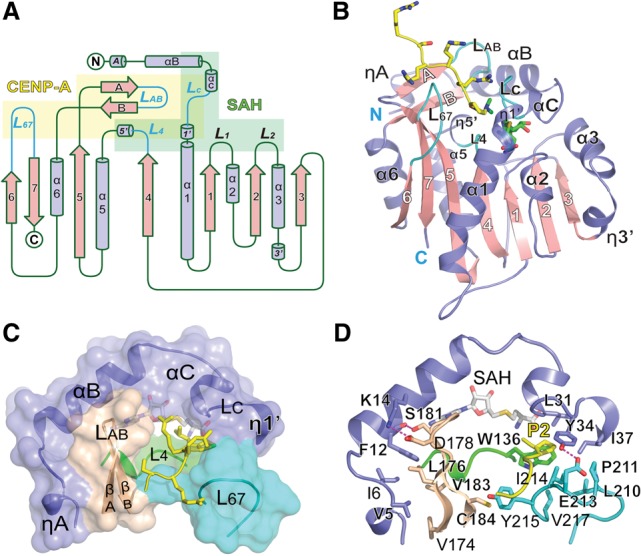
The α-N-dimethylated G1 is positioned at the catalytic center with one of its methyl groups pointing to the sulfur atom of SAH at a distance of ∼3.5 Å, representing a post-methyl transfer state of the catalytic cycle (Fig. 4A). In the catalytic pocket, SAH is sandwiched by αB–αC segments (W20–M30) and the L4 loop (W136–G139) and is further anchored by two pairs of hydrogen bonds with R74 and D91 (Fig. 4A). A network of water molecules occupying the βAB channel is notably stabilized by highly coordinated hydrogen-bonding interactions involving the following residues: I175, L176, D177, D180, and S182 from the βAB hairpin; W136, G139, and H140 from L4–η5′; D167 and N168 from β5; Y215 from L67; and me2G1 and R3 from the inserted CENP-A peptide (Fig. 4A). These waters are connected from the inner core of the active pocket all through to the solvent-exposed protein surface (Fig. 4B), which can serve as an ideal path for the release of protons generated by the methyl transfer reaction (Hammes-Schiffer and Tully 1994).
Figure 4.
Active site and catalytic mechanism. (A) Stereo view of the NRMT1 active site. Key residues involved in water network coordination and catalytic reaction are shown as slate-blue sticks. Substrate CENP-A and SAH are depicted as yellow and green sticks, respectively. Waters are shown as red balls. NRMT1 backbone is represented as a gray tube. (B) Cutaway view of the catalytic pocket. Note the deep insertion of CENP-A within the pocket. (Blue mesh) Fo–Fc omit map of waters contoured at the 2.5 σ level. (C) Geometry of the catalytic transition state with modeled SAM and α-N-monomethylated CENP-A. SAM is modeled by superimposition with SAH (green) in the crystal structure. Monomethylated CENP-A is generated by simple removal of a methyl group of the dimethylated peptide. Fo–Fc omit maps (blue mesh) contoured at the 2.5 σ level are calculated around SAH, CENP-A, and water molecules to indicate their positions in the crystal structure. (D) Schematic drawing of the transition state. For a stepwise catalytic process, please see Supplemental Figure 4. (E) MALDI-TOF MS analysis of CENP-A1-20 peptide products catalyzed by NRMT1 after 2 h. (F) TSA thermal melting curves of NRMT1 in complex with CENP-A peptides in different methylation states. The melting curve of the SAH-bound NRMT1 binary complex is colored violet and is shown as a blank control. All other curves are of NRMT1 ternary complexes with additional CENP-A peptide added to the binary complex.
The SN2 reaction has been proposed as a common mechanism for SAM-MTase, in which the methyl-acceptor group is deprotonated usually by a general base in the first position (Guo and Guo 2007). However, we were not able to identify any adjacent acidic or proton acceptor residue that may directly deprotonate the α-amino of CENP-A. Instead, we observed four water molecules that mediate a hydrogen-bonding network and connect the CENP-A α-amine to residues H140, D167, and D180 of NRMT1 (Fig. 4C). To explore the catalytic importance of these residues, we generated single-point mutants H140A, D167N, D167Q, and D180N and analyzed their activity using an in vitro methylation assay. The catalytic activities of H140A and D180N decreased threefold to fourfold, while D167N and D167Q completely lost their activities (Fig. 3A). We hypothesized that the deprotonation of CENP-A α-amine might be accomplished through the water chain by a “hydrogen-hopping” mechanism (Hammes-Schiffer and Tully 1994). D167 is deeply buried within the active pocket (Fig. 4A), rendering it a driving force for CENP-A α-amine deprotonation mediated by the “Wat3–Wat2–Wat1” water chain (Fig. 4C). The complete activity loss of D167N and D167Q suggests a dominating role of acidic D167 in this deprotonation process.
In order to fulfill a reaction cycle, the proton released by the methyl transfer reaction has to be exchanged to solvent. H140, D177, and D180 that line up the acidic surface of the βAB channel may facilitate this process in concert with the coordinated water network (Fig. 4A). Interestingly, D177N and D177N/D180N mutants displayed a pronounced activity drop (∼10-fold) compared with H140A (approximately threefold) and D180N (approximately fourfold) even though residue D180 but not D177 directly participates in CENP-A R3 recognition (Fig. 4A). These results suggest a critical role of acidic D177 in proton release for the sake of enzymatic turnover.
Based on the above structural and biochemical studies, we proposed a catalytic reaction model (Supplemental Fig. 4) in which the following occurs: (1) The reaction is initialized by water-mediated deprotonation of the α-amine driven by D167. (2) The resulting lone pair electron of α-amine executes a nucleophilic attack at the methyl group of SAM. (3) Formation of the transition state can be further stabilized by C–H…O hydrogen bonding between the main chain carbonyl of M30 and the methyl group (Fig. 4D). (4) After the methyl transfer, the protonated D167 releases the proton to solvent via the coordinated water network. Interestingly, the methyl donor SAM is stabilized within a deep pocket by stacking with the W20 and M30 side chains. Point mutants of W20A, W20M, and W20Y significantly decreased NRMT1 activity (Fig. 3A), suggesting the importance of the stacking interaction between the W20 side chain and the SAM sugar ring.
State-specific trimethylation of CENP-A by NRMT1
The α-amino group of CENP-A can undergo monomethylation, dimethylation, and trimethylation. To explore the methylation state specificity of NRMT1, we performed an in vitro methylation assay by incubating CENP-A peptide 1–20 with NRMT1 in the presence of SAM and analyzed the product by MS. After 2 h of incubation, the trimethylated CENP-A peptide peak was clearly detected. Interestingly, the product existed predominantly in the trimethylated form with almost no monomethylated or dimethylated intermediates even though nearly half of the CENP-A peptide was still unmethylated (Fig. 4E). The observed methylation pattern suggests that NRMT1 catalyzes CENP-A trimethylation in a processive manner.
We reasoned that the processivity of NRMT1 might relate to its binding preference to peptide substrates. We next performed TSA analysis to evaluate the methylation state dependency of NRMT1–CENP-A interactions. As expected, the Tm of the NRMT1–CENP-A complexes can be sorted in an order of me2 (52.5°C) > me3 (50°C) > me1 (49°C) > me0 (48°C) (Fig. 4F), suggesting that dimethylated CENP-A is a better substrate for NRMT1 than the monomethylated form and that monomethylated CENP-A is a better substrate than the unmodified one. Such substrate preference partly explains the observed processivity of NRMT1, as the methylated product will serve as a better substrate for the next cycle of methylation until a full methylation state is achieved. Trimethylated CENP-A is the final product catalyzed by NRMT1. As reflected by the reduced Tm (Fig. 4F), the complex of NRMT1 with the trimethylated CENP-A peptide is less stable, likely due to the bulkier size of the trimethyl group. Conceivably, this less stable association is good for product release and enzymatic turnover of NRMT1.
Enzymatic profiling of histone peptides by NRMT1
In order to explore the histone substrate specificity of NRMT1, we synthesized a series of nine-residue peptides corresponding to the N-terminal sequence motif of all known human histones and fruit fly histone H2B (DmH2B) and subjected them to methylation by NRMT1. MS analysis of the products revealed that out of 29 synthesized human histone peptides, only CENP-A could be methylated, demonstrating the high selectivity of NRMT1 to CENP-A (Supplemental Table 2). In fruit flies, the NRMT1 homolog dNTMT has been reported to dimethylate DmH2B (Villar-Garea et al. 2012). Our enzymatic assay confirmed dimethylation of DmH2B by human NRMT1 over an N-terminal sequence of “1PPKTSGKAA9” (Fig. 5A), suggesting enzymatic functional conservation of NRMT1 across species.
Figure 5.
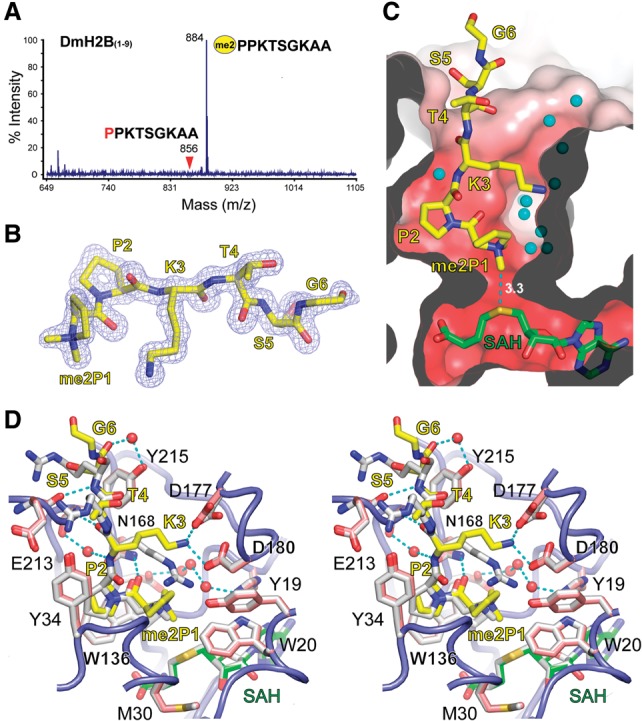
Ternary structure of NRMT1 bound to DmH2B and SAH. (A) MALDI-TOF MS spectrum of the dimethylated DmH2B peptide generated by NRMT1. (B) Fo–Fc omit map of α-N-dimethylated H2B contoured at the 2.5 σ level. (C) Cutaway view of the NRMT1 active pocket with bound DmH2B peptide, SAH, and waters. The figure is in the same orientation as Figure 4B. The active pocket is shown as an electrostatic potential surface. Red denotes a negatively charged surface. (D) Stereo view of superimposed NRMT1 ternary complexes bound to CENP-A/SAH (white) and DmH2B (yellow)/SAH (green). Key residues involved in DmH2B recognition are depicted as pink sticks, with the associated NRMT1 backbone shown as a blue tube.
Human histone H2B has very diverse N-terminal sequence motifs; however, none of them can be methylated by NRMT1. Sequence analysis revealed that, except for CENP-A, human H2B and other histones do not harbor the “Xaa–Pro–Lys/Arg” signature motif required for NRMT1 recognition. Particularly, most human H2B has a glutamate or arginine residue at position 2 that has been shown to abolish the methylation capacity of CENP-A (Fig. 3B). In contrast, the N-terminal motifs of DmH2B (“P–P–K”) and human CENP-A (“G–P–R”) well obey the “Xaa–Pro–Lys/Arg” rule. Collectively, this suggests that it is the recognition motif but not the histone type that governs the histone substrate specificity of NRMT1 and its orthologs.
Structural basis for fruit fly H2B recognition by NRMT1
We next crystallized the complex of NRMT1 bound to the DmH2B 1–9 peptide in the presence of SAM and solved the structure at 1.5 Å (Supplemental Table 1). Although unmodified peptide and SAM were used for crystallization, the dimethylated DmH2B and SAH were captured in the crystal structure, which represents a full methylation state after two cycles of methyl transfer. Based on the electron density, we were able to model residues 1–6 of the DmH2B peptide with two newly added methyl groups clearly traceable in the Fo–Fc omit map (Fig. 5B).
Similar to human CENP-A, DmH2B is docked in the L67 channel of an acidic substrate-binding pocket. At the center of the pocket, dimethylated proline 1 is deeply buried, with one of its methyl groups pointing to the sulfur group of SAH (Fig. 5C). Structure-based superimposition revealed a highly conserved recognition mode of both DmH2B and CENP-A by NRMT1 (Fig. 5D). Despite the sequence discrepancy, especially in residues 4–6 (1PPKTSG6 vs. 1GPRRRS6), DmH2B peptide adopts nearly the same main chain conformation as CENP-A and interacts with NRMT1 through direct or water-mediated hydrogen bonds. Correspondingly, all key residues required for peptide substrate recognition in NRMT1 are essentially unperturbed (Fig. 5D, cf. pink and white sticks). DmH2B has a lysine instead of an arginine at position 3. As an adaptation, the more flexible K3 forms direct hydrogen bonds with both D180 and D177, accompanied by slight side chain rotation of D177 (Fig. 5D). The observed ion pair interactions explain the preference of basic long side chain residues at position 3 of the NRMT1 substrate. Proline 2 of DmH2B is anchored in the same P2 pocket that has been described for CENP-A, again underscoring the importance of this consensus proline. Moreover, we observed no steric clash when P1 of DmH2B was positioned in the active center, endorsing the necessity of the small side chain residue at position 1.
Evolutionary conservation of histone α-N-modification
Human NRMT1 substrates include RCC1, CENP-A, and CENP-B. To our knowledge, RCC1, CENP-A, and CENP-B are all centromere related proteins. Strikingly, N-terminal sequence alignment from yeast to humans (Supplemental Table S3) suggested coevolution of NRMT1 recognition motifs in RCC1, CENP-A, and CENP-B, in which sequences “1SPKRIA6” of RCC1, “1GPRRRS6” of CENP-A, and “1GPKRRQ6” of CENP-B co-occur in mammals but are all missing in lower organisms. In contrast, the NRMT1 recognition motif of histone H2B is conserved from ciliates to insects but is lost in mammals. Remarkably, yeast and chicken orthologs of the above proteins do not harbor an NRMT1 recognition motif, suggesting that NRMT1 may exert its cellular function in these organisms through other protein substrates. The coevolution of α-N-modifications of RCC1, CENP-A, and CENP-B in mammals suggests an acquired modification-dependent regulation centered on centromere function and mitosis in adaptation to the increased organismal complexity. The conservation of histone H2B α-N-methylation in insects and ciliates suggests an important yet largely unexplored cellular function in these organisms, although such an H2B-centered regulation seemingly did not develop in mammals during evolution.
Materials and methods
Protein production and crystallographic studies
Wild-type and mutant human NRMT1 were recombinantly produced in Escherichia coli and purified as His-tagged proteins. Crystallization was performed via vapor diffusion method, and diffraction data were collected at Shanghai Synchrotron Radiation Facility BL17U under cryo conditions. Structures were determined by molecular replacement and refined by the program Phenix.
Enzymatic and binding assays
Radiometric filter assay and MALDI-TOF MS were used to measure the enzymatic activity of NRMT1 and its mutants. ITC and TSA were applied to measure binding between NRMT1 and its substrates.
Detailed descriptions of the Materials and Methods are provided in the Supplemental Material.
Accession codes
Coordinates have been deposited into Protein Data Bank under accession codes 5CVD for NRMT1–CENP-A–SAH and 5CVE for NRMT1–DmH2B–SAH ternary complexes, respectively.
Supplementary Material
Acknowledgments
We sincerely thank Dr. Jiahuai Han at Xiamen University for sharing NRMT1 cDNA. We thank the staff members at beamline BL17U of the Shanghai Synchrotron Radiation Facility, Dr. S. Fan at Tsinghua Center for Structural Biology for his assistance in data collection, and the China National Center for Protein Sciences Beijing for providing facility support. This work was supported by grants from the Major State Basic Research Development Program in China (2015CB910503 and 2011CB96303 to H.L.).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.270926.115.
References
- Bailey AO, Panchenko T, Sathyan KM, Petkowski JJ, Pai PJ, Bai DL, Russell DH, Macara IG, Shabanowitz J, Hunt DF, et al. 2013. Posttranslational modification of CENP-A influences the conformation of centromeric chromatin. Proc Natl Acad Sci 110: 11827–11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford MT, Clarke SG. 2009. Protein arginine methylation in mammals: who, what, and why. Mol Cell 33: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmueller E, Gehrig PM, Gruissem W. 2007. Charactorization of post-translational modifications of histone H2B-variants isolated from Arabidopsis thaliana. J Proteome Res 6: 3655–3668. [DOI] [PubMed] [Google Scholar]
- Black JC, Van Rechem C, Whetstine JR. 2012. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell 48: 491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagna-Slater V, Mok MW, Nguyen KT, Feher M, Najmanovich R, Schapira M. 2011. Structural chemistry of the histone methyltransferases cofactor binding site. J Chem Inf Model 51: 612–623. [DOI] [PubMed] [Google Scholar]
- Desrosiers R, Tanguay RM. 1988. Methylation of Drosophila histones at proline, lysine, and arginine residues during heat-shock. J Biol Chem 263: 4686–4692. [PubMed] [Google Scholar]
- Guo H-B, Guo H. 2007. Mechanism of histone methylation catalyzed by protein lysine methyltransferase SET7/9 and origin of product specificity. Proc Natl Acad Sci 104: 8797–8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammes-Schiffer S, Tully JC. 1994. Proton transfer in solution: molecular dynamics with quantum transitions. J Chem Phys 101: 4657. [Google Scholar]
- Jenuwein T, Allis CD. 2001. Translating the histone code. Science 293: 1074–1080. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Swindells MB. 2011. LigPlot+: multiple ligand–protein interaction diagrams for drug discovery. J Chem Inf Model 51: 2778–2786. [DOI] [PubMed] [Google Scholar]
- Nomoto M, Kyogoku Y, Iwai K. 1982. N-trimethylalanine, a novel blocked N-terminal residue of Tetrahymena histone-H2B. J Biochem 92: 1675–1678. [DOI] [PubMed] [Google Scholar]
- Okada T, Ohzeki J-I, Nakano M, Yoda K, Brinkley WR, Larionov V, Masumoto H. 2007. CENP-B controls centromere formation depending on the chromatin context. Cell 131: 1287–1300. [DOI] [PubMed] [Google Scholar]
- Petkowski JJ, Schaner Tooley CE, Anderson LC, Shumilin IA, Balsbaugh JL, Shabanowitz J, Hunt DF, Minor W, Macara IG. 2012. Substrate specificity of mammalian N-terminal α-amino methyltransferase NRMT. Biochemistry 51: 5942–5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooley CE, Petkowski JJ, Muratore-Schroeder TL, Balsbaugh JL, Shabanowitz J, Sabat M, Minor W, Hunt DF, Macara IG. 2010. NRMT is an α-N-methyltransferase that methylates RCC1 and retinoblastoma protein. Nature 466: 1125–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar-Garea A, Forne I, Vetter I, Kremmer E, Thomae A, Imhof A. 2012. Developmental regulation of N-terminal H2B methylation in Drosophila melanogaster. Nucleic Acids Res 40: 1536–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb KJ, Lipson RS, Al-Hadid Q, Whitelegge JP, Clarke SG. 2010. Identification of protein N-terminal methyltransferases in yeast and humans. Biochemistry 49: 5225–5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.