In this study, Rast-Somssich et al. investigated morphological differences between C. hirsuta, which has complex leaves with leaflets, and its relative, A. thaliana, which has simple leaves. By transferring single genes from one species into another under their endogenous regulatory elements, the authors show that leaf form can be modified in the recipient species, extending our knowledge of how paralogous genes are regulated in a complex eukaryote.
Keywords: KNOXI genes, regulatory evolution, CUP-SHAPED COTYLEDON, Cardamine hirsuta, pleiotropy, compound leaf
Abstract
Two interrelated problems in biology are understanding the regulatory logic and predictability of morphological evolution. Here, we studied these problems by comparing Arabidopsis thaliana, which has simple leaves, and its relative, Cardamine hirsuta, which has dissected leaves comprising leaflets. By transferring genes between the two species, we provide evidence for an inverse relationship between the pleiotropy of SHOOTMERISTEMLESS (STM) and BREVIPEDICELLUS (BP) homeobox genes and their ability to modify leaf form. We further show that cis-regulatory divergence of BP results in two alternative configurations of the genetic networks controlling leaf development. In C. hirsuta, ChBP is repressed by the microRNA164A (MIR164A)/ChCUP-SHAPED COTYLEDON (ChCUC) module and ChASYMMETRIC LEAVES1 (ChAS1), thus creating cross-talk between MIR164A/CUC and AS1 that does not occur in A. thaliana. These different genetic architectures lead to divergent interactions of network components and growth regulation in each species. We suggest that certain regulatory genes with low pleiotropy are predisposed to readily integrate into or disengage from conserved genetic networks influencing organ geometry, thus rapidly altering their properties and contributing to morphological divergence.
One approach to understand the genetic basis for evolutionary change is to identify genes that underlie morphological diversity and investigate how those evolved and how their diversification influenced morphogenesis. In this context, there is considerable interest in determining whether genes or genetic changes underlying morphological differences between species share unifying features, which would indicate that evolution is, to an extent, predictable (Williams et al. 2008; Gompel and Prud'homme 2009; Stern and Orgogozo 2009; Chan et al. 2010; Prud'homme et al. 2011). Current evidence suggests that morphological evolution often results from changes in gene expression of key developmental regulators. Such mutations facilitate morphological change while minimizing the potentially adverse effects of pleiotropy; i.e., the phenomenon by which a single gene influences multiple traits. In this way, regulatory evolution, constrained by pleiotropy, can drive morphological change in specific traits without reducing organismal fitness (Williams et al. 2008; Stern and Orgogozo 2009; Chan et al. 2010; Prud'homme et al. 2011). However, the precise influence of pleiotropy in determining the relative evolutionary potential of different genes remains unclear. Paralogous genes with overlapping functions but different levels of pleiotropy offer an attractive opportunity to investigate this problem. If the evolutionary potential of such genes is highly constrained by their pleiotropy, then we expect the more pleiotropic gene to incur a higher fitness penalty when diversifying. Consequently, we expect this gene to evolve variants that make only modest contributions to trait diversity. In contrast to this, the less pleiotropic paralog with comparable developmental function would be less constrained, and we expect this gene to evolve variants that make a greater contribution to trait diversification.
It is clear that regulatory divergence supports trait evolution and can contribute to the assembly of new genetic modules influencing morphology (Raff 1996; Doebley and Lukens 1998; Carroll 2005; Gompel et al. 2005; Arnoult et al. 2013; Rebeiz et al. 2015). However, the specific impact of diversification at the individual gene level on genetic network architecture remains poorly understood. One well-established possibility is module reuse, whereby broadly conserved genetic interactions are redeployed in space and/or time due to altered gene expression, resulting in morphological diversity (Arthur 2002; Carroll 2008; Mallarino and Abzhanov 2012). Alternatively, the modified expression of a developmental regulator could result in a more radical reorganization of genetic networks through novel genetic interactions, thus amplifying the potential for regulatory changes at a single locus to generate morphological diversity. This possibility remains underexplored owing to the relative paucity of comparative experimental systems that allow developmentally fine-grained investigation into how species-specific genetic variants cause phenotypic diversity.
Leaves of seed plants provide attractive opportunities to study these problems because they show a tremendous degree of heritable, morphological variation (Bar and Ori 2014, 2015). Leaf shapes can be classified as simple (if the blade is entire, as in the model species Arabidopsis thaliana) or dissected, also referred to as compound (if the blade is divided into distinct leaflets, as in Cardamine hirsuta) (Efroni et al. 2010). Leaves initiate as entire structures at the flanks of the pluripotent shoot apical meristem (SAM), but, in some species, elaboration of novel axes of cell proliferation results in leaflet formation (Ori et al. 2007; Barkoulas et al. 2008). Additionally, the margins of both simple and compound leaves can elaborate less-pronounced incisions, referred to as serrations or lobes depending on their depth (Fig. 1A; Blein et al. 2008; Hasson et al. 2011; Rubio-Somoza et al. 2014).
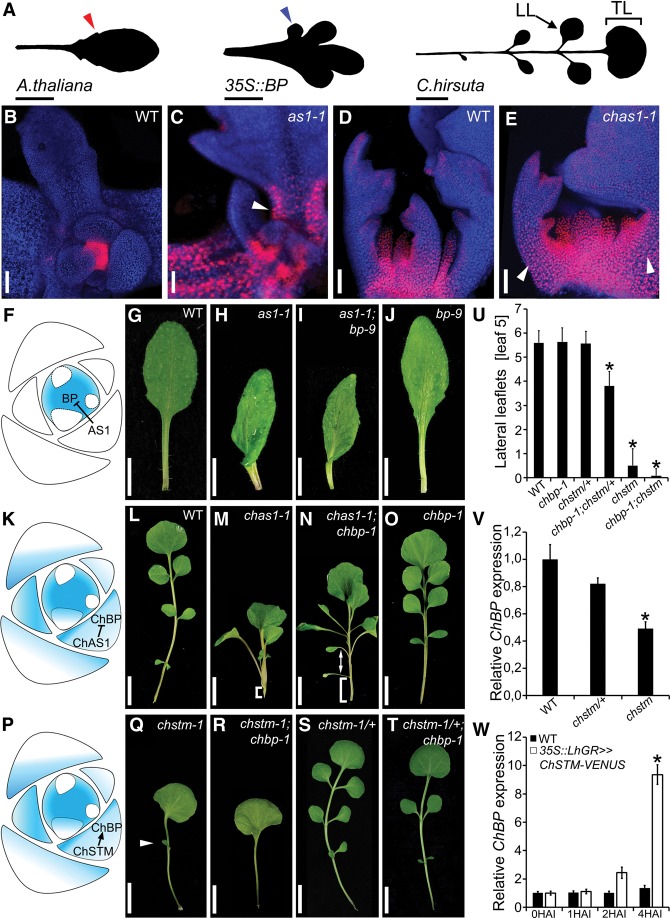
Figure 1.
BREVIPEDICELLUS (BP) repression by AS1 is conserved between A. thaliana and C. hirsuta but has different phenotypic significance in the two species. (A) Leaf 5 silhouettes of A. thaliana wild type with marginal serrations (red arrowhead); plants expressing BP under the 35S promoter, causing the formation of marginal lobes (blue arrowhead); and C. hirsuta wild type with lateral (LL) and terminal (TL) leaflets. (B–E) BP::VENUS (B,C) and ChBP::VENUS (D,E) expression (red) combined with chlorophyll autofluorescence (blue) in A. thaliana (B,C) and C. hirsuta (D,E) wild-type and as1-1 mutant plants. (C,E) Note the broadened BP/ChBP expression in as1-1 and chas1-1 mutants relative to the respective wild type (arrowheads). (F) Cartoon of an A. thaliana shoot apex: AS1 restricts BP expression from the leaf primordia. (G–J) Leaf 5 of A. thaliana wild type (G) and as1-1 (H), as1-1;bp-9 (I), and bp-9 (J) mutants. (K) Cartoon of a C. hirsuta shoot apex: Both ChAS1 and ChBP are expressed in leaves of C. hirsuta. Nevertheless, the repressive interaction is conserved. (L–O) Leaf 5 of C. hirsuta wild type (L) and chas1-1 (M), chas1-1;chbp-1 (N), and chbp-1 (O) mutants. Note the suppression of petiole growth arrest (bracket in M,N) and leaflet positioning defects along the proximodistal axis (arrow in N) in chas1-1;chbp-1 compared with chas1-1 mutant leaves. (P) Cartoon of a C. hirsuta shoot apex: ChSTM promotes ChBP expression during leaflet development. (Q–T) Leaf 5 of C. hirsuta chstm-1 (Q; arrowhead indicates a rare leaflet), chstm-1;chbp-1 (R), chstm-1/+ (S), and chstm-1/+;chbp-1 (T) mutants. (U) Quantification of lateral leaflets on leaf 5 of plants with the indicated genotype. n ≥ 25. (V,W) ChBP transcript levels in C. hirsuta chstm-1/+ and chstm-1 mutants (V; 14 d after germination [14DAG]; n = 3) and upon induction of ChSTM misexpression with 10 mM DEX in 35S::LhGR>>ChSTM-VENUS C. hirsuta seedlings (W; 14DAG; n = 3). Error bars in U–W indicate standard deviation. (HAI) Hours after induction; (asterisk) statistically significant difference from wild-type (U,V) or uninduced (W) samples (P ≤ 0.05, Student's t-test).
Regulators of leaflet development have been identified in several taxa, but the genetic basis for species-specific leaflet formation remains poorly understood (Blein et al. 2008, 2010; Ben-Gera and Ori 2012; Bar and Ori 2014, 2015). So far, genetic variation in two pathways has been causally connected to differences between simple and dissected leaves. The first involves local growth restriction that promotes leaflet separation and requires the REDUCED COMPLEXITY (RCO) HD-ZIP I gene, which was discovered in C. hirsuta (Vlad et al. 2014) RCO evolved through duplication in the Brassicaceae family, and its species-specific activity in leaf diversity results from its unique expression pattern at the base of initiating leaflets. Conversely, loss of RCO from the A. thaliana genome contributed to leaf simplification in this species (Sicard et al. 2014; Vlad et al. 2014). The second and more extensively studied pathway involves differential expression of class I KNOTTED1-LIKE HOMEOBOX (KNOXI) homeodomain proteins (Hareven et al. 1996; Bharathan et al. 2002; Hay and Tsiantis 2006; Blein et al. 2008; Shani et al. 2009; Furumizu et al. 2015). In most simple-leafed species, including A. thaliana, KNOXI proteins are confined to the meristem, where they prevent differentiation (Jackson et al. 1994; Lincoln et al. 1994; Smith et al. 1995; Long et al. 1996). Conversely, in many dissected-leafed species, including C. hirsuta, KNOXI proteins also accumulate in leaves, where they promote leaflet development (Bharathan et al. 2002; Hay and Tsiantis 2006, 2010). This differential expression of KNOXI genes between simple and dissected leaves results from cis-regulatory divergence of KNOXI loci (Hay and Tsiantis 2006). However, the regulatory logic underlying KNOXI-dependent diversification of leaf morphology remains poorly understood. For example, it is untested whether KNOXI genes are sufficient to increase the complexity of simple leaves when expressed from their native regulatory sequences that confer expression in dissected leaves. Such experiments are important, as they are the best available test to evaluate the contribution of genes or regulatory sequences to trait diversification between related species (Chan et al. 2010; Arnoult et al. 2013; Stern and Frankel 2013; Vlad et al. 2014; Rebeiz et al. 2015). Moreover, although upstream components of the KNOXI pathway have been identified (Timmermans et al. 1999; Tsiantis et al. 1999; Byrne et al. 2000; Ori et al. 2000; Ge et al. 2014), it is unclear how the correct KNOXI expression domain in dissected leaves is precisely delimited.
Here we studied the contribution of SHOOTMERISTEMLESS (STM) and BREVIPEDICELLUS (BP)—two redundantly acting, paralogous KNOXI genes—to leaf shape diversity between C. hirsuta and A. thaliana. Using comparative genetics and cross-species gene transfer assays, we show that the less pleiotropic gene, BP, has a higher potency to modify leaf form. We found that the cis-regulatory properties of BP that underlie its species-specific expression also influence its genetic interactions with conserved regulators of leaf development. Specifically, in the C. hirsuta leaf, ChBP is concurrently regulated by the microRNA164A (MIR164A)/ChCUP-SHAPED COTYLEDON (ChCUC) module and ChASYMMETRIC LEAVES1 (ChAS1), thus creating a regulatory linkage between MIR164A/CUC/AS1 that does not occur in A. thaliana leaves (Ori et al. 2000; Hay and Tsiantis 2006). We show that this particular regulatory architecture creates novel developmental boundaries that influence leaf shape. Our findings illustrate how the cis-regulatory properties of an individual gene may have been influenced by gene pleiotropy to create novel regulatory interactions with considerable impact on leaf morphology.
Results
Repression of ChBP expression in dissected leaves of C. hirsuta
We first compared the regulation of BP between A. thaliana and C. hirsuta. In A. thaliana, the AS1 MYB protein prevents BP transcription in simple leaf primordia (Waites et al. 1998; Byrne et al. 2000, 2002; Ori et al. 2000; Guo et al. 2008). In contrast, ChBP is transcribed in dissected C. hirsuta leaves despite being negatively regulated by ChAS1 (Hay and Tsiantis 2006). To analyze BP and ChBP expression at cellular resolution, we constructed fluorescent reporter gene fusions in both species. We observed ectopic expression of both BP::VENUS and ChBP::VENUS in as1 mutant leaves of A. thaliana and C. hirsuta, respectively (Fig. 1B–E, arrowheads). Moreover, we found that both reporter genes showed broadened and elevated expression in as1 leaves of A. thaliana, suggesting that their divergent cis-regulatory properties do not affect their negative regulation by AS1 (Supplemental Fig. S1). Thus, the AS1/BP interaction is conserved but has different developmental significance in the two species. In A. thaliana, AS1 excludes BP transcription from leaves to safeguard leaf development from inappropriate expression of a meristem gene (Ori et al. 2000; Byrne et al. 2002). In C. hirsuta, ChBP is expressed in leaves due to its cis-regulatory properties, and ChAS1 defines its expression pattern and dose (Fig. 1F,K).
To understand the consequences of this differential deployment of the AS1/BP module for leaf development, we compared the significance of this repressive interaction for morphology in the two species. In A. thaliana, the leaf phenotypes of as1;bp double mutants do not deviate appreciably from as1 single mutants (Fig. 1G–J) because other genes, including BP paralogs, contribute to the as1 mutant phenotype (Byrne et al. 2000; Ori et al. 2000; Ikezaki et al. 2010). We reasoned that chbp loss-of-function alleles might condition stronger suppression of the chas1 phenotype because both genes are active in the leaf of C. hirsuta. To test this hypothesis in an unbiased fashion, we conducted a genetic screen for suppressors of chas1, from which we recovered a loss-of-function chbp allele (Fig. 1L–O; Supplemental Fig. S2A,B). Quantification of the chas1;chbp double-mutant phenotype revealed that repression of growth along the proximodistal axis of chas1 leaves is strongly ChBP-dependent, indicating that ectopic ChBP expression contributes to the chas1 mutant leaf phenotype. Conversely, reanalysis of as1;bp double mutants in A. thaliana Col-0 showed only very subtle effects on proximodistal leaf growth, as did as1;bp double mutants in the A. thaliana Ler ecotype, confirming that this double-mutant phenotype is not allele- or background-specific (Table 1).
Table 1.
Quantification of leaf length, petiole length, petiole cell length, and petiole cell number on leaf 5 of A. thaliana and C. hirsuta wild-type and mutant plants
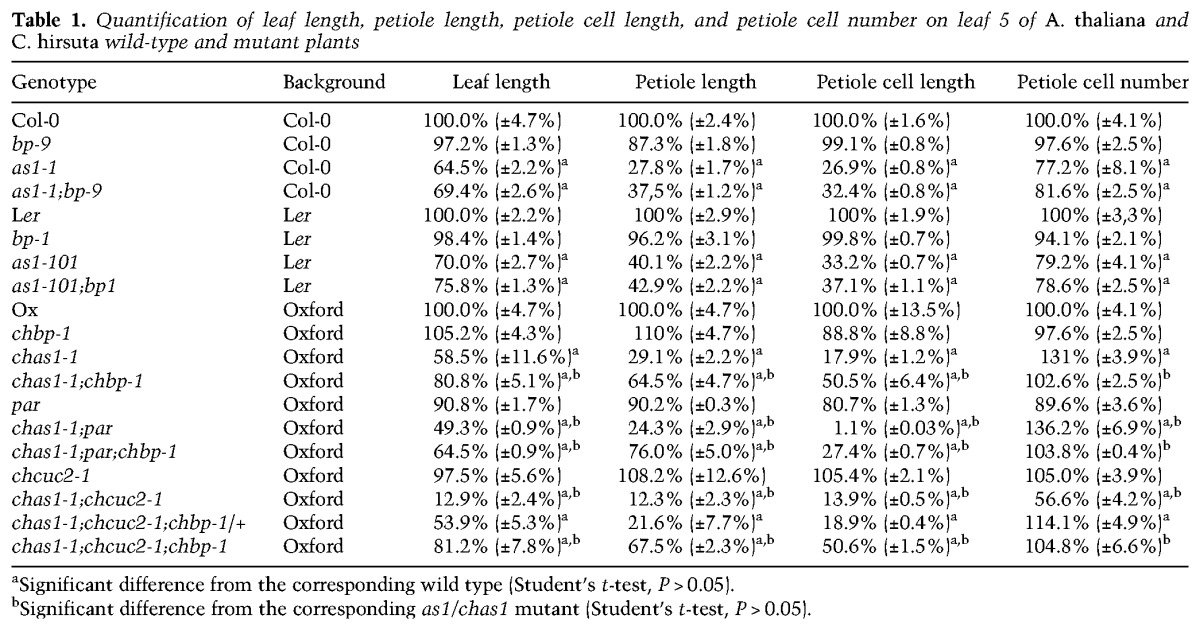
We next investigated whether the reduced length of chas1 and as1 mutant leaves reflects a reduction in cell proliferation or cell expansion. To this end, we analyzed epidermal cell size and number along the adaxial leaf surface. We found that leaf epidermal cells of chas1 and as1 mutants fail to elongate, particularly in the leaf petiole. This defect is strongly suppressed in C. hirsuta chas1;chbp but not in A. thaliana as1;bp double mutants (Table 1). Thus, ChAS1-dependent regulation of ChBP expression is required to define the correct timing of leaf cell elongation and differentiation and, consequently, growth along the proximodistal axis. These observations indicate that ChBP accounts for a considerable proportion of ChAS1 function in C. hirsuta leaves, suggesting that this interaction is more important for leaf growth and development in C. hirsuta than in A. thaliana.
ChBP and ChSTM act redundantly to promote leaflet formation
To further investigate the function of the ChAS1/ChBP module in C. hirsuta leaf development, we evaluated the role of ChBP in leaflet formation. chbp single mutants do not show leaflet number or positioning defects (Fig. 1O), indicating that other genes likely act redundantly with ChBP in C. hirsuta leaflet formation. Previous work indicated that ChSTM is required for leaflet formation and that cis-regulatory differences contribute to ChSTM expression in C. hirsuta leaves and exclusion of STM expression from A. thaliana leaves (Hay and Tsiantis 2006). Consequently, we postulated that ChBP and ChSTM might act redundantly to promote leaflet production. This would be in line with redundant functions of the related genes BP and STM in A. thaliana SAM maintenance and Rough sheath1 and Knotted1 in maize shoot development (Byrne et al. 2002; Bolduc et al. 2014). We tested this hypothesis with a hypomorphic chstm allele that we isolated from a genetic screen for mutants with reduced leaflet formation. Both the homozygous chstm single mutant and the chstm;chbp double mutant had meristem defects but retained some ability to make leaves (Supplemental Fig. S2D–G). However, the frequency of leaf formation was significantly lower in chstm-1;chbp than in chstm mutants. Furthermore, chstm/+ had a dose-dependent effect on leaflet formation in a chbp background, as chstm/+;chbp plants produced significantly fewer lateral leaflets than wild type without influencing SAM function (Fig. 1P–U; Supplemental Fig. S2H–J,L). Thus, ChBP acts redundantly with ChSTM to promote meristem function and leaflet formation. The basis of this redundancy is likely multifaceted, as ChSTM is a positive regulator of ChBP expression in leaves (Fig. 1V,W; Supplemental Fig. S2M), and previous work indicated that STM and BP physically interact (Smith and Hake 2003).
Although chstm mutants show leaflet defects, chbp mutants only show such defects in a chstm/+ background. This observation indicates that while ChSTM and ChBP act redundantly to promote leaflet formation, there is a stricter requirement for ChSTM function. STM has a broader role than BP throughout development in both A. thaliana and C. hirsuta, as seen from the pronounced SAM and organogenic defects and infertility found in stm but not bp mutants (Supplemental Fig. S2B–E; Endrizzi et al. 1996; Byrne et al. 2002; Hay et al. 2002; Smith and Hake 2003). In conclusion, the two KNOXI genes ChSTM and ChBP redundantly promote leaflet development, but ChSTM has a more central role in the process and more pleiotropic effects during development.
The ability of ChBP and ChSTM to alter A. thaliana leaf shape: evidence for a tradeoff between gene pleiotropy and potency
We next investigated how the difference in pleiotropy of ChBP and ChSTM associates with the sufficiency of each of the two loci to alter leaf shape from simple to more complex. To this end, we introduced two transgenes, ChBP::ChBP-VENUS (ChBP-V) and ChSTM::ChSTM-VENUS (ChSTM-V), into wild-type A. thaliana plants and evaluated the relative potency of each gene to shift the morphology of the recipient species (A. thaliana with simple leaves) to that of the species of origin (C. hirsuta with dissected leaves). Because both transgenes complemented their respective loss-of-function phenotypes in C. hirsuta (Supplemental Fig. S2K,N–R), we reasoned that they drive KNOXI expression in A. thaliana from a cis-regulatory context faithful to their native one in C. hirsuta. Morphological analysis of the resulting transgenic lines demonstrated that expression of both ChBP-V and ChSTM-V was sufficient to shift the A. thaliana simple leaf to a more complex form. However, ChBP-V was considerably more potent than ChSTM-V in altering A. thaliana leaf morphology despite the more stringent requirement for ChSTM in C. hirsuta leaflet development (Fig. 2A,B).
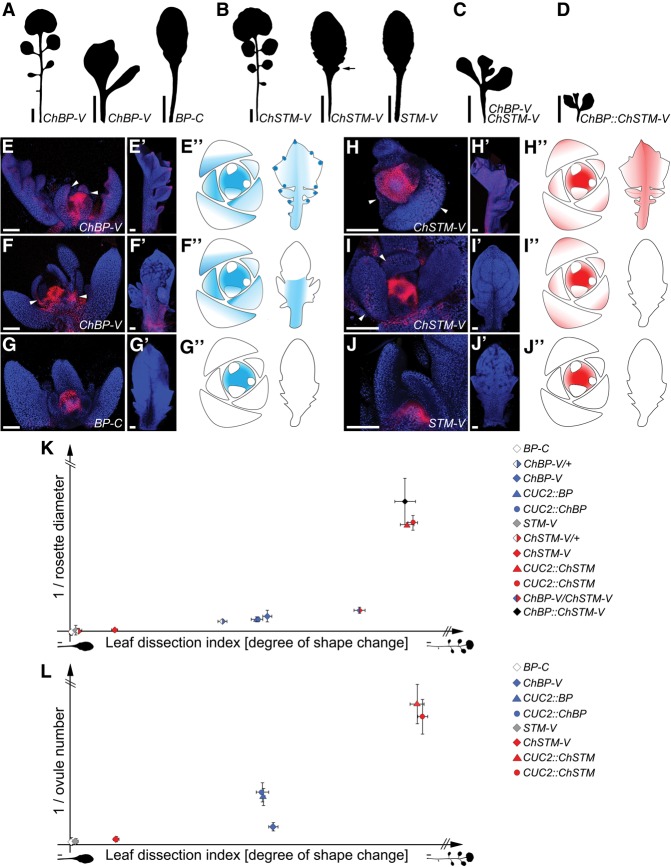
Figure 2.
ChBP is less pleiotropic but more potent in altering A. thaliana leaf shape than ChSTM. (A–D) Leaf 5 silhouettes of transgenic C. hirsuta (first silhouette in A,B) and A. thaliana (second and third silhouettes in A,B; C,–D) lines expressing ChBP::ChBP-VENUS (ChBP-V), BP::BP-CFP (BP-C), ChSTM::ChSTM-VENUS (ChSTM-V), STM::STM-VENUS (STM-V), both ChBP-V and ChSTM-V (C), or ChBP::ChSTM-VENUS (ChBP-ChSTM-V) (D). (E–J′′) Maximum intensity projections of confocal stacks showing reporter gene expression (red) combined with chlorophyll autofluorescence (blue) (E–J′) and cartoons of a shoot apex and a leaf (500 µm) summarizing the observed expression in transgenic C. hirsuta and A. thaliana lines (E′′–J′′). ChBP-V expression in C. hirsuta (E–E′′) and A. thaliana (F–F′′). (G–G′′) BP-C expression in A. thaliana. ChSTM-V expression in C. hirsuta (H–H′′) and A. thaliana (I–I′′). (J–J′′) STM-V expression in A. thaliana. Expression of ChBP-V and ChSTM-V is detectable in the A. thaliana SAM and leaves (arrowheads in E,F,H,I indicate leaf-specific expression), but ChSTM-V expression is not sustained after the leaf reaches a size of 400 µm (I′). (K,L) Diagrams depicting the degree of leaf shape change (calculated as leaf dissection index) versus the reduction in rosette diameter (K) or ovule number (L) caused by each transgene (Supplemental Fig. S3P–R). Genotypes are indicated in the key. To evaluate the effect of transgene zygosity in ChBP-V/ChSTM-V plants, the ChBP-V and ChSTM-V homozygous lines were backcrossed to wild type (ChBP-V/+ and ChSTM-V/+) and analyzed in the F1. Bars: A–D, 1 cm; E–J′, 100 µm.
We investigated whether the differences in potency to alter leaf shape between ChBP-V and ChSTM-V could be explained by differences in expression of the two transgenes. We found ChBP-V expression in the SAM and leaves of both C. hirsuta and A. thaliana (Fig. 2E–E′′,F–F′′), while an A. thaliana BP::BP-CFP (BP-C) reporter generated expression only in the SAM (Fig. 2G–G′′). Analysis of ChSTM-V showed expression in the SAM and developing leaf primordia in both species (Fig. 2H–H′′,I–I′′), whereas the A. thaliana promoter drove expression only in the SAM (Fig. 2J–J′′). However, in contrast to C. hirsuta, ChSTM-V expression in A. thaliana was not detectable after leaf primordia grew beyond a size of 400 µm (Fig. 2I′). This observation could be explained by factors active in C. hirsuta but not A. thaliana leaves to maintain ChSTM-V expression or repressors present only in A. thaliana that down-regulate ChSTM-V expression at later stages of leaf development. This premature cessation of ChSTM-V expression likely explains its weaker effect on A. thaliana leaf development. In contrast to ChSTM-V, the regulatory information in ChBP-V is sufficient to drive sustained expression throughout leaf development in a pattern similar to that observed at the adaxial site of C. hirsuta leaves and endow A. thaliana with a complex leaf partly resembling that of the donor species C. hirsuta. In conclusion, ChSTM and ChBP act redundantly to promote leaflet formation in C. hirsuta, and it is the least pleiotropic of these genes, ChBP, that is more potent in changing A. thaliana leaf form in a cross-species gene transfer assay.
These results suggest an inverse relationship between the pleiotropy of ChSTM and ChBP and the ability of these genomic loci to alter A. thaliana leaf shape, which is largely determined by their expression properties. To compare the ability of each protein to increase leaf complexity, we expressed STM/ChSTM and BP/ChBP in the restricted CUC2 marginal domain of A. thaliana lateral organs. Each transgene considerably increased leaf complexity and caused the formation of leaflet-like structures, confirming that the subtle phenotype of the ChSTM-V construct in A. thaliana is not a result of reduced protein function (Supplemental Fig. S3A–K). We revealed an enhancement of the ChBP-V leaf phenotype in A. thaliana by additional expression of the ChSTM-V transgene, which further indicates that both genes have independent effects on leaf morphology, albeit with different severities (Fig. 2C; Supplemental Fig. S3L). To directly test the contribution of regulatory sequences to the potency of these KNOXI genes to influence leaf development, we expressed ChSTM-VENUS from the ChBP promoter. We found that this transgene considerably increased A. thaliana leaf complexity compared with ChSTM-V but also severely perturbed plant growth (Fig. 2D; Supplemental Fig. S3M). In fact, the change in leaf shape in the ChBP::ChSTM-V and CUC2::STM/ChSTM lines is associated with severely arrested leaf and plant growth, but this is not the case in ChBP-V and CUC2::ChBP lines (Supplemental Fig. S3N–Q). Thus, ChSTM/STM-mediated changes in leaf geometry are associated with a higher penalty for the normal progression of plant development. This observation is visualized in Figure 2K, where the change in A. thaliana leaf shape in response to each BP or STM transgene is quantified relative to the arrest of plant rosette growth caused by this transgene. Comparable results were obtained by quantifying ovule number in selected genotypes (Fig. 2L; Supplemental Fig. S3R). In conclusion, comparative consideration of the consequences of STM/ChSTM and BP/ChBP gain of function shows that the increased leaf complexity caused by STM/ChSTM is concomitant with broader developmental defects. Together with the more pervasive defects caused by loss of STM/ChSTM compared with BP/ChBP, our results indicate that STM orthologs are more pleiotropic than their BP paralogs. We propose that lower pleiotropy may have allowed ChBP to evolve a higher level of evolutionarily relevant cis-regulatory activity in leaves. This activity is revealed by the ChBP locus being more potent than ChSTM in modifying A. thaliana leaf form toward a complex shape in our cross-species gene transfer experiments. Consistent with the view that STM evolved in a more constrained fashion than BP, crucifer STM sequences are more conserved than their BP counterparts in both coding and noncoding regions (Supplemental Fig. S3S–X; Aguilar-Martinez et al. 2015).
Dissection of the genetic networks influencing ChBP expression
Our results show that regulation of BP expression may be a central process on which evolution acts to influence leaf morphology. To gain insight into ChBP regulation in dissected leaves, we sought to identify additional mutants with increased leaflet number phenotypes conditioned by elevated ChBP expression (Hay and Tsiantis 2006). We isolated the parsley (par) mutant and found broadened ChBP expression in leaves, suggesting that PAR may influence leaf shape by controlling ChBP expression (Fig. 3A–C; Supplemental Fig. S4A–F). Through a map-based cloning approach, we showed that PAR corresponds to ChMIR164A (Supplemental Fig. S4G). In Arabidopsis, mir164A influences leaf development via delimiting the expression domain of CUC transcription factors (Nikovics et al. 2006). CUC proteins promote the formation of auxin peaks that underlie formation of serrations and leaflets in the leaf margins of A. thaliana and C. hirsuta (Hay et al. 2006; Barkoulas et al. 2008; Bilsborough et al. 2011; Rubio-Somoza et al. 2014). Consistent with this, we observed increased ChCUC2 expression, additional convergence points of the PINFORMED1 (PIN1) auxin efflux carrier, and auxin activity maxima in par mutant leaves associated with the position of ectopic intercalary leaflet formation along the rachis (Supplemental Fig. S4H–N). The regulation of ChCUC2 expression and auxin homeostasis is therefore a conserved function of MIR164A action in A. thaliana and C. hirsuta. However, mir164a mutants of A. thaliana do not misexpress BP in leaves (Nikovics et al. 2006; Bilsborough et al. 2011). Thus, the regulation of ChBP in C. hirsuta is likely a species-specific function of the PAR (ChMIR164A/ChCUC) module. This idea is supported by two further observations: that ChBP expression is strongly reduced in a 35S:MIR164B;CUC3RNAi transgenic line (Fig. 3A,D) where expression of ChCUC1–3 genes is reduced (Nikovics et al. 2006) and that ChBP expression is increased upon induced CUC2 misexpression (Supplemental Fig. S4O).
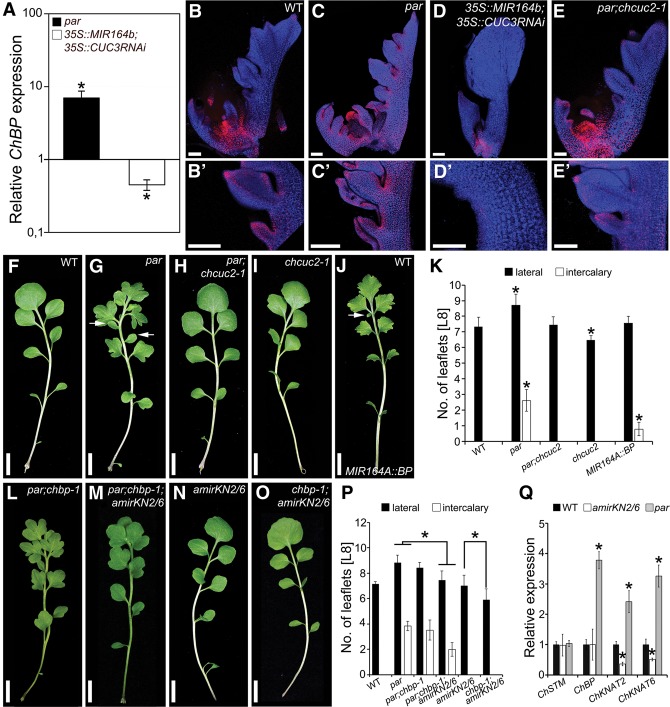
Figure 3.
Ectopic ChBP expression contributes to the par mutant phenotype. (A) ChBP transcript level in par and 35S::MIR164b;CUC3RNAi (Nikovics et al. 2006) relative to wild-type (set as 1) leaves. (B–E′) ChBP::VENUS expression (red) combined with chlorophyll autofluorescence (blue) in C. hirsuta wild-type (B,B′), par (C,C′), 35S::MIR164B;CUC3RNAi (D,D′), and par;chcuc2-1 (E,E′) leaves. Shown are maximum intensity projections of confocal stacks. (F–J) Leaf 8 of C. hirsuta wild-type (F), par (G), par;chcuc2-1 (H), and chcuc2-1 (I) plants and transgenic plants expressing MIR164A::BP (J). (K) Quantification of lateral and intercalary leaflet number on leaf 8 of plants with the indicated genotype. n ≥ 25. (L–O) Leaf 8 of par;chbp-1 (L), par;chbp-1;amirKN2/6 (M), amirKN2/6 (N), and chbp-1;amirKN2/6 (O) plants. (P) Quantification of lateral and intercalary leaflet number on leaf 8 of plants with the indicated genotype. n ≥ 25. (Q) Relative expression of ChSTM, ChBP, ChKNAT2, and ChKNAT6 in par and amirKN2/6 leaves compared with wild type (n = 3). Bars: B–E′, 100 μm; F–O, 1 cm. Error bars in K, P, and Q indicate standard deviation. (Arrows) Intercalary leaflets; (asterisks) statistically significant difference from wild-type (K,Q) or the indicated genotype (P) (P ≤ 0.05, Student's t-test).
The above observations indicate that a key difference between the gene regulatory networks (GRNs) controlling leaf shape in A. thaliana and C. hirsuta is that ChBP is expressed in C. hirsuta leaves and regulated by PAR/ChCUC2. Three lines of evidence support this view and underscore the significance of PAR/ChCUC2-mediated restriction of ChBP for leaf morphology. First, through a mutant screen, we isolated a chcuc2 allele as an extragenic suppressor of par that is sufficient to revert the ChBP expression and leaf phenotype to a near wild-type pattern (Fig. 3E–I; Supplemental Fig. S4P–U). This indicates that elevated ChCUC2 expression is a major contributor to both the morphological defects and the elevated/broadened ChBP expression seen in par. Second, ChBP expression in the MIR164A domain is sufficient to mimic the leaf lobing and increased lateral and intercalary leaflet numbers found in par (Fig. 3J,K). Thus, a ChCUC-dependent increase in ChBP expression likely contributes to the par phenotype. Third, we found that two additional KNOXI genes, ChKNAT2 and ChKNAT6, contribute redundantly with ChBP to the par leaf phenotype. Simultaneous silencing of KNAT2 and KNAT6 through an artificial microRNA (amirKN2/6) combined with the chbp allele resulted in a significant reduction of lateral and intercalary leaflet number compared with par single or par;chbp double mutants (Fig. 3L,M). Furthermore, these three KNOXI genes act redundantly to define leaflet number during wild-type leaflet development (Fig. 3N–P). This is in line with the increased ChBP, ChKNAT2, and ChKNAT6 but not ChSTM transcript levels in par mutant leaves (Fig. 3Q). In conclusion, the PAR/ChCUC2 module regulates ChBP (and ChKNAT2/6) expression during C. hirsuta leaf development. Our genetics show that PAR acts via ChCUC2 and that ChBP/ChKNAT2/6 account for a considerable proportion of this activity. This regulation is pivotal for defining leaflet number and position. Nevertheless, leaflets remain lobed in par;chbp;amirKN2/6 mutants, indicating that other genes downstream from ChCUC2 are required to fully restore the par phenotype (Fig. 3M). Candidate genes include components of the auxin homeostasis machinery that account for the altered distribution of PIN1 and auxin activity in par mutants (Supplemental Fig. S3J,K).
Investigation of genetic interactions within the ChBP/ChCUC GRN
Our results point to two repressors influencing ChBP expression in the C. hirsuta leaf: ChAS1 and PAR (ChMIR164A), with PAR acting via ChCUC2. To test how those two repressors interact, we studied chas1;par double mutants. We found enhanced leaflet lobing and increased leaflet numbers compared with either single mutant as well as the presence of tertiary leaflets, a phenotype not observed in either single mutant (Fig. 4A–Q). This finding indicates that ChAS1 and PAR/ChCUC2 act in parallel pathways to regulate leaflet development and suggests that these two pathways converge on regulation of ChBP.
Figure 4.
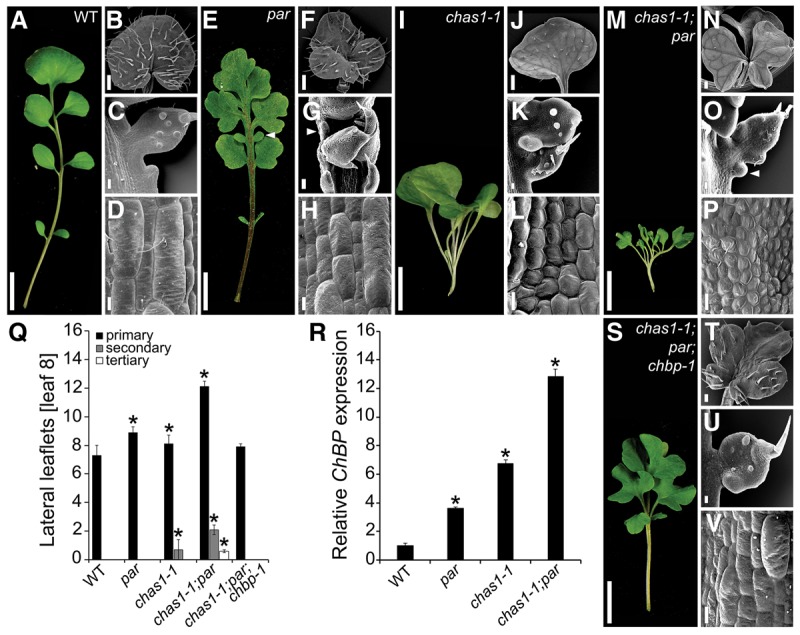
The ChAS1 and PAR/ChCUC pathways converge on ChBP regulation. (A–P) C. hirsuta wild-type (A–D), par (E–H), chas1-1 (I–L), and chas1-1;par (M–P). For each genotype, rosette leaf 5 (A,E,I,M) and scanning electron micrographs of the terminal leaflet (B,F,J,N), lateral leaflet (C,G,K,O), and epidermal cells on the adaxial surface of the leaf petiole (D,H,L,P) are shown. (Q) Quantification of lateral leaflet number (primary, secondary, and tertiary) on leaf 8 of plants with the indicated genotype. n ≥ 15. (R) ChBP transcript level in C. hirsuta wild-type and par, chas1-1, and chas1-1;par. n = 3. (S–V) Rosette leaf 5 (S), terminal leaflet (T), lateral leaflet (U), and leaf petiole adaxial epidermal cells (V) of the chas1-1;par;chbp-1 mutant. Bars: A,E,I,M,S, 1 cm; B,F,J,N,T, 500 µm; C,G,K,O,U, 100 µm; D,H,L,P,V, 20 µm. Error bars in Q and R indicate standard deviation. The asterisks in Q and R indicate statistically significant difference from wild type (P ≤ 0.05, Student's t-test).
Consistent with this view, the level of ChBP expression is substantially higher in chas1;par double-mutant than in chas1 or par single-mutant leaves (Fig. 4R). This increase in ChBP misexpression also correlates with an enhanced repression of leaf growth along the proximodistal axis of chas1;par leaves and is accompanied by a further reduction in epidermal cell length, suggesting a further delay in cell differentiation (Table 1). We found that all phenotypic defects observed in chas1;par mutants are at least partially suppressed in chas1;par;chbp leaves, indicating that the enhancement of chas1 by par largely depends on ChBP (Fig. 4Q–V; Table 1). Thus, ChBP emerges as a key convergence point through which the upstream repressors ChAS1 and PAR act. It is noteworthy that the chas1 phenotype is suppressible by chbp, whereas the par phenotype is not. This higher contribution of ChBP to the chas1 versus par phenotypes may reflect the broader misexpression of ChBP in chas1 than in par (Figs. 1D, 3C).
Based on these findings in C. hirsuta, we hypothesized that CUC2 expression in the simple leaves of A. thaliana would predispose the leaf GRN to place BP under the influence of the MIR164A/CUC2 module if BP is expressed in leaves. We tested this hypothesis by crossing mir164a-4 and as1 mutants in A. thaliana and found an enhanced leaf phenotype in the double mutant (Supplemental Fig. S5A–H). Furthermore, the ectopic BP expression in the leaves of the as1;mir164a-4 double mutants strictly depends on CUC2, as, in as1;mir164a;cuc2 triple mutants, BP::GUS (β-glucuronidase) expression is again confined to the SAM. Thus, a single genetic event causing BP expression in leaves can in principle be sufficient to allow MIR164A/CUC2-dependent BP regulation (Supplemental Fig. S5I–J). In this scenario, a pre-existing module can “capture” a new gene—in this case, BP—when its expression diversifies.
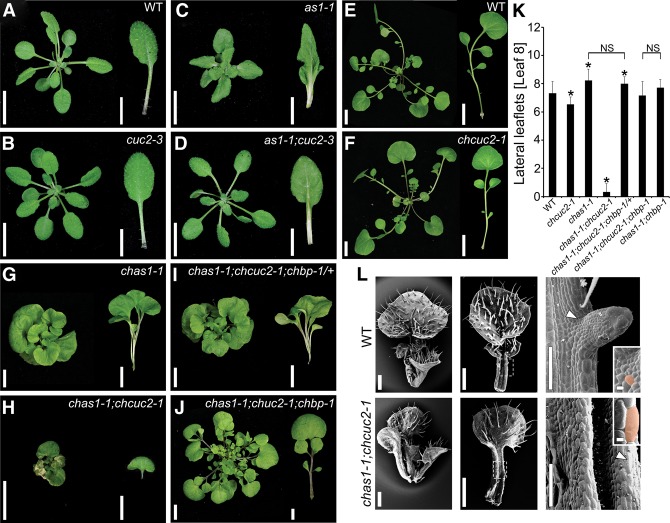
Together, our results provide evidence that, in the C. hirsuta leaf, ChAS1 and ChCUC2 become interconnected through their combined input on ChBP expression. Thus, ChBP expression in the leaf creates a node in the C. hirsuta leaf GRNs that does not exist in A. thaliana. To evaluate the consequences of this alterative GRN organization on genetic interactions between ChAS1 and ChCUC2 during leaf development, we studied the chas1;chcuc2 double-mutant phenotype. In contrast to A. thaliana, where cuc2 suppresses as1 (Fig. 5A–D; Bilsborough et al. 2011), we observed severely increased growth defects in C. hirsuta chas1;chcuc2 double mutants that are characterized by a strong reduction in rosette diameter, decreased leaf and petiole length, and a failure to initiate lateral leaflets (Fig. 5E–H). Notably, this genetic interaction between ChAS1 and ChCUC2 is ChBP-dependent in a dose-sensitive fashion. Specifically, the chas1;chcuc2;chbp/+ mutant phenotype was suppressed with respect to chas1;chcuc2 and resembled chas1 mutants, and chas1;chcuc2;chbp mutants were indistinguishable from chas1;chbp mutants (Fig. 5I–K). Thus, the C. hirsuta leaf context reveals a combined requirement of ChAS1 and ChCUC2 for leaf growth that is species-specific and ChBP-dependent.
Figure 5.
Genetic interactions between AS1 and CUC2 in A. thaliana and C. hirsuta. (A–D) Rosettes and leaf 5 of A. thaliana wild-type (A) and cuc2-3 (B), as1-1 (C), and as1-1;cuc2-3 (D) mutant plants. (E–J) Rosettes and leaf 5 of C. hirsuta wild-type (E) and chcuc2-1 (F), chas1-1 (G), chas1-1;chcuc2-1 (H), chas1-1;chcuc2-1;chbp-1/+ (I), and chas1-1;chcuc2-1;chbp-1 (J) mutant plants. (K) Quantification of lateral leaflet number on leaf 8 of plants with the indicated genotype. Asterisks indicate significant differences from wild type. n ≥ 15. (L) Scanning electron micrographs of a vegetative shoot, the fifth developing rosette leaf (1000 µm), and the leaf margin of wild type (top panel) and chas1-1;chcuc2-1 mutants (bottom panel). The insets show typical cells in the boundary (wild type; arrowhead) or marginal (as1-1;chcuc2-1; arrowhead) region, indicated in orange. Bars: A–J, 1 cm; L, 100 µm. Error bars in K indicate standard deviation. The asterisks in K indicate significant difference from wild type (P ≤ 0.05, Student's t-test). (NS) No significant difference.
We found that the reduction in plant size and leaf length in chas1;chcuc2 double mutants was attributable to a reduction in cell number rather than cell elongation (Table 1), suggesting that ChAS1 and ChCUC2 are together required for cell proliferation in the leaf. It is known that CUC genes influence growth in multiple developmental boundaries (Aida et al. 1997; Vroemen et al. 2003; Breuil-Broyer et al. 2004; Wang et al. 2015), while AS1 and its orthologs regulate growth redundantly with other, partially uncharacterized factors (Waites et al. 1998). Therefore, one possibility is that ChCUC2 and ChAS1 act together to regulate BP expression and promote formation of boundary domains required for leaflet development and leaf growth. Consistent with this view, the small cells that typically mark the boundary between leaflet and rachis are absent in chas1;chcuc2 leaves (Fig. 5L, insets).
To further investigate this hypothesis that ChCUC2 and ChAS1 may jointly regulate leaflet boundary formation via promoting and repressing the expression of ChBP, respectively, we determined the relative expression patterns of ChAS1 and ChCUC2 during C. hirsuta leaf development. As shown previously, ChAS1 mRNA is detectable in leaf primordia but not in the SAM (Hay and Tsiantis 2006). We obtained higher-resolution information on ChAS1 expression relative to developing leaflets and found that ChAS1 transcripts accumulate in a central domain of the rachis and leaflets in C. hirsuta leaves (Fig. 6A–D; Supplemental Fig. S6). We observed that ChAS1 and ChCUC2 expression domains are near complementary during leaflet formation, while ChBP and ChCUC2 domains overlap (Fig. 6E–L). These observations are consistent with the suggestion borne out of genetics that ChAS1 and ChCUC2 promote C. hirsuta leaf growth through their opposing inputs on ChBP across a developmental boundary that delimits leaflets (Fig. 6M). In order to refine this model, it will be important to colocalize expression of all three genes at cellular resolution. Notably, the complex interactions at the boundary of C. hirsuta leaflets are reminiscent of so-called paradoxical interactions underlying boundary development in animal systems (Sprinzak et al. 2010; Hart and Alon 2013) and highlight the need for further study into the cellular basis of developmental boundary function in plants (Rossmann et al. 2015).
Figure 6.
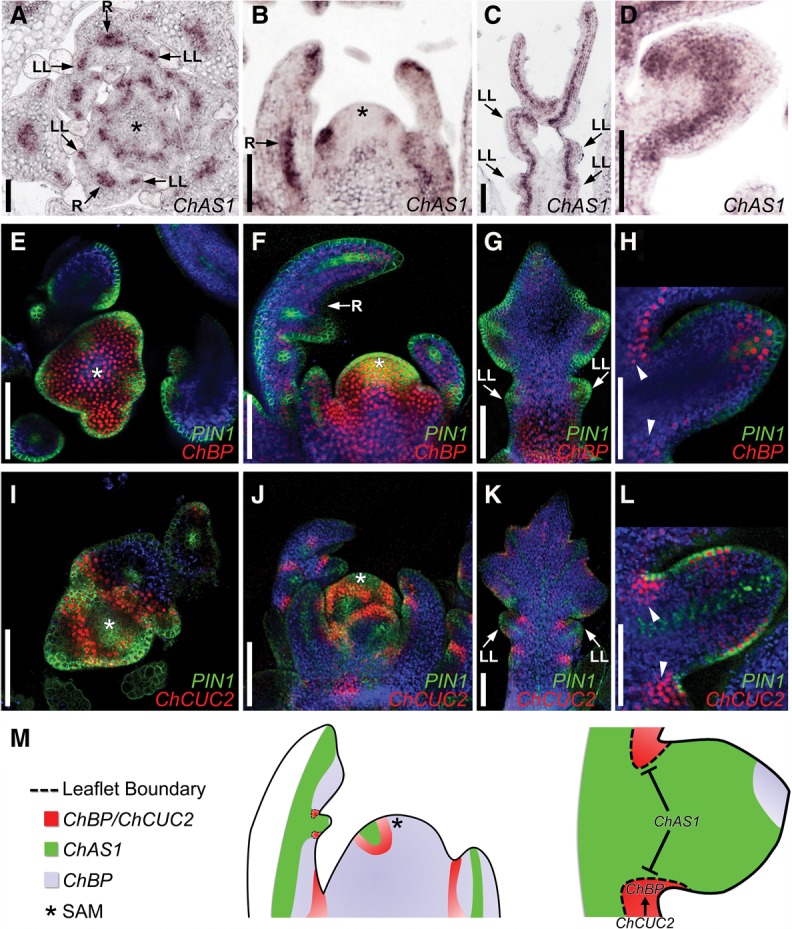
ChCUC and ChAS1 define boundary domains of ChBP expression. (A–D) ChAS1 expression in the C. hirsuta SAM and leaves analyzed by RNA in situ hybridization. ChBP::VENUS; PIN1::PIN1-GFP expression (E–H) and ChCUC2g-VENUS; PIN1::PIN1-GFP (I–L) in the C. hirsuta SAM and young leaves. (Red) Venus fluorescence; (green) GFP fluorescence; (blue) chlorophyll autofluorescence. Shown are transverse (A,E,I) and longitudinal (B,F,J) sections through the SAM; leaf 5 at a length of 750 µm (C,G,K); and a close-up of developing lateral leaflets (D,H,L). (M) Cartoon summarizing the observed ChAS1, ChBP, and ChCUC2 expression patterns. In leaf primordia, ChBP is expressed in the adaxial side and at the margin of the rachis (F,G) as well as at the tip of developing leaflets (H). (H,L) This expression pattern is near complementary to that of ChAS1 and overlaps with that of ChCUC2 in the leaf rachis margin at leaflet boundaries (arrowheads). This results in the formation of boundary domains (outlined with dotted lines) that are highly sensitive to ChBP dose and required to promote leaf growth. Bars, 100 µm. (R) Leaf rachis; (LL) lateral leaflet. Asterisks mark the SAM.
Discussion
Our work highlights how in-depth genetic analysis of development in a comparative context is a key tool for understanding the genetic basis for morphological diversity. We showed that two paralogous homeobox genes, ChSTM and ChBP, share redundant functions in C. hirsuta leaflet formation and found that cis-regulatory change in the less pleiotropic gene of the two, ChBP, had a more active role in generating diverse leaf morphologies. These comparisons provide an opportunity to visualize tradeoffs between the ability of a gene to cause morphological change and its pleiotropy and provide empirical evidence for a prediction that narrowly pleiotropic regulators and, in particular, transcription factors might be key drivers of plant diversity (Doebley and Lukens 1998).
Leaf margin morphology depends on the action of a small GRN comprising MIR164A, CUC genes, and auxin activity components (Supplemental Fig. S7; Blein et al. 2008; Kawamura et al. 2010; Bilsborough et al. 2011). Previous work indicated that tinkering with this GRN through alterations in KNOX activity might contribute to the evolution of divergent leaf morphologies (Ori et al. 2000; Hake and Ori 2002; Hay and Tsiantis 2006; Barkoulas et al. 2008; Blein et al. 2008; Bilsborough et al. 2011). However, the evolutionary changes underlying such diversity and their impact on the architecture of the GRN were unknown. Here we show that cis-regulatory divergence of BP provides a direct mechanistic path for creating alternate GRN architectures between simple and complex leaves. Specifically, we found that ChBP expression in the C. hirsuta leaf renders it a mediator of mir164A/ChCUC activity and a functionally critical target of ChAS1. This network architecture, which is not seen in A. thaliana, creates a species-specific interaction between ChAS1 and ChCUC2 that supports leaflet formation and influences leaf growth broadly. Thus, we provide an example of how regulatory evolution of a single low pleiotropy gene, BP, can contribute to substantial rewiring of a developmental network regulating leaf shape.
We propose that the flexible integration or disengagement of weakly pleiotropic regulators, such as BP, from conserved genetic networks provides a path through which regulatory evolution can alter molecular circuitries that influence organ growth. Such network rewiring may help complex organisms readily evolve morphological diversity by overcoming potential fitness penalties caused by pleiotropy (Stern and Orgogozo 2008). In the future, it will be interesting to test these possibilities by evaluating the relative pleiotropy and capacity for network reorganization of other genes that are sufficient to account for trait diversity between species. Overall, our work indicates that the interplay between pleiotropy and regulatory evolution underpins morphological change in not only metazoans, where stereotypical body plans are laid down during embryogenesis, but also seed plants, where organogenesis occurs post-embryonically and shows considerable plasticity (Steeves and Sussex 1989; Carroll et al. 2004). This interplay may therefore reflect a fundamental property of morphological evolution rather than lineage-specific constraints associated with metazoan organogenesis (Arthur 2004).
Materials and methods
Plant material and growth conditions
The origins of mutant alleles and transgenic lines used in this study were as follows: chas1-1 (Hay and Tsiantis 2006), as1-1 (CS3374, Arabidopsis Biological Resource Center), bp-9 (Smith and Hake 2003), as1-101 (Sun et al. 2002), bp-1 (Venglat et al. 2002), cuc2-3 (Hibara et al. 2006), mir164a-4 (Nikovics et al. 2006), 35S::MIR164b (Blein et al. 2008), 35S::MIR164b;35S::CUC3RNAi (Blein et al. 2008), 35S::CUC2-GR (Bennett et al. 2010), 35S::STM-GR (Gallois et al. 2002), DR5rev::VENUS (Barkoulas et al. 2007), PIN1::PIN1-GFP (Heisler et al. 2005), CUC2::CUC2-VENUS (Heisler et al. 2005), and BP::GUS (Hay and Tsiantis 2006). Plants were grown on soil under long-day conditions (18 h light; 20°C). With the exception of the chstm/+ allele, which was maintained as a segregating pool, generation of double or triple mutants was performed by crossing of homozygous plants. These crossings were genotyped in the F2 populations and phenotyped in the F3 generation. Analysis of reporter gene expression was performed in the F2 and confirmed in the F3 and F4 generations after genetic crossing.
Ethane methyl sulfonate (EMS) mutagenesis
For EMS mutagenesis, C. hirsuta wild-type (Oxford; Hay and Tsiantis 2006) or mutant seeds (chas1-1, chas1-1;chstm-1/+, or par) were mutagenized by agitation with 0.2% EMS (Sigma) for 10 h, washed in dH2O, sown on soil, and harvested in pools of five plants. M2 plants (total numbers are given below) were subsequently screened for leaf phenotypes or suppression of leaf phenotypes. Mutant characterization was performed after backcrossing to wild type at least twice. A detailed description of the chstm-1, chbp-1, par, and chcuc2-1 mutant isolation and complementation is provided in the Supplemental Material.
Binary constructs and plant transformation
All constructs were transformed into C. hirsuta and A. thaliana by floral dip (Clough and Bent 1998) using Agrobacterium tumefaciens strain GV3101. For a detailed description of how the constructs were generated, see the Supplemental Material. For each construct, a minimum of 15 independent transgenic lines were self-pollinated to obtain T2 seeds. The progeny of at least five independent T1 lines were analyzed in each case.
Quantitative real time RT–PCR (qRT–PCR) analysis
The RNeasy plant minikit (Qiagen) was used for RNA extraction. Total RNA (1 µg) was treated with DNase I and transcribed into cDNA using SuperScript II reverse transcriptase and an oligo(dt) primer (Invitrogen). qRT–PCR was performed in triplicate from two independent RNA extractions using the SYBR Green PCR master mix (Applied Biosystems) and an ABI Prism 7300 sequence detection system (Applied Biosystems). The primer efficiency and expression level were determined as described (Pfaffl 2001). Expression levels were normalized to the reference gene GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (GAPDH). With the exception of the genes ChKNAT2 (5′-TGGCTATCTTGCGCTGCTAC-3′ and 5′-TGCAAGAGGCCTTTCAGTTT-3′) and ChKNAT6 (5′-CGGAGATCAGAAGAAACGATGA-3′ and 5′-GCGAGGATACGATGGATGAC-3′), all primers sequences used have been published previously (Kougioumoutzi et al. 2013).
RNA in situ hybridization
RNA in situ hybridizations on 15-µm sections through fixed and paraffin-embedded shoot apices of 2- to 3-wk-old short-day grown plants were performed as described (Vlad et al. 2014). Digoxigenin-labeled antisense RNA probes to C. hirsuta ChAS1 were generated using cDNA templates obtained after amplification with the primer combinations 5′-AGTAGTGAGAGTGTGTTCTTGTC-3′ and 5′-CCAAGCTTCTAATACGACTCACTATAGGGAGATCTAATCTGCAACCCATG-3′ (the T7 RNA polymerase-binding motif is underlined). To cover the entire hybridization pattern, several consecutive sections were registered, and minimal projections were generated using the image processing package Fijii (Supplemental Fig. S6; Schindelin et al. 2012). The signal was observed and images were acquired with a Zeiss Axiophot light microscope and a Leica DFC 490 digital camera.
Phenotypic analysis and estimation of pleiotropy
Quantification of lateral, intercalary, axillary, or secondary leaflets was done with at least 15 individual wild-type or mutant plants, and each experiment was repeated at least twice. To obtain leaf silhouettes, fully developed leaves were flattened onto clear adhesive on white paper and then digitally scanned. Leaf length, area, and perimeter were calculated from silhouettes using Fijii software (Schindelin et al. 2012). The petiole cell length and number was measured as described (Rast and Simon 2012; Vlad et al. 2014).
The degree of reduction of plant rosette diameter and, for selected genotypes, the total number of intact ovules per silique were used as pleiotropy estimates. Plant rosette diameter was calculated from photographs using Fiji software (n ≥ 15). Siliques 6–10 of the first side shoot (n ≥ 45) of at least 10 independent plants were collected, and ovules were counted. An average number of ovules per silique was then calculated from this sample. Both rosette diameter and seed number were then plotted as 1/x against the leaf dissection index (perimeter squared)/(4π × area) (Bilsborough et al. 2011).
Scanning electron microscopy (SEM), confocal laser scanning microscopy, and light microscopy
SEM and confocal laser-scanning microscopy were carried out as described (Bilsborough et al. 2011). SEM samples were analyzed using a JSM-5510 microscope (Jeol). For the analysis of fluorescence reporter expression, seedlings were mounted and observed in water without fixation. Confocal imaging was performed using a Leica TCS SP5 II microscope and a 10× objective (HC PL Fluotar 10 9 0.30) or a water dipping 20× objective (HCX APOLU-V-I 0 9 0.5) or a Zeiss LSM 780 upright microscope and water immersion objective (AP 20×/0.8 M27). Visualization of VENUS expression was performed using a 488-nm argon laser and a 657- to 743-nm filter for the chlorophyll autofluorescence and a 505- to 550-nm bandpass filter. Maximal projections were generated from stacks of five to 30 sections. GUS activity was detected as described in Hay and Tsiantis (2006). Imaging of GUS-stained samples and agarose prints was performed using a Leica DFC 490 digital camera mounted on a Zeiss Axiophot light microscope. Images were processed and analyzed using Fijii and Adobe Photoshop software.
Supplementary Material
Acknowledgments
We thank A. Hudson and N. Gompel for helpful discussion on AS1/KNOX interactions and pleiotropy; M. Byrne for discussions on as1 genetics; E. Rabbinowitsch, J. Baker, Z. Lewis, R. Pabst, R. Berndtgen, and E. Schmelzer for technical assistance; M. Barkoulas for help with microscopy; and A. Hudson, S. McCormick, Richard Smith, and members of the Tsiantis laboratory for comments on the manuscript. This work was supported by Biotechnology and Biological Sciences Research Council grants BB/H011455/1 (to M.T.) and BB/H006974/1 (to M.T. and A.H.), Deutsche Forschungsgemeinschaft “Adaptomics” grants TS 229/1-1 (to M.T. and A.H.) and SFB 680 (to M.T.), the Gatsby Charitable Foundation (M.T.), Human Frontier Science Program grant RGP0047/2010 (to M.T.), and a core grant from the Max Planck Society (to M.T.) M.T. also acknowledges support of the Cluster of Excellence on Plant Sciences. M.I.R.-S., S.B., H.J., A.H., and M.T. designed and carried out the experiments; C.C., D.V., M.K., G.B., and R.D.I. generated and characterized genetic material; C.O. and M.G.H. generated the BP::BP-CFP construct; R.E. generated bioinformatics tools; P.L. donated the 35S::MIR164b and 35S::CUC3RNAi transgenes before publication; and P.H. contributed to the RNA in situ hybridization experiment. M.I.R.-S. and M.T. organized the data analysis and wrote the paper with input from A.H. M.T. conceived and directed the study.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.269050.115.
Freely available online through the Genes & Development Open Access option.
References
- Aguilar-Martinez JA, Uchida N, Townsley B, West DA, Yanez A, Lynn N, Kimura S, Sinha N. 2015. Transcriptional, posttranscriptional, and posttranslational regulation of SHOOT MERISTEMLESS gene expression in Arabidopsis determines gene function in the shoot apex. Plant Physiol 167: 424–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M. 1997. Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9: 841–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnoult L, Su KF, Manoel D, Minervino C, Magrina J, Gompel N, Prud'homme B. 2013. Emergence and diversification of fly pigmentation through evolution of a gene regulatory module. Science 339: 1423–1426. [DOI] [PubMed] [Google Scholar]
- Arthur W. 2002. The emerging conceptual framework of evolutionary developmental biology. Nature 415: 757–764. [DOI] [PubMed] [Google Scholar]
- Arthur W. 2004. Biased embryos and evolution. University Press, Cambridge, UK. [Google Scholar]
- Bar M, Ori N. 2014. Leaf development and morphogenesis. Development 141: 4219–4230. [DOI] [PubMed] [Google Scholar]
- Bar M, Ori N. 2015. Compound leaf development in model plant species. Curr Opin Plant Biol 23C: 61–69. [DOI] [PubMed] [Google Scholar]
- Barkoulas M, Galinha C, Grigg SP, Tsiantis M. 2007. From genes to shape: regulatory interactions in leaf development. Curr Opin Plant Biol 10: 660–666. [DOI] [PubMed] [Google Scholar]
- Barkoulas M, Hay A, Kougioumoutzi E, Tsiantis M. 2008. A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nat Genet 40: 1136–1141. [DOI] [PubMed] [Google Scholar]
- Ben-Gera H, Ori N. 2012. Auxin and LANCEOLATE affect leaf shape in tomato via different developmental processes. Plant Signal Behav 7: 1255–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett T, van den Toorn A, Sanchez-Perez GF, Campilho A, Willemsen V, Snel B, Scheres B. 2010. SOMBRERO, BEARSKIN1, and BEARSKIN2 regulate root cap maturation in Arabidopsis. Plant Cell 22: 640–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharathan G, Goliber TE, Moore C, Kessler S, Pham T, Sinha NR. 2002. Homologies in leaf form inferred from KNOXI gene expression during development. Science 296: 1858–1860. [DOI] [PubMed] [Google Scholar]
- Bilsborough GD, Runions A, Barkoulas M, Jenkins HW, Hasson A, Galinha C, Laufs P, Hay A, Prusinkiewicz P, Tsiantis M. 2011. Model for the regulation of Arabidopsis thaliana leaf margin development. Proc Natl Acad Sci 108: 3424–3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blein T, Pulido A, Vialette-Guiraud A, Nikovics K, Morin H, Hay A, Johansen IE, Tsiantis M, Laufs P. 2008. A conserved molecular framework for compound leaf development. Science 322: 1835–1839. [DOI] [PubMed] [Google Scholar]
- Blein T, Hasson A, Laufs P. 2010. Leaf development: what it needs to be complex. Curr Opin Plant Biol 13: 75–82. [DOI] [PubMed] [Google Scholar]
- Bolduc N, Tyers RG, Freeling M, Hake S. 2014. Unequal redundancy in maize knotted1 homeobox genes. Plant Physiol 164: 229–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuil-Broyer S, Morel P, de Almeida-Engler J, Coustham V, Negrutiu I, Trehin C. 2004. High-resolution boundary analysis during Arabidopsis thaliana flower development. Plant J 38: 182–192. [DOI] [PubMed] [Google Scholar]
- Byrne ME, Barley R, Curtis M, Arroyo JM, Dunham M, Hudson A, Martienssen RA. 2000. Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408: 967–971. [DOI] [PubMed] [Google Scholar]
- Byrne ME, Simorowski J, Martienssen RA. 2002. ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129: 1957–1965. [DOI] [PubMed] [Google Scholar]
- Carroll SB. 2005. Evolution at two levels: on genes and form. PLoS Biol 3: e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. 2008. Evo–devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134: 25–36. [DOI] [PubMed] [Google Scholar]
- Carroll SB, Grenier JK, Weatherbee SD. 2004. From DNA to diversity, molecular genetics and the evolution of animal design, 2nd ed Blackwell Publishing, Maiden, MA. [Google Scholar]
- Chan YF, Marks ME, Jones FC, Villarreal G Jr, Shapiro MD, Brady SD, Southwick AM, Absher DM, Grimwood J, Schmutz J, et al. 2010. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science 327: 302–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method forAgrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Doebley J, Lukens L. 1998. Transcriptional regulators and the evolution of plant form. Plant Cell 10: 1075–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I, Eshed Y, Lifschitz E. 2010. Morphogenesis of simple and compound leaves: a critical review. Plant Cell 22: 1019–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endrizzi K, Moussian B, Haecker A, Levin JZ, Laux T. 1996. The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J 10: 967–979. [DOI] [PubMed] [Google Scholar]
- Furumizu C, Alvarez JP, Sakakibara K, Bowman JL. 2015. Antagonistic roles for KNOX1 and KNOX2 genes in patterning the land plant body plan following an ancient gene duplication. PLoS Genet 11: e1004980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallois JL, Woodward C, Reddy GV, Sablowski R. 2002. Combined SHOOT MERISTEMLESS and WUSCHEL trigger ectopic organogenesis in Arabidopsis. Development 129: 3207–3217. [DOI] [PubMed] [Google Scholar]
- Ge L, Peng J, Berbel A, Madueno F, Chen R. 2014. Regulation of compound leaf development by PHANTASTICA in Medicago truncatula. Plant Physiol 164: 216–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompel N, Prud'homme B. 2009. The causes of repeated genetic evolution. Dev Biol 332: 36–47. [DOI] [PubMed] [Google Scholar]
- Gompel N, Prud'homme B, Wittkopp PJ, Kassner VA, Carroll SB. 2005. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature 433: 481–487. [DOI] [PubMed] [Google Scholar]
- Guo M, Thomas J, Collins G, Timmermans MC. 2008. Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell 20: 48–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake S, Ori N. 2002. Plant morphogenesis and KNOX genes. Nat Genet 31: 121–122. [DOI] [PubMed] [Google Scholar]
- Hareven D, Gutfinger T, Parnis A, Eshed Y, Lifschitz E. 1996. The making of a compound leaf: genetic manipulation of leaf architecture in tomato. Cell 84: 735–744. [DOI] [PubMed] [Google Scholar]
- Hart Y, Alon U. 2013. The utility of paradoxical components in biological circuits. Mol Cell 49: 213–221. [DOI] [PubMed] [Google Scholar]
- Hasson A, Plessis A, Blein T, Adroher B, Grigg S, Tsiantis M, Boudaoud A, Damerval C, Laufs P. 2011. Evolution and diverse roles of the CUP-SHAPED COTYLEDON genes in Arabidopsis leaf development. Plant Cell 23: 54–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A, Tsiantis M. 2006. The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat Genet 38: 942–947. [DOI] [PubMed] [Google Scholar]
- Hay A, Tsiantis M. 2010. KNOX genes: versatile regulators of plant development and diversity. Development 137: 3153–3165. [DOI] [PubMed] [Google Scholar]
- Hay A, Kaur H, Phillips A, Hedden P, Hake S, Tsiantis M. 2002. The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr Biol 12: 1557–1565. [DOI] [PubMed] [Google Scholar]
- Hay A, Barkoulas M, Tsiantis M. 2006. ASYMMETRIC LEAVES1 and auxin activities converge to repress BREVIPEDICELLUS expression and promote leaf development in Arabidopsis. Development 133: 3955–3961. [DOI] [PubMed] [Google Scholar]
- Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz EM. 2005. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol 15: 1899–1911. [DOI] [PubMed] [Google Scholar]
- Hibara K, Karim MR, Takada S, Taoka K, Furutani M, Aida M, Tasaka M. 2006. Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 18: 2946–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikezaki M, Kojima M, Sakakibara H, Kojima S, Ueno Y, Machida C, Machida Y. 2010. Genetic networks regulated by ASYMMETRIC LEAVES1 (AS1) and AS2 in leaf development in Arabidopsis thaliana: KNOX genes control five morphological events. Plant J 61: 70–82. [DOI] [PubMed] [Google Scholar]
- Jackson D, Veit B, Hake S. 1994. Expression of maize Knotted1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120: 405–413. [Google Scholar]
- Kawamura E, Horiguchi G, Tsukaya H. 2010. Mechanisms of leaf tooth formation in Arabidopsis. Plant J 62: 429–441. [DOI] [PubMed] [Google Scholar]
- Kougioumoutzi E, Cartolano M, Canales C, Dupre M, Bramsiepe J, Vlad D, Rast M, Dello Ioio R, Tattersall A, Schnittger A, et al. 2013. SIMPLE LEAF3 encodes a ribosome-associated protein required for leaflet development in Cardamine hirsuta. Plant J 73: 533–545. [DOI] [PubMed] [Google Scholar]
- Lincoln C, Long J, Yamaguchi J, Serikawa K, Hake S. 1994. A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6: 1859–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK. 1996. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69. [DOI] [PubMed] [Google Scholar]
- Mallarino R, Abzhanov A. 2012. Paths less traveled: evo–devo approaches to investigating animal morphological evolution. Annu Rev Cell Dev Biol 28: 743–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikovics K, Blein T, Peaucelle A, Ishida T, Morin H, Aida M, Laufs P. 2006. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 18: 2929–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori N, Eshed Y, Chuck G, Bowman JL, Hake S. 2000. Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127: 5523–5532. [DOI] [PubMed] [Google Scholar]
- Ori N, Cohen AR, Etzioni A, Brand A, Yanai O, Shleizer S, Menda N, Amsellem Z, Efroni I, Pekker I, et al. 2007. Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat Genet 39: 787–791. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud'homme B, Minervino C, Hocine M, Cande JD, Aouane A, Dufour HD, Kassner VA, Gompel N. 2011. Body plan innovation in treehoppers through the evolution of an extra wing-like appendage. Nature 473: 83–86. [DOI] [PubMed] [Google Scholar]
- Raff RA. 1996. The shape of life: genes, development and the evolution of animal form. University of Chicago Press, Chicago. [Google Scholar]
- Rast MI, Simon R. 2012. Arabidopsis JAGGED LATERAL ORGANS acts with ASYMMETRIC LEAVES2 to coordinate KNOX and PIN expression in shoot and root meristems. Plant Cell 24: 2917–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz M, Patel NH, Hinman VH. 2015. Unraveling the tangled skein: the evolution of transcriptional regulatory networks in development. Annu Rev Genomics Hum Genet 16: 103–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann S, Kohlen W, Hasson A, Theres K. 2015. Lateral suppressor and Goblet act in hierarchical order to regulate ectopic meristem formation at the base of tomato leaflets. Plant J 81: 837–848. [DOI] [PubMed] [Google Scholar]
- Rubio-Somoza I, Zhou CM, Confraria A, Martinho C, von Born P, Baena-Gonzalez E, Wang JW, Weigel D. 2014. Temporal control of leaf complexity by miRNA-regulated licensing of protein complexes. Curr Biol 24: 2714–2719. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shani E, Burko Y, Ben-Yaakov L, Berger Y, Amsellem Z, Goldshmidt A, Sharon E, Ori N. 2009. Stage-specific regulation of Solanum lycopersicum leaf maturation by class 1 KNOTTED1-LIKE HOMEOBOX proteins. Plant Cell 21: 3078–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicard A, Thamm A, Marona C, Lee YW, Wahl V, Stinchcombe JR, Wright SI, Kappel C, Lenhard M. 2014. Repeated evolutionary changes of leaf morphology caused by mutations to a homeobox gene. Curr Biol 24: 1880–1886. [DOI] [PubMed] [Google Scholar]
- Smith HM, Hake S. 2003. The interaction of two homeobox genes, BREVIPEDICELLUS and PENNYWISE, regulates internode patterning in the Arabidopsis inflorescence. Plant Cell 15: 1717–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LG, Jackson D, Hake S. 1995. Expression of knotted1 marks shoot meristem formation during maize embryogenesis. Dev Genet 16: 344–348. [Google Scholar]
- Sprinzak D, Lakhanpal A, Lebon L, Santat LA, Fontes ME, Anderson GA, Garcia-Ojalvo J, Elowitz MB. 2010. Cis-interactions between Notch and Delta generate mutually exclusive signalling states. Nature 465: 86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeves TA, Sussex IM. 1989. Patterns in plant development, 2nd ed Cambridge University Press, Cambridge, UK. [Google Scholar]
- Stern DL, Frankel N. 2013. The structure and evolution of cis-regulatory regions: the shavenbaby story. Philos Trans R Soc Lond B Biol Sci 368: 20130028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL, Orgogozo V. 2008. The loci of evolution: how predictable is genetic evolution? Evolution 62: 2155–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL, Orgogozo V. 2009. Is genetic evolution predictable? Science 323: 746–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Zhou Q, Zhang W, Fu Y, Huang H. 2002. ASYMMETRIC LEAVES1, an Arabidopsis gene that is involved in the control of cell differentiation in leaves. Planta 214: 694–702. [DOI] [PubMed] [Google Scholar]
- Timmermans MC, Hudson A, Becraft PW, Nelson T. 1999. ROUGH SHEATH2: a Myb protein that represses knox homeobox genes in maize lateral organ primordia. Science 284: 151–153. [DOI] [PubMed] [Google Scholar]
- Tsiantis M, Schneeberger R, Golz JF, Freeling M, Langdale JA. 1999. The maize rough sheath2 gene and leaf development programs in monocot and dicot plants. Science 284: 154–156. [DOI] [PubMed] [Google Scholar]
- Venglat SP, Dumonceaux T, Rozwadowski K, Parnell L, Babic V, Keller W, Martienssen R, Selvaraj G, Datla R. 2002. The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proc Natl Acad Sci 99: 4730–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad D, Kierzkowski D, Rast MI, Vuolo F, Dello Ioio R, Galinha C, Gan X, Hajheidari M, Hay A, Smith RS, et al. 2014. Leaf shape evolution through duplication, regulatory diversification, and loss of a homeobox gene. Science 343: 780–783. [DOI] [PubMed] [Google Scholar]
- Vroemen CW, Mordhorst AP, Albrecht C, Kwaaitaal MA, de Vries SC. 2003. The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 15: 1563–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waites R, Selvadurai HR, Oliver IR, Hudson A. 1998. The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell 93: 779–789. [DOI] [PubMed] [Google Scholar]
- Wang Q, Hasson A, Rossmann S, Theres K. 2015. Divide et impera: boundaries shape the plant body and initiate new meristems. New Phytol 10.1111/nph.13641. [DOI] [PubMed] [Google Scholar]
- Williams TM, Selegue JE, Werner T, Gompel N, Kopp A, Carroll SB. 2008. The regulation and evolution of a genetic switch controlling sexually dimorphic traits in Drosophila. Cell 134: 610–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.