Twaroski et al. identified naked cuticle homolog 1 (NKD1) as being directly regulated by FGFR activity during the transition from endoderm to hepatic progenitor cell. Loss of NKD1 suppresses the formation of hepatic progenitor cells from human induced pluripotent stem cells, and this phenotype can be rescued by using a pharmacological antagonist of canonical WNT signaling.
Keywords: FGF signaling, liver development, iPSC-derived hepatocytes, WNT, HNF4a
Abstract
Fibroblast growth factors (FGFs) are required to specify hepatic fate within the definitive endoderm through activation of the FGF receptors (FGFRs). While the signaling pathways involved in hepatic specification are well understood, the mechanisms through which FGFs induce hepatic character within the endoderm are ill defined. Here we report the identification of genes whose expression is directly regulated by FGFR activity during the transition from endoderm to hepatic progenitor cell. The FGFR immediate early genes that were identified include those encoding transcription factors, growth factors, and signaling molecules. One of these immediate early genes encodes naked cuticle homolog 1 (NKD1), which is a repressor of canonical WNT (wingless-type MMTV integration site) signaling. We show that loss of NKD1 suppresses the formation of hepatic progenitor cells from human induced pluripotent stem cells and that this phenotype can be rescued by using a pharmacological antagonist of canonical WNT signaling. We conclude that FGF specifies hepatic fate at least in large part by inducing expression of NKD1 to transiently suppress the canonical WNT pathway.
Lineage tracing has revealed that the parenchymal cells of the liver derive from ventral and lateral portions of the foregut endoderm (Tremblay and Zaret 2005; Zaret 2008). In mouse embryos, specification of the hepatic progenitor cells occurs when the embryo has generated seven to eight somite pairs (approximately embryonic day 8.25 [E8.25]) (Gualdi et al. 1996). At this stage of development, closure of the anterior intestinal portal positions the ventral face of the endoderm in close apposition to mesodermal cells that give rise to the heart, epicardium, and diaphragm (for reviews, see Zaret 2008; Lemaigre 2009; Zorn and Wells 2009; Si-Tayeb et al. 2010a). Explant culture studies performed using avian embryos demonstrated that the mesoderm was the source of factors that were necessary and sufficient for hepatic specification (LeDouarin 1968, 1975; Houssaint 1980). The identity of the factors released from the mesoderm remained obscure until molecular studies using mouse embryos revealed that fibroblast growth factor1 (FGF1) and FGF2 could substitute for the cardiac mesoderm to induce the onset of hepatocyte differentiation from the endoderm (Jung et al. 1999). Subsequent studies have shown that FGF acts in a concentration-dependent manner, with relatively low concentrations inducing hepatic differentiation and higher concentrations inducing lung development (Serls et al. 2005). Since the original discovery in mouse embryos, several reports have demonstrated that FGFs play integral roles in regulating hepatic cell fate in evolutionarily distinct species, including Xenopus, chicks, and zebrafish (Jung et al. 1999; Chen et al. 2003; Zhang et al. 2004; Shin et al. 2011; Shifley et al. 2012).
In addition to FGFs, both bone morphogenetic proteins (BMPs) and wingless-type MMTV integration site (WNT) proteins have roles in regulating the onset of hepatic development. BMPs are secreted from the septum transversum mesenchyme to activate expression of transcription factors that are required for hepatocyte gene expression (Rossi et al. 2001). The role of WNTs appears to be more complex and dynamic, with WNTs having developmental stage-specific effects (McLin et al. 2007; Li et al. 2008). Studies in Xenopus have shown that WNT signaling promotes hepatogenesis following specification of the hepatic progenitor cells (McLin et al. 2007). However, in contrast to the role of WNTs after the hepatic progenitors are formed, at early somite stages, WNT antagonizes expression of the transcription factor hematopoietically expressed homeobox (Hhex), which is required for formation of hepatocytes. These studies imply that specific antagonists of WNT signaling, which may include secreted frizzled-related protein 5 (Sfrp5), regulate the threshold of WNT activity in the anterior foregut to allow the endoderm to adopt a hepatic fate (Li et al. 2008; Zhang et al. 2013). Similar results have been obtained using mouse embryos and human embryonic stem cells (hESCs), suggesting that the temporally regulated inhibition of WNT signaling during hepatic specification is evolutionarily conserved (Han et al. 2011). Moreover, cocultures of endoderm and endothelial cells have suggested that the endothelial cells may be the source of factors that suppress WNT activity in the anterior endoderm of mouse embryos (Han et al. 2011).
Although the signaling cascades that respond to FGFs are well understood, how the activation of FGF receptors (FGFRs) ultimately induces the endoderm to adopt a hepatic fate remains unclear. Given that FGFR activation ultimately controls changes in gene expression, it seems likely that events occurring downstream from FGF action will include the induction of liver-enriched transcription factors. The relative paucity of information explaining how FGFs mechanistically control hepatic development in part reflects the difficulty in performing molecular and biochemical analyses on the nascent hepatic endoderm. Several groups have shown that human induced pluripotent stem cells (hiPSCs) and hESCs can be differentiated into cells with hepatocyte characteristics by the sequential addition of growth factors to mimic hepatogenesis (Cai et al. 2007; Agarwal et al. 2008; Hay et al. 2008; Basma et al. 2009; Song et al. 2009; Si-Tayeb et al. 2010b; Sullivan et al. 2010). The generation of hepatocyte-like cells from human pluripotent stem cells using the better protocols is efficient, reproducible, and synchronous. In addition, when differentiations are performed under wholly defined culture conditions, the procedure offers a model system that can be manipulated to explore the role of specific proteins in establishing hepatic cell fate (Si-Tayeb et al. 2010b; Delaforest et al. 2011; Mallanna and Duncan 2013). Since most protocols include FGF2 in the cocktail of growth factors used to induce the production of hepatic progenitor cells from iPSC-derived endoderm, we attempted to use this dynamic culture model of hepatocyte differentiation to define the molecular basis for FGF's control of hepatic fate. We reveal that FGF signaling directly regulates expression of a cadre of transcription factors as well as the WNT signaling inhibitor naked cuticle homolog 1 (NKD1). Moreover, deletion of NKD1 inhibits hepatic progenitor cell formation from the endoderm, a phenotype that can be rescued by an antagonist of WNT signaling. Based on these studies, we conclude that FGF controls the specification of hepatic progenitors from hiPSCs at least in large part by inhibiting canonical WNT signaling.
Results
FGFR signaling is required for specification of hepatic progenitor cells during hiPSC differentiation
FGFs have been shown to be required for the initiation of hepatic development in several divergent species (Jung et al. 1999; Chen et al. 2003; Zhang et al. 2004; Shin et al. 2011; Shifley et al. 2012). Based on such studies, most protocols used to generate hepatocyte-like cells from hiPSCs include the addition of FGF1 or FGF2, commonly along with BMP4, to induce hepatic specification of the endoderm (Cai et al. 2007; Agarwal et al. 2008; Hay et al. 2008; Basma et al. 2009; Song et al. 2009; Si-Tayeb et al. 2010b; Sullivan et al. 2010). However, whether FGF signaling is essential for hepatic progenitor cell formation during hiPSC differentiation has not been determined. Unfortunately, it is not feasible to generate FGFR knockout cell lines because FGFs are required to maintain the pluripotency of human pluripotent stem cells (Ludwig et al. 2006a,b; Lanner and Rossant 2010). Moreover, oligonucleotide array analyses (Delaforest et al. 2011) of endogenous mRNA levels encoding 15 FGFs during iPSC differentiation revealed that several FGFs, including FGF2, are expressed at the endodermal and hepatic progenitor cell stages of differentiation (data not shown). With this in mind, we believed that simply removing exogenous FGF during the differentiation protocol would be unlikely to yield a definitive answer.
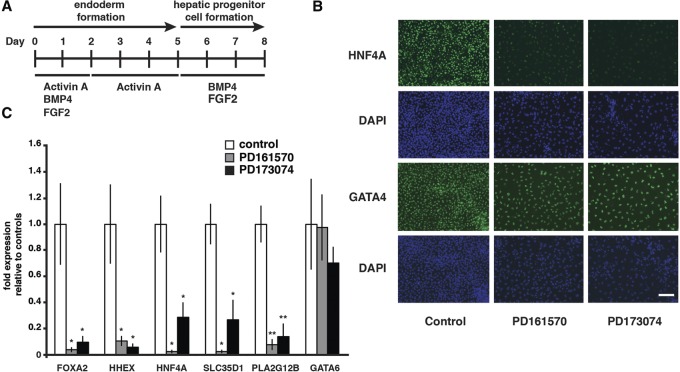
As an alternative approach, we chose to block FGF signaling during the conversion of the endoderm to a hepatic fate using pharmacological inhibitors of the FGFRs. iPSCs were induced to differentiate into endoderm by addition of BMP4/FGF2/Activin A for 2 d followed by Activin A for 3 d as described previously (Fig. 1A; Mallanna and Duncan 2013). At this stage, typically 80%–90% of cells express endodermal markers, including GATA4, CXCR4, and SOX17 (D'Amour et al. 2005; McLean et al. 2007; Si-Tayeb et al. 2010b). The endoderm cells were then induced to adopt a hepatic fate by the addition of BMP4 and FGF2 (Mallanna and Duncan 2013) in the presence or absence of 2.5 µM FGFR inhibitor PD161570 or PD173074. Immunocytochemistry and real-time quantitative PCR (RT-qPCR) were then used to examine the impact of the inhibitors on the expression of markers that are characteristically expressed in hepatic progenitor cells. As shown in Figure 1B, in control differentiations, the cells were small and densely packed and robustly expressed the nuclear receptor hepatocyte nuclear factor 4α (HNF4A); however, the inclusion of FGFR inhibitors dramatically altered cell morphology and reduced the level of HNF4a protein to close to undetectable. In contrast to HNF4A, the level of the transcription factor GATA4, which is expressed in the endoderm prior to hepatic specification, was similar under all culture conditions. Quantification by RT-qPCR of a subset of mRNAs that are expressed in hepatic progenitor cells revealed a substantial reduction in the levels of HNF4A, Forkhead box A2 (FOXA2), HHEX, solute carrier family 35 (UDP-GlcA/UDP-GalNAc transporter) member D1 (SLC35D1), and phospholipase A2 group XIIB (PLA2G12B) mRNAs; however, there was no reduction in expression of GATA6 mRNA, which, like GATA4, is found in the endoderm prior to hepatic specification (Fig. 1C). We demonstrated previously that depletion of HNF4A in human pluripotent stem cells prevents hepatic specification (Delaforest et al. 2011). Given that the addition of PD161570 or PD173074 results in a dramatic reduction in expression of HNF4A as well as several other hepatic progenitor cell markers, we conclude that FGFR signaling acts upstream of HNF4A and is therefore essential for the production of hepatic progenitor cells from hiPSC-derived endoderm.
Figure 1.
Signaling through the FGFR is required for formation of hepatic progenitor cells from hiPSC-derived endoderm. (A) Overview of the procedure used to differentiate hepatic progenitor cells from hiPSCs (Mallanna and Duncan 2013). (B) Micrographs showing the result of immunocytochemistry to detect HNF4A and GATA4 following the differentiation of iPSCs to hepatic progenitor cells (day 8 of differentiation) in the absence (control) or presence of 2.5 µM FGFR inhibitors PD161570 and PD173074. The FGFR inhibitors were included along with FGF2 and BMP4 for 3 d during the transition of endoderm to hepatic progenitor cells. Cells were counterstained with DAPI to show the presence of cell nuclei. Bar, 100 µM. (C) Bar graph showing relative levels of mRNAs encoding hepatic markers FOXA2, HHEX, HNF4A, SLC35D1, and PLA2G12B and the endodermal marker GATA6 at day 8 of control differentiations or differentiations that included PD161570 and PD173074 during the conversion of the endoderm to a hepatic fate. Significance was calculated using Student's t-test. n = 3. (*) P < 0.05; (**) P < 0.01.
Identification of direct targets of FGFR signaling in definitive endoderm
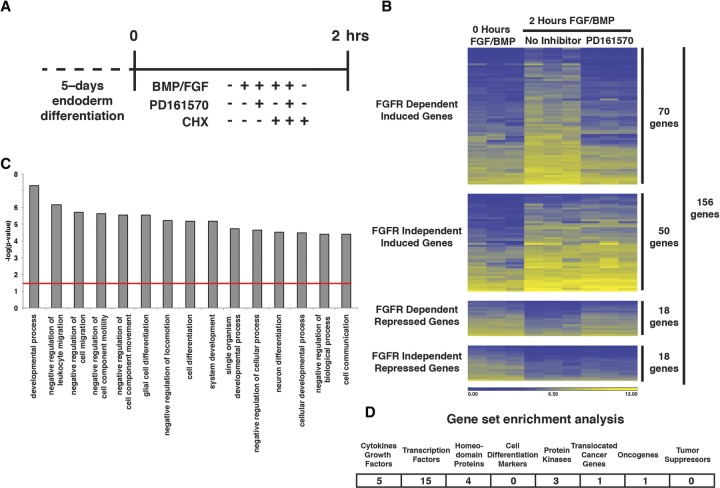
We assumed that FGF signaling would most likely mediate hepatic specification by ultimately regulating gene expression. To understand the molecular mechanism through which FGF regulates hepatic fate, we performed a series of experiments to identify those genes whose expression was controlled as a direct consequence of FGFR signaling (FGF immediate early genes) (Fig. 2A). First, RNA levels were determined by oligonucleotide array analyses after the formation of endoderm (0 h) and 2 h after the addition of FGF2 and BMP4 in the presence or absence of 2.5 µM FGFR inhibitor PD161570. BMP4 was included because it is necessary for specification of hepatic progenitor cells (Rossi et al. 2001). We identified 156 genes whose expression changed by >2.4-fold (z-score = less than −2 or >2) 2 h after addition of FGF2/BMP4 (Supplemental Table S1). By examining the effect of PD161570 on RNA levels, we could group the genes into four distinct clusters (Fig. 2B). We defined the expression of a gene as being dependent on FGFR activity if the change in expression from that found in the endoderm was twofold or less in the presence of PD161570. Addition of FGF2/BMP4 for 2 h induced expression of 120 genes, and, of those, the expression of 70 was dependent on FGFR activity as defined by this parameter. The expression of 36 genes was down-regulated after 2 h of FGF2/BMP4 treatment, and the inhibition of 18 was dependent on the FGFR. Gene ontology analyses revealed that genes whose change in expression was FGFR-dependent fell into a broad area of functional groups, including those associated with the regulation of development and cell differentiation and movement, which would be consistent with a role for FGF signaling in specifying hepatic fate (Fig. 2C). Interestingly, gene set enrichment analysis (GSEA) (Subramanian et al. 2005) revealed that, of the 88 FGFR-dependent genes examined, 15 encoded transcription factors, four of which contained a homeodomain (Fig. 2D). This implies that one function of FGF is to establish the transcription factor networks that are necessary to promote cell type-specific expression profiles.
Figure 2.
Identification of genes whose expression rapidly responds to treatment of endoderm with FGF2. (A) Schematic showing the experimental design used to identify FGFR immediate early genes. The levels of mRNAs were determined by oligonucleotide array analyses performed on three independently differentiated samples (n = 3 biological replicates) for each condition. (B) Heat map illustrating changes in gene expression that occur in response to addition of 10 ng/mL FGF2 and 20 ng/mL BMP4 in the presence or absence of FGFR inhibitor PD161570. (C) Bar graph showing Ingenuity Pathway Analysis (IPA) classification of functions (X-axis) associated with 88 genes whose expression was changed in response to FGFR signaling. The probability of each function (Y-axis) occurring by chance (red line) was determined by Fisher's exact test. (D) Table showing the classification of protein types identified by GSEA.
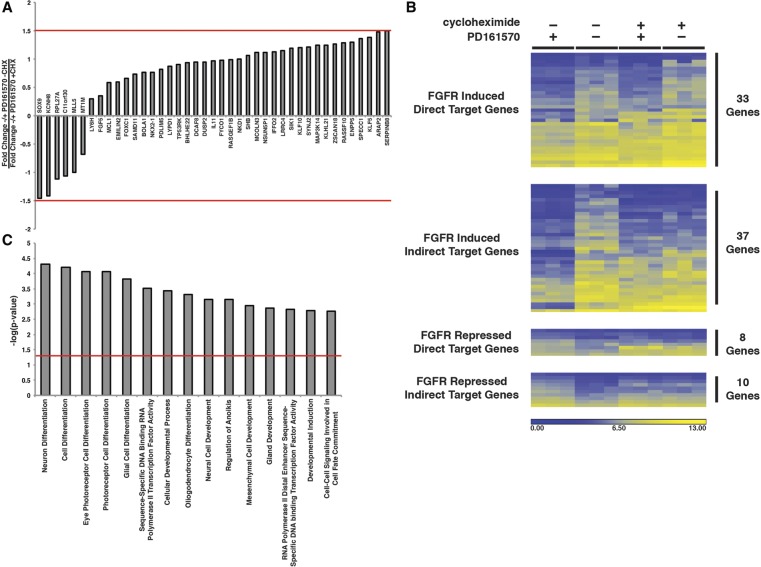
Although we performed all analyses at 2 h after addition of FGF, we recognized that expression of some genes could be regulated indirectly via the action of the transcription factors that were themselves induced by FGF. To definitively identify genes that are directly regulated by FGFR signaling, we performed transcriptional profiling on endoderm samples that were cultured for 2 h with FGF/BMP in the presence and absence of PD161570 along with cycloheximide to inhibit translation (Supplemental Fig. S1). Expression of direct targets should be either increased or decreased by PD161570, and any impact of the FGFR inhibitor should be independent of the presence of cycloheximide (Supplemental Table S1). A gene was considered a direct target when treatment with cycloheximide reduced the FGFR inhibitor-mediated change of expression ≤1.5-fold (Fig. 3A). As shown in Figure 3B, analyses of the 88 genes whose expression was affected by FGFR signaling (Fig. 2) revealed that the expression of 41 was affected independently of cycloheximide. Of the 41 genes whose expression was directly affected by FGFR signaling, the expression of 33 was increased, and the expression of eight was decreased. Gene ontology and literature searches revealed that the 41 genes that were directly dependent on FGF for expression had diverse functions, with many being associated with cell differentiation and development (Fig. 3C). GSEAs revealed that eight of the genes encoded transcription factors (Supplemental Fig. S2A). Based on the mechanisms underlying FGFR signaling, analyses of regulatory networks by Ingenuity Pathway Analysis (IPA), as expected, revealed that several FGFR immediate early genes were closely integrated into a network that incorporated RAS–RAF–MAPK and PI3K–AKT signaling (Supplemental Fig. S2B).
Figure 3.
Identification of FGF-induced immediate early genes. (A) Bar graph showing the impact of cycloheximide on the expression of genes that are classified as being directly regulated by FGFR signaling. Immediate early genes were defined as those whose change in expression was affected by ≤1.5-fold in the presence of cycloheximide (red line). (B) Heat map illustrating the impact of 2.5 µM PD161570 and 100 µM cycloheximide on mRNA levels encoded by genes that are regulated through FGFR signaling. The levels of mRNAs were determined by oligonucleotide array analyses performed on three independently differentiated samples (n = 3 biological replicates) for each condition. (C) Bar graph showing IPA classification of functions (X-axis) associated with 41 genes whose expression was considered an immediate early response to FGFR signaling. The probability of each function (Y-axis) occurring by chance (red line) was determined by Fisher's exact test.
NKD1 is a direct target of FGFR signaling
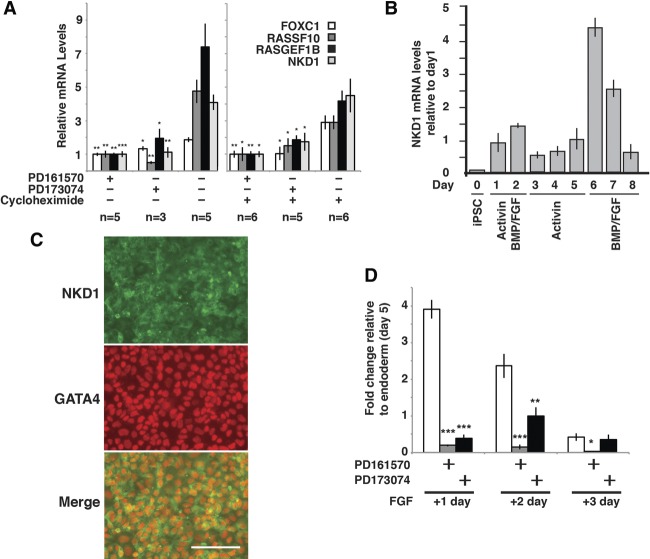
To confirm the validity of the screen, we performed RT-qPCR to determine the RNA levels encoded by four selected genes—RASGEF1B, FOXC1, RASSF10, and NKD1—whose expression we predicted to be directly regulated through the FGFR. As shown in Figure 4A, the level of RNA expressed from each gene 2 h after addition of FGF was inhibited by the addition of both PD161570 and PD173074, and the inclusion of cycloheximide did not prevent the induction by FGF (−FGFR inhibitors). These data indicate that the experimental design was appropriate to identify FGFR-regulated immediate early genes during the formation of hepatic progenitor cells.
Figure 4.
NKD1 is transiently induced in the endoderm by FGFR signaling. (A) Bar graphs displaying the relative levels of FOXC1, RASSF10, RASGEF1B, and NKD1 RNA determined by RT-qPCR 2 h after addition of FGF2 and BMP4 to iPSC-derived endoderm in the presence and absence of 2.5 µM FGFR inhibitors PD161570 and PD173074 in the presence and absence of 100 µM cycloheximide. Significance of changes compared with no FGFR inhibitor was determined by Student's t-test. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. (B) Bar graph showing changes in the level of NKD1 mRNA throughout the differentiation process measured by RT-qPCR. Error bars indicate SEM. n = 3 biological replicates. Note that the highest level of NKD1 mRNA occurs shortly after the addition of FGF2 (day 6). (C) Immunocytochemistry reveals the presence of NKD1 protein (green) predominantly in the cytoplasm and associated with membranes in cells expressing the endoderm marker GATA4 (red) in the nucleus. Bar, 100 µM. (D) Bar graphs showing NKD1 mRNA levels determined by RT-qPCR in differentiations performed in the absence or presence of PD161570 or PD173074 during the conversion of the endoderm to hepatic progenitor cells. Student's t-test, (*) P ≤ 0.05; (**) P ≤ 0.01; (***) P ≤ 0.001.
Of the FGF targets that could potentially control hepatic progenitor cell formation, NKD1 appeared to be a particularly provocative candidate because it acts as an intracellular WNT signaling inhibitor by repressing the action of the Dishevelled family of proteins (Yan et al. 2001; Gao and Chen 2010). As discussed above, several studies have shown that WNT signaling must be transiently blocked in order for hepatic specification to progress (McLin et al. 2007; Li et al. 2008). Given these prior findings, we hypothesized that FGF could control specification of the hepatic progenitor cells by inducing the expression of NKD1 during the endoderm-to-hepatic transition in order to effect a temporally regulated inhibition of WNT signaling.
We first examined the expression profile of NKD1 during each stage of the differentiation procedure by RT-qPCR (Fig. 4B). NKD1 mRNA was close to undetectable in pluripotent stem cells (day 0) but increased slightly as the cells differentiated toward an endoderm fate (days 1–5). One day after the addition of FGF2/BMP4 to the endoderm (day 6), the level of NKD1 mRNA sharply increased approximately fourfold compared with that found in the endoderm and then gradually declined over the next 2 d. We examined the distribution of NKD1 protein by immunocytochemistry to determine whether NKD1 was present specifically within the endoderm following addition of FGF. As shown in Figure 4C, anti-NKD1 immunoreactivity was detected predominantly throughout the cytoplasm and associated cell membranes of FGF2/BMP4-treated cells and the cells costained with an antibody that recognizes GATA4, which is characteristically expressed in the endodermal population. Finally, we confirmed that the induction of NKD1 throughout hepatic progenitor cell formation was dependent on FGFR activity using FGFR inhibitors (Fig. 4D). As before, NKD1 was induced approximately fourfold after the addition of FGF2/BMP4 compared with hiPSC-derived definitive endoderm, and the levels then declined gradually over time. However, when the cells were treated with FGF2/BMP4 in the presence of either PD161570 or PD173074, this induction of NKD1 expression was severely inhibited (Fig. 4D). Since induction of NKD1 also occurs in the presence of cycloheximide (Fig. 4A), these data cumulatively demonstrate that expression of NKD1 is induced in the endoderm during hepatic specification and is a direct target of FGFR activation.
FGF-induced expression of NKD1 is essential for hepatic progenitor cell formation
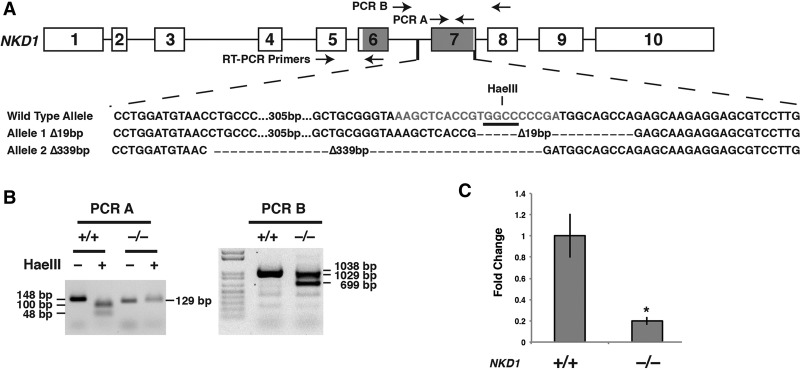
If FGF mediates the generation of hepatic progenitor cells by activating the expression of NKD1, we predicted that loss of NKD1 function should inhibit the differentiation of the endoderm toward a hepatic fate. To test this hypothesis, we first generated NKD1−/− iPSCs using CRISPR–Cas9 (Cong et al. 2013; Ding et al. 2013; Mali et al. 2013). We chose to target NKD1 exon 7 because this exon encodes the Dishevelled (DVL1)-interacting domain, which is necessary for NKD1 function (Fig. 5A; Katoh 2001; Zhang et al. 2007). We identified an iPSC clone by DNA sequence and PCR analyses that harbored deletions of 19 and 339 base pairs (bp) (Fig. 5B) in its two NKD1 alleles. We referred to this compound heterozygous line as NKD1−/−. NKD1 mRNA levels were examined by RT-qPCR using oligonucleotides that recognized exon 5, which lies outside of the targeted region (Fig. 5A). After 1 d of FGF2/BMP4 treatment, which corresponds to day 6 of differentiation when NKD1 is maximally expressed (Fig. 4B), NKD1 mRNA was reduced to background levels in the NKD1−/− cells (Fig. 5C). Given that the deletions in the NKD1−/− cells were within exon 7, downstream from the primers used for RT-qPCR, the observed reduction in NKD1 mRNA likely reflects nonsense-mediated decay of the transcript (Baker and Parker 2004).
Figure 5.
Generation of NKD1−/− hiPSCs using CRISPR-CAS9. (A) Schematic illustrating the strategy used to introduce deletions into NKD1 exon 7. NKD1 intron–exon structure is shown with exons 6 and 7, which encode DVL1-interacting domains, shaded in gray. The guide sequence is presented in red along with the position of a diagnostic HaeIII site (underlined) and PCR primers used in the genotype analyses. The relative position of indels found in NKD1−/− iPSCs are also indicated in the sequence. (B) PCR analyses reveal deletions in NKD1 exon 7 of NKD1−/− hiPSCs. The amplicon from control (+/+) cells contains a HaeIII site in exon 7 that is deleted in the NKD−/− cells (left panel) and a 339-bp deletion can be identified in amplicons generated using oligonucleotide set B (right panel). Expected sizes of all products are shown in base pairs. All indels were confirmed by DNA sequencing. (C) Bar graphs showing the results of RT-qPCR analyses on control (+/+) and NKD1−/− (−/−) cells at day 6 of differentiation. t-test, (*) P ≤ 0.05 compared with control cells. n = 2.
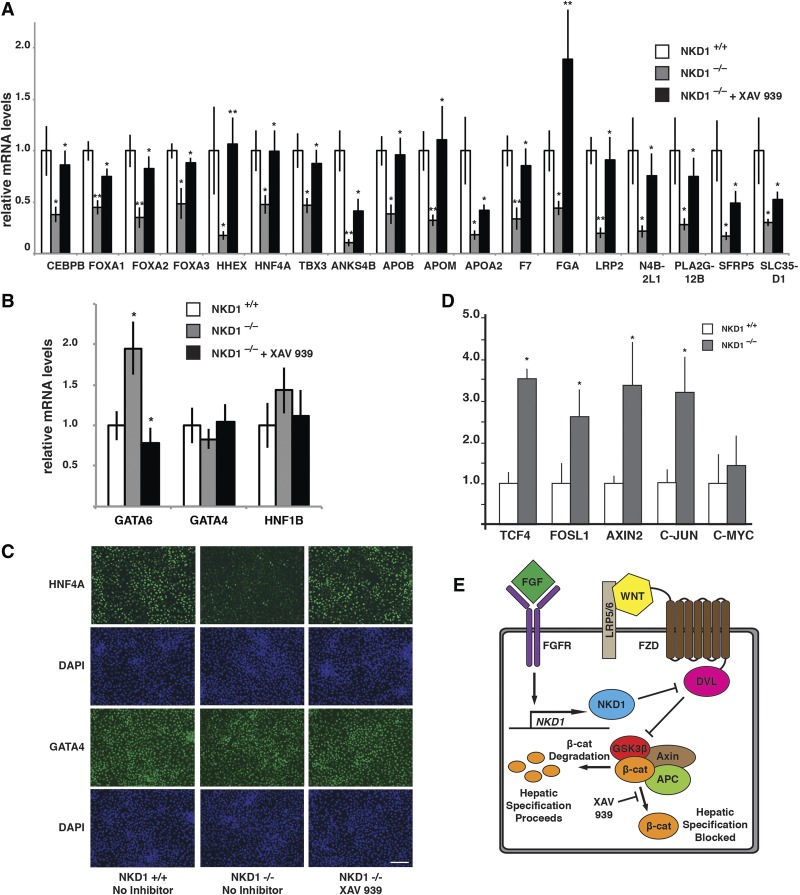
We next addressed whether NKD1 was necessary for hepatic progenitor cell formation by comparing expression of characteristic hepatic markers following the differentiation of NKD1+/+ and NKD1−/− iPSCs (Fig. 6). The mRNA levels of 11 markers of hepatic fate (ANKS4B, APOB, APOM, APOA2, F7, FGA, LRP2, N4B2L1, PLA2G12B, SFRP5, and SLC35D1) and seven transcription factors with known roles in liver development (CEBPB, FOXA1, FOXA2, FOXA3, HHEX, HNF4A, and TBX3) were strikingly and significantly decreased in NKD1−/− compared with NKD1+/+ hepatic progenitor cells at day 8 of differentiation (Fig. 6A). In contrast to the majority of markers examined, GATA6 and GATA4, which are expressed in the endoderm, and HNF1B, which is required for specification of hepatic progenitor cells in mouse embryos, were unaffected by the loss of NKD1, implying that regulation of these genes during hepatic progenitor cell formation is by a distinct mechanism (Fig. 6B). Finally, we felt it was important to confirm that HNF4A protein was depleted in NKD1−/− cells by immunostaining because HNF4A has such a key role in regulating hepatic progenitor cell fate. While HNF4A was readily detected in control NKD1+/+ cells, it was severely depleted in NKD1−/− cells by day 8 of differentiation (Fig. 6C). Consistent with the observation that only low levels of NKD1 mRNA were detected in the endoderm prior to hepatic specification, mRNAs encoding the endodermal proteins FOXA1, FOXA2, FOXA3, GATA4, SOX17, and CXCR4 were similar in day 5 endoderm derived from both control and NKD1−/− iPSCs (Supplemental Fig. S3A). Similar to analyses of mRNA levels, there was no observed effect on endodermal expression of GATA4, FOXA2, and SOX17 proteins (Supplemental Fig. S3B). These data imply that loss of NKD1 does not affect endoderm character but, in contrast, is necessary for the conversion of the endoderm to a hepatic fate.
Figure 6.
NKD1 is required for formation of hepatic progenitor cells. (A,B) Bar graphs showing RT-qPCR analyses of the levels of mRNAs encoding characteristic hepatic markers and liver enriched transcription factors in hepatic progenitor cells derived from control (+/+) or NKD1−/− (−/−) hiPSCs in the absence or presence of XAV 939. Significance was determined between control versus NKD1−/− cells and NKD1−/− versus NKD1−/− + XAV 939 using Student's t-test n = 5. (*) P < 0.05; (**) P < 0.01. (C) Micrographs showing immunocytochemistry to detect HNF4A and GATA4 in day 8 hepatic progenitor cells derived from control, NKD1−/−, and NKD1−/− XAV 939 hiPSCs. Cells were counterstained with DAPI to reveal nuclei. Bar, 100 µM. (D) Bar graph showing the results of RT-qPCR analyses to determine the mRNA levels of β-catenin-regulated genes in day 6 hepatic progenitor cells derived from control (NKD1+/+) or NKD1−/− iPSCs. Significance was calculated using Student's t-test. n = 4. (*) P < 0.05. (E) Schematic summarizing the major conclusion of this study. FGF directly induces expression of the canonical WNT inhibitor NKD1 to suppress WNT signaling that inhibits the conversion of the endoderm to a hepatic fate.
NKD1 acts by inhibiting WNT signaling through its direct interaction with DVL1, which is required for WNT-mediated signal transduction through the frizzled family of proteins (Stewart 2014). This mechanism of action would therefore imply that hepatic progenitor cell formation is blocked in NKD1−/− cells due to the inappropriate activation of WNT signaling pathways during the endoderm-to-hepatic transition. To test whether this implication was correct, we attempted to rescue the formation of hepatic progenitor cells by inhibiting WNT signaling in NKD1−/− cells using a WNT signaling antagonist, XAV 939. XAV 939 is a small molecule inhibitor of tankyrase, which antagonizes WNT signaling by promoting β-catenin degradation. We chose XAV 939 because, by promoting β-catenin degradation, this small molecule should phenocopy the mechanism of action of NKD1. As presented in Figure 6A, the inclusion of XAV 939 between days 6 and 7 of the differentiation of NKD1−/− cells (a time frame that coincides with maximal expression of NKD1 mRNA) returned the expression of the majority of hepatic mRNAs back to that found in control NKD1+/+ cells and resulted in an increase in HNF4A protein expression (Fig. 6C). In contrast to the effect on markers whose expression was depleted by the loss of NKD1, XAV 939 had no effect on expression of GATA4 or HNF1B (Fig. 6B). A partial rescue of hepatic marker expression was also obtained using an independent WNT inhibitor called IWR1 (Supplemental Fig. S4). Given that NKD1 is an inhibitor of WNT signaling, we predicted that expression of canonical WNT targets should be increased in NKD1−/− cells. We therefore examined the impact of loss of NKD1 on mRNA levels encoded by known β-catenin target genes TCF4, AXIN2, C-JUN, C-MYC, and FOSL1 by RT-qPCR. As predicted, all of these mRNAs were increased in hepatic progenitor cells derived from NKD1−/− iPSCs compared with control iPSCs (Fig. 6D), although the impact on C-MYC was modest. Based on these results, we conclude that FGF2 mediates the formation of hepatic progenitor cells primarily by antagonizing canonical WNT signaling, effected by its induction of NKD1 expression (Fig. 6E).
Discussion
FGF signaling has been studied extensively and shown to play critical and diverse roles in development, cell differentiation, metabolism, and tissue injury (Ornitz and Itoh 2015). Paracrine FGF signaling is primarily mediated by the interaction of FGFs with FGFR tyrosine kinases, which results in the activation of the RAS–RAF–MAPK, PI3K–AKT, STAT, and PLCγ signaling cascades (Itoh and Ornitz 2011). Studies in mouse embryos have shown that the regulation of hepatic specification of the endoderm by FGF predominantly occurs through induction of MAPK (Calmont et al. 2006). Whether this is conserved in human cells remains to be determined. The FGF pathway is highly regulated at multiple levels, including interactions of FGFs with heparan sulfate proteoglycans (which regulate FGF bioavailability), differential splicing of FGFRs, modulation of signaling activity by dual specificity phosphatases (DUSPs) and Sprouty proteins, and binding of FGF to cofactors and decoy receptors (for review, see Dailey et al. 2005; Lanner and Rossant 2010; Ornitz and Itoh 2015). While FGF signaling has been shown to regulate specification of hepatic fate in diverse organisms, the complexity associated with FGF regulation has made it challenging to understand the specific mechanism through which FGFs convert endoderm to hepatic progenitor cells.
In the present study, we exploited human pluripotent stem cells—a model that allows synchronous and reproducible production of hepatic progenitor cells—in order to identify FGFR-regulated immediate early genes that might mediate the conversion of the endoderm to a hepatic fate. Using relatively stringent selection criteria, we identified 41 genes that exhibited a change in expression that was (1) induced in response to FGF/BMP treatment for 2 h, (2) inhibited by FGFR antagonists, and (3) independent of inhibition of protein synthesis by cycloheximide. We therefore conclude that the observed change in expression of these 41 genes is an immediate early response of the endoderm to FGFR signaling.
Examination of these 41 genes using GSEA revealed that eight encoded transcription factors (BHLHE22, C11orf30, FOXC1, KLF10, KLF5, NKX2-1, SOX9, and ZSCAN18), which is consistent with the view that FGF plays an early role in establishing the transcriptional regulation of genes that define hepatic progenitor cells. Although little is known about the role of these factors during hepatic specification, several have been implicated in cell differentiation in other systems. BHLHE22 is a member of the basic helix–loop–helix family of transcription factors that typically bind to E-boxes to mediate a diverse array of cellular functions; this family includes NEUROD, eHAND/dHAND, and MYOD, known to have roles in neuronal (Jan and Jan 1993), cardiac (Srivastava and Olson 2000), and myogenic (Weintraub et al. 1991) development. FOXC1 is a member of the forkhead family of transcription factors (which includes FOXA1/A2) that act as pioneer transcription factors that are essential for hepatic specification (Lee et al. 2005; Zaret and Carroll 2011). FOXC1 has been implicated in growth of cerebellum (Haldipur et al. 2014) and corneal vascular development (Seo et al. 2012). Finally, SOX9 is an HMG-box transcription factor that has been implicated in regulating development of various stem cell compartments, including the liver (Jo et al. 2014). It is unlikely that any of these transcription factors are individually essential for hepatic specification because defects in liver development have not been reported in the relevant mouse knockout strains. Alternatively, it seems more likely that these targets could coordinate the transition of the endoderm to a hepatic fate as an FGF-responsive transcription factor cluster, and we believe that the stem cell differentiation system should allow future dissection of such contributions.
In addition to transcription factors, several genes encoding cell signaling proteins were shown to be regulated through FGFR activation. These include ARAP2, ENPP5, NKD1, RASGEF1B, RASSF10, and SHB. Our attention was drawn to NKD1 because it is a negative regulator of WNT signaling (Yan et al. 2001), and previous studies have shown that WNT signaling must be transiently inhibited to specify the hepatic lineage (McLin et al. 2007; Li et al. 2008).
NKD was first discovered in a Drosophila melanogaster mutagenesis screen and was called naked cuticle (nkd) (Jürgens 1984; Martizez Arias et al. 1988). Mutation of nkd resulted in elevated levels of β-catenin despite normal levels of Wg (the fly homolog of Wnt protein) (Zeng et al. 2000). Subsequently, Nkd was shown to inactivate the scaffolding protein DVL1 (Yan et al. 2001). DVL1 has been described as the hub of intracellular WNT signaling (Gao and Chen 2010). It promotes canonical WNT pathways primarily by being recruited to Frizzled receptors and preventing the proteolytic degradation of β-catenin; however, its mechanisms of action are diverse and likely affect multiple aspects of the WNT pathway (Gao and Chen 2010). In addition to NKD1, several proteins that inhibit or promote the activity of Dishevelled activity have been described (Gao and Chen 2010).
To assess the role of NKD1 in hepatic differentiation, we used CRISPR/Cas9 to inactivate both alleles of NKD1 in our hiPSCs. When NKD1−/− cells were induced to differentiate, we found that the expression of hepatic markers was reduced substantially. Importantly, this block to differentiation was reversed when XAV 939, a small molecule inhibitor of WNT signaling, was included in the medium for 48 h during the endoderm-to-hepatic transition. Both NKD1 and XAV 939 inhibit canonical WNT signaling by promoting the degradation of β-catenin—NKD1 by inhibiting DVL1, and XAV 939 by stabilizing AXIN through tankyrase inhibition. The fact that both of these WNT signaling inhibitors act by targeting β-catenin degradation through independent mechanisms provides confidence that the loss of hepatic progenitors during the differentiation of NKD1−/− iPSCs is indeed due to a failure to inhibit canonical WNT signaling. Several independent studies support this conclusion. Studies using Xenopus laevis provide compelling evidence that WNT/β-catenin activity must be transiently suppressed in the anterior endoderm to allow development of the liver and pancreas (McLin et al. 2007). Importantly, complete repression of WNT activity is also incompatible with hepatic specification, and thus a minimum threshold of WNT signaling must be maintained (Zhang et al. 2013). Suppression of canonical WNT signaling is required, however, because β-catenin indirectly inhibits expression of a transcription factor hhex (McLin et al. 2007; Li et al. 2008), which is essential to establish the parenchymal cells of the liver (Bogue et al. 2000; Keng et al. 2000; Martinez Barbera et al. 2000; Wallace et al. 2001). Consistent with these data, we found that HHEX is significantly reduced during the differentiation of NKD1−/− iPSCs, and this expression is re-established in the presence of XAV 939. Interestingly, RNA sequencing (RNA-seq) studies in mouse embryos have revealed gene signatures that suggest that noncanonical WNT signaling in the endoderm is required to distinguish pancreatic from hepatic fate (Rodriguez-Seguel et al. 2013). However, the relationship between the activation of noncanonical and the suppression of canonical WNT signaling during hepatic progenitor cell formation remains unclear.
We believe that the evidence for a role for NKD1 in suppressing canonical WNT signaling during the formation of hiPSC-derived hepatic progenitor cells is compelling. However, it is important to recognize that mice with compound mutations in Nkd1 and Nkd2 are viable and have no reported defects in liver development, although Nkd1−/−;Nkd2−/− mice do have reduced litter sizes (Zhang et al. 2007). Since over a dozen proteins have been reported to inhibit DVL1 (Gao and Chen 2010), it is possible that the absence of a developmental phenotype in Nkd1−/−;Nkd2−/− mice may reflect functional redundancy. If any DVL1 inhibitors are differentially expressed in mice versus humans, then differences in the requirement for NKD1 between the species could be observed. However, we favor an alternative explanation, which is that secreted inhibitors of WNT signaling that are expressed during the hiPSC differentiation process (Delaforest et al. 2011), such as Cerberus and SFRPs, are removed during medium changes. The removal of secreted inhibitors in culture would increase the dependence on cell-autonomous suppressors of the WNT pathway, such as NKD1, facilitating their identification as key contributors to the differentiation of iPSCs in screens such as ours.
In summary, we identified immediate early genes that respond to FGFR activity during the formation of hepatic progenitor cells from hiPSC-derived endoderm. Among these genes are several that encode transcription factors, signaling molecules, and growth factors. We predict that several of these products will coordinate the transition of the endoderm to a hepatic fate and prepare the cell and its local environment for subsequent stages of differentiation. Among the immediate early genes that were induced by FGFR activation was NKD1, which encodes an intracellular inhibitor of canonical WNT signaling. We demonstrated that mutation of NKD1 prevents the formation of hepatic progenitor cells in a fashion similar to inhibition of FGFRs. Replacing NKD1 with a pharmacological inhibitor of canonical WNT signaling can circumvent the block in hepatic specification. We therefore conclude that one mechanism through which FGFR activation mediates hepatic fate is to transiently suppress WNT activity by inducing expression of NKD1. We believe that further analyses of FGFR immediate early genes will provide additional novel insights into the fundamental molecular mechanisms that control hepatic cell fate.
Materials and methods
Cell culture
Human iPS-K3 cells (Si-Tayeb et al. 2010b) were maintained in mTeSR (Ludwig et al. 2006a) supplemented with 1% KOSR (LifeTech) and 4 ng/mL zbFGF (Ludwig et al. 2006a) on an E-cadherin-IgG Fc fusion protein matrix (Nagaoka et al. 2010) under 4% O2/5% CO2. For maintenance, cells were passaged every 4–6 d using Versene/EDTA (Lonza). Cells were plated for differentiation after dispersal using Accutase (Millipore) and plated onto 12-well tissue culture-treated plates precoated with 2 mg/mL Matrigel (LifeTech). Three subconfluent 100-mm plates of iPSCs were sufficient for differentiation in one 12-well plate. One day after cell seeding, differentiation was initiated as described in a step-by-step protocol published previously (Mallanna and Duncan 2013).
Immunostaining
Cultured cells were fixed with 4% PFA for 30 min at room temperature and then made permeable with 0.5% Triton X-100 in PBS for 15 min. Cells were blocked with 3% BSA in PBS for 30 min and incubated overnight at 4°C with primary antibodies diluted in 1% BSA in PBS. The antibodies used were HNF4A (1:500; Santa Cruz Biotechnology, sc-6556), GATA4 (1:250; Santa Cruz Biotechnology, sc-1237), and NKD1 (1:250; Abcam, ab133650). Cells were washed in 1% BSA in PBS and incubated with secondary antibody (Molecular Probes, A21206, Alexa fluor anti-rabbit 488 nm; A11058, Alexa fluor anti-goat 594 nm; and A11055, Alexa fluor anti-goat 488) and DAPI for 1 h at room temperature and washed with PBS. Images were processed using Adobe Photoshop to optimize brightness and contrast, with all control and experimental images being treated identically.
RT-qPCR analysis
RNA was isolated from hiPSCs using RNeasy minikits (Qiagen). Genomic DNA was removed using 1 µL of RNase-free DNase I per 5 µg of RNA. First strand cDNA was synthesized using M-MLV RT with dNTPs and random hexamer primers. RT-qPCR was performed on an Applied Biosystems StepOne Plus real-time PCR machine using PrimeTime (Integrated DNA Technologies) assays or a Bio-Rad CFX-384 real-time PCR machine using Power SYBR Green (LifeTech) following the manufacturers’ protocols. PrimeTime assays and SYBR Green primers are listed in Supplemental Table S2.
Oligonucleotide array analysis
Total RNA was isolated from three independent differentiations for each condition using the RNeasy minikit (Qiagen). Biotinylated cRNA was generated using the IVT Express kit and hybridized to GeneChip Primeview human gene expression arrays (Affymetrix). Images were acquired using a GeneChip Scanner 3000 (Affymetrix), and data analysis was conducted using the Partek Genomics suite statistical analysis software (Partek). Genes induced after 2 h of 10 ng/mL FGF2 treatment were identified using a Z-score cutoff of less than −2 or more than +2, which corresponded to a 2.4-fold change. Genes changed between 2 h of 10 ng/mL FGF2 without inhibitor and 2 h of 10 ng/mL FGF2 with 2.5 µM PD161570 were identified. We defined expression of a gene as being dependent on FGFR activity if the change in expression from that found in the endoderm was twofold or less in the presence of PD161570. Changes that were independent of protein synthesis were identified following 2 h of 10 ng/mL FGF2 with and without 2.5 µM PD161570 in the presence and absence of 100 µM cycloheximide. A gene was considered a direct target when treatment with cycloheximide reduced the FGFR inhibitor-mediated change of expression ≤1.5-fold. Original .CEL files are available from the Gene Expression Omnibus (accession no. GSE69533).
CRISPR/Cas9 genome editing
CRISPRs were designed to target exon 7 of NKD1 following the procedure described by Zhang and colleagues (Ran et al. 2013). A guide sequence for the NKD1 DVL1-interacting domain (AAGCTCACCGTGGCCCCCGA) was cloned into pSPCas9(BB)-2A-Puro (Addgene plasmid ID 48139) (Ran et al. 2013). The construct was introduced using electroporation into pluripotent hiPSCs (iPS-K3 cells) (Si-Tayeb et al. 2010c). Transfected cells were cultured on Matrigel in Y27632 (StemRD) for 24 h, and transiently transfected cells were enriched by culture in 1 µg/mL puromycin for 2 d. Cells were maintained in 1% KOSR/mTeSR for a further 2 wk without selection. Individual colonies were collected and divided for cell line expansion and PCR analysis of genomic DNA using QuickExtract DNA extraction solution (EpiBio). The targeted region of NKD1 was amplified (PCR A) using Herculase fusion polymerase (Agilent) and oligonucleotides that recognized sequence within exon 7 (For, GAGGAATGAGAGTCCATTTC; Rev, TCAGTGGGCTTGGTCTCTGC). The amplicon was digested with HaeIII restriction enzyme (New England Biolabs) to detect indels. A second independent PCR (PCR B) was performed using oligonucleotides that spanned exon 7 (For, GAGGAATGAGAGTCCATTTC; Rev, TCAGTGGGCTTGGTCTCTGC) to detect a larger deletion. The nucleotide sequence of each amplicon was established (Retrogen) in order to confirm the identities of the indels.
Supplementary Material
Acknowledgments
Amy Ludwig-Kubinski and Jixuan Li provided excellent technical support. This work was supported by gifts from the Marcus Family, the Phoebe R. and John D. Lewis Foundation, the Sophia Wolf Quadracci Memorial Fund, and the Advancing a Healthier Wisconsin Fund and by National Institutes of Health grants DK102716, HG006398, and HD082570.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.268961.115.
References
- Agarwal S, Holton KL, Lanza R. 2008. Efficient differentiation of functional hepatocytes from human embryonic stem cells. Stem Cells 26: 1117–1127. [DOI] [PubMed] [Google Scholar]
- Baker KE, Parker R. 2004. Nonsense-mediated mRNA decay: terminating erroneous gene expression. Curr Opin Cell Biol 16: 293–299. [DOI] [PubMed] [Google Scholar]
- Basma H, Soto-Gutiérrez A, Yannam GR, Liu L, Ito R, Yamamoto T, Ellis E, Carson SD, Sato S, Chen Y, et al. 2009. Differentiation and transplantation of human embryonic stem cell-derived hepatocytes. Gastroenterology 136: 990–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogue CW, Ganea GR, Sturm E, Ianucci R, Jacobs HC. 2000. Hex expression suggests a role in the development and function of organs derived from foregut endoderm. Dev Dyn 219: 84–89. [DOI] [PubMed] [Google Scholar]
- Cai J, Zhao Y, Liu Y, Ye F, Song Z, Qin H, Meng S, Chen Y, Zhou R, Song X, et al. 2007. Directed differentiation of human embryonic stem cells into functional hepatic cells. Hepatology 45: 1229–1239. [DOI] [PubMed] [Google Scholar]
- Calmont A, Wandzioch E, Tremblay KD, Minowada G, Kaestner KH, Martin GR, Zaret KS. 2006. An FGF response pathway that mediates hepatic gene induction in embryonic endoderm cells. Dev Cell 11: 339–348. [DOI] [PubMed] [Google Scholar]
- Chen Y, Jürgens K, Hollemann T, Claußen M, Ramadori G, Pieler T. 2003. Cell-autonomous and signal-dependent expression of liver and intestine marker genes in pluripotent precursor cells from Xenopus embryos. Mech Dev 120: 277–288. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey L, Ambrosetti D, Mansukhani A, Basilico C. 2005. Mechanisms underlying differential responses to FGF signaling. Cytokine Growth Factor Rev 16: 233–247. [DOI] [PubMed] [Google Scholar]
- D'Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, Baetge EE. 2005. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol 23: 1534–1541. [DOI] [PubMed] [Google Scholar]
- Delaforest A, Nagaoka M, Si-Tayeb K, Noto FK, Konopka G, Battle MA, Duncan SA. 2011. HNF4A is essential for specification of hepatic progenitors from human pluripotent stem cells. Development 138: 4143–4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Regan SN, Xia Y, Oostrom LA, Cowan CA, Musunuru K. 2013. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell 12: 393–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Chen YG. 2010. Dishevelled: the hub of Wnt signaling. Cell Signal 22: 717–727. [DOI] [PubMed] [Google Scholar]
- Gualdi R, Bossard P, Zheng M, Hamada Y, Coleman JR, Zaret KS. 1996. Hepatic specification of the gut endoderm in vitro: cell signaling and transcriptional control. Genes Dev 10: 1670–1682. [DOI] [PubMed] [Google Scholar]
- Haldipur P, Gillies GS, Janson OK, Chizhikov VV, Mithal DS, Miller RJ, Millen KJ. 2014. Foxc1 dependent mesenchymal signalling drives embryonic cerebellar growth. Elife 3: e03962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Dziedzic N, Gadue P, Keller GM, Gouon-Evans V. 2011. An endothelial cell niche induces hepatic specification through dual repression of Wnt and Notch signaling. Stem Cells 29: 217–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay DC, Zhao D, Fletcher J, Hewitt ZA, McLean D, Urruticoechea-Uriguen A, Black JR, Elcombe C, Ross JA, Wolf R, et al. 2008. Efficient differentiation of hepatocytes from human embryonic stem cells exhibiting markers recapitulating liver development in vivo. Stem Cells 26: 894–902. [DOI] [PubMed] [Google Scholar]
- Houssaint E. 1980. Differentiation of the mouse hepatic primordiam. I. An analysis of tissue interactions in hepatocyte differentiation. Cell Diff 9: 269–279. [DOI] [PubMed] [Google Scholar]
- Itoh N, Ornitz DM. 2011. Fibroblast growth factors: from molecular evolution to roles in development, metabolism and disease. J Biochem 149: 121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan YN, Jan LY. 1993. HLH proteins, fly neurogenesis, and vertebrate myogenesis. Cell 75: 827–830. [DOI] [PubMed] [Google Scholar]
- Jo A, Denduluri S, Zhang B, Wang Z, Yin L, Yan Z, Kang R, Shi LL, Mok J, Lee MJ, et al. 2014. The versatile functions of Sox9 in development, stem cells, and human diseases. Genes Dis 1: 149–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J, Zheng M, Goldfarb M, Zaret KS. 1999. Initiation of mammalian liver development from endoderm by fibroblast growth factors. Science 284: 1998–2003. [DOI] [PubMed] [Google Scholar]
- Jürgens G. 1984. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. Roux's Arch Dev Biol 193: 283–295. [DOI] [PubMed] [Google Scholar]
- Katoh M. 2001. Molecular cloning, gene structure, and expression analyses of NKD1 and NKD2. Int J Oncol 19: 963–969. [PubMed] [Google Scholar]
- Keng VW, Yagi H, Ikawa M, Nagano T, Myint Z, Yamada K, Tanaka T, Sato A, Muramatsu I, Okabe M, et al. 2000. Homeobox gene Hex is essential for onset of mouse embryonic liver development and differentiation of the monocyte lineage. Biochem Biophys Res Commun 276: 1155–1161. [DOI] [PubMed] [Google Scholar]
- Lanner F, Rossant J. 2010. The role of FGF/Erk signaling in pluripotent cells. Development 137: 3351–3360. [DOI] [PubMed] [Google Scholar]
- LeDouarin N. 1968. Synthese du glycogene dans les hepatocytes en voie de differentiation: role des mesenchymes homologue et heterologues. Dev Biol 17: 101–114. [DOI] [PubMed] [Google Scholar]
- LeDouarin NM. 1975. An experimental analysis of liver development. Med Biol 53: 427–455. [PubMed] [Google Scholar]
- Lee CS, Friedman JR, Fulmer JT, Kaestner KH. 2005. The initiation of liver development is dependent on Foxa transcription factors. Nature 435: 944–947. [DOI] [PubMed] [Google Scholar]
- Lemaigre FP. 2009. Mechanisms of liver development: concepts for understanding liver disorders and design of novel therapies. Gastroenterology 137: 62–79. [DOI] [PubMed] [Google Scholar]
- Li Y, Rankin SA, Sinner D, Kenny AP, Krieg PA, Zorn AM. 2008. Sfrp5 coordinates foregut specification and morphogenesis by antagonizing both canonical and noncanonical Wnt11 signaling. Genes Dev 22: 3050–3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig TE, Bergendahl V, Levenstein ME, Yu J, Probasco MD, Thomson JA. 2006a. Feeder-independent culture of human embryonic stem cells. Nat Methods 3: 637–646. [DOI] [PubMed] [Google Scholar]
- Ludwig TE, Levenstein ME, Jones JM, Berggren WT, Mitchen ER, Frane JL, Crandall LJ, Daigh CA, Conard KR, Piekarczyk MS, et al. 2006b. Derivation of human embryonic stem cells in defined conditions. Nat Biotechnol 24: 185–187. [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. 2013. RNA-guided human genome engineering via Cas9. Science 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallanna SK, Duncan SA. 2013. Differentiation of hepatocytes from pluripotent stem cells. Curr Protoc Stem Cell Biol 26: 1G.4.1–1G.4.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez Barbera JP, Clements M, Thomas P, Rodriguez T, Meloy D, Kioussis D, Beddington RS. 2000. The homeobox gene Hex is required in definitive endodermal tissues for normal forebrain, liver and thyroid formation. Development 127: 2433–2445. [DOI] [PubMed] [Google Scholar]
- Martizez Arias A, Baker NE, Ingham PW. 1988. Role of segment polarity genes in the definition and maintenance of cell states in the Drosophila embryo. Development 103: 157–170. [DOI] [PubMed] [Google Scholar]
- McLean AB, D'Amour KA, Jones KL, Krishnamoorthy M, Kulik MJ, Reynolds DM, Sheppard AM, Liu H, Xu Y, Baetge EE, et al. 2007. Activin a efficiently specifies definitive endoderm from human embryonic stem cells only when phosphatidylinositol 3-kinase signaling is suppressed. Stem Cells 25: 29–38. [DOI] [PubMed] [Google Scholar]
- McLin VA, Rankin SA, Zorn AM. 2007. Repression of Wnt/β-catenin signaling in the anterior endoderm is essential for liver and pancreas development. Development 134: 2207–2217. [DOI] [PubMed] [Google Scholar]
- Nagaoka M, Si-Tayeb K, Akaike T, Duncan SA. 2010. Culture of human pluripotent stem cells using completely defined conditions on a recombinant E-cadherin substratum. BMC Dev Biol 10: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz DM, Itoh N. 2015. The fibroblast growth factor signaling pathway. Wiley Interdiscip Rev Dev Biol 4: 215–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. 2013. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8: 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Seguel E, Mah N, Naumann H, Pongrac IM, Cerda-Esteban N, Fontaine JF, Wang Y, Chen W, Andrade-Navarro MA, Spagnoli FM. 2013. Mutually exclusive signaling signatures define the hepatic and pancreatic progenitor cell lineage divergence. Genes Dev 27: 1932–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi JM, Dunn NR, Hogan BL, Zaret KS. 2001. Distinct mesodermal signals, including BMPs from the septum transversum mesenchyme, are required in combination for hepatogenesis from the endoderm. Genes Dev 15: 1998–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S, Singh HP, Lacal PM, Sasman A, Fatima A, Liu T, Schultz KM, Losordo DW, Lehmann OJ, Kume T. 2012. Forkhead box transcription factor FoxC1 preserves corneal transparency by regulating vascular growth. Proc Natl Acad Sci 109: 2015–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serls AE, Doherty S, Parvatiyar P, Wells JM, Deutsch GH. 2005. Different thresholds of fibroblast growth factors pattern the ventral foregut into liver and lung. Development 132: 35–47. [DOI] [PubMed] [Google Scholar]
- Shifley ET, Kenny AP, Rankin SA, Zorn AM. 2012. Prolonged FGF signaling is necessary for lung and liver induction in Xenopus. BMC Dev Biol 12: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D, Lee Y, Poss KD, Stainier DYR. 2011. Restriction of hepatic competence by Fgf signaling. Development 138: 1339–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si-Tayeb K, Lemaigre FP, Duncan SA. 2010a. Organogenesis and development of the liver. Dev Cell 18: 175–189. [DOI] [PubMed] [Google Scholar]
- Si-Tayeb K, Noto FK, Nagaoka M, Li J, Battle MA, Duris C, North PE, Dalton S, Duncan SA. 2010b. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology 51: 297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si-Tayeb K, Noto FK, Sepac A, Sedlic F, Bosnjak ZJ, Lough JW, Duncan SA. 2010c. Generation of human induced pluripotent stem cells by simple transient transfection of plasmid DNA encoding reprogramming factors. BMC Dev Biol 10: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Cai J, Liu Y, Zhao D, Yong J, Duo S, Song X, Guo Y, Zhao Y, Qin H, et al. 2009. Efficient generation of hepatocyte-like cells from human induced pluripotent stem cells. Cell Res 19: 1233–1242. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Olson EN. 2000. A genetic blueprint for cardiac development. Nature 407: 221–226. [DOI] [PubMed] [Google Scholar]
- Stewart DJ. 2014. Wnt signaling pathway in non-small cell lung cancer. J Natl Cancer Inst 106: djt356. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan GJ, Hay DC, Park IH, Fletcher J, Hannoun Z, Payne CM, Dalgetty D, Black JR, Ross JA, Samuel K, et al. 2010. Generation of functional human hepatic endoderm from human induced pluripotent stem cells. Hepatology 51: 329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay KD, Zaret KS. 2005. Distinct populations of endoderm cells converge to generate the embryonic liver bud and ventral foregut tissues. Dev Biol 280: 87–99. [DOI] [PubMed] [Google Scholar]
- Wallace KN, Yusuff S, Sonntag JM, Chin AJ, Pack M. 2001. Zebrafish hhex regulates liver development and digestive organ chirality. Genesis 30: 141–143. [DOI] [PubMed] [Google Scholar]
- Weintraub H, Dwarki VJ, Verma I, Davis R, Hollenberg S, Snider L, Lassar A, Tapscott SJ. 1991. Muscle-specific transcriptional activation by MyoD. Genes Dev 5: 1377–1386. [DOI] [PubMed] [Google Scholar]
- Yan D, Wallingford JB, Sun TQ, Nelson AM, Sakanaka C, Reinhard C, Harland RM, Fantl WJ, Williams LT. 2001. Cell autonomous regulation of multiple Dishevelled-dependent pathways by mammalian Nkd. Proc Natl Acad Sci 98: 3802–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS. 2008. Genetic programming of liver and pancreas progenitors: lessons for stem-cell differentiation. Nat Rev Genet 9: 329–340. [DOI] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. 2011. Pioneer transcription factors: establishing competence for gene expression. Genes Dev 25: 2227–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Wharton KAJ, Mack JA, Wang K, Gadbaw M, Suyama K, Klein PS, Scott MP. 2000. naked cuticle encodes an inducible antagonist of Wnt signalling. Nature 403: 789–795. [DOI] [PubMed] [Google Scholar]
- Zhang W, Yatskievych TA, Baker RK, Antin PB. 2004. Regulation of Hex gene expression and initial stages of avian hepatogenesis by Bmp and Fgf signaling. Dev Biol 268: 312–326. [DOI] [PubMed] [Google Scholar]
- Zhang S, Cagatay T, Amanai M, Zhang M, Kline J, Castrillon DH, Ashfaq R, Oz OK, Wharton KAJ. 2007. Viable mice with compound mutations in the Wnt/Dvl pathway antagonists nkd1 and nkd2. Mol Cell Biol 27: 4454–4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Rankin SA, Zorn AM. 2013. Different thresholds of Wnt–Frizzled 7 signaling coordinate proliferation, morphogenesis and fate of endoderm progenitor cells. Dev Biol 378: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn AM, Wells JM. 2009. Vertebrate endoderm development and organ formation. Annu Rev Cell Dev Biol 25: 221–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.