In this study, Lee et al. show that SREBP and MDT-15 prevent the life-shortening effects of a glucose-rich diet by regulating fat-converting processes in C. elegans. By using a genome-wide screen, they found that inhibition of SREBP or MDT-15 further accelerated aging in glucose-rich conditions, and up-regulation of SREBP or MDT-15 reversed accelerated aging.
Keywords: aging, glucose, fat, SREBP, MDT-15, C. elegans
Abstract
Glucose-rich diets shorten the life spans of various organisms. However, the metabolic processes involved in this phenomenon remain unknown. Here, we show that sterol regulatory element-binding protein (SREBP) and mediator-15 (MDT-15) prevent the life-shortening effects of a glucose-rich diet by regulating fat-converting processes in Caenorhabditis elegans. Up-regulation of the SREBP/MDT-15 transcription factor complex was necessary and sufficient for alleviating the life-shortening effect of a glucose-rich diet. Glucose feeding induced key enzymes that convert saturated fatty acids (SFAs) to unsaturated fatty acids (UFAs), which are regulated by SREBP and MDT-15. Furthermore, SREBP/MDT-15 reduced the levels of SFAs and moderated glucose toxicity on life span. Our study may help to develop strategies against elevated blood glucose and free fatty acids, which cause glucolipotoxicity in diabetic patients.
Glucose is an essential energy source for many cellular processes and should be tightly regulated to maintain organismal health. Abnormal glucose metabolism is linked to obesity and pathological conditions, such as diabetes mellitus and cardiovascular diseases. Glucose reduces the life spans of model organisms, including Caenorhabditis elegans, by influencing various factors that regulate aging (Schulz et al. 2007; Lee et al. 2009; Schlotterer et al. 2009).
Glucose and fat metabolisms are tightly linked. Excessive amounts of dietary carbohydrates, including glucose, are stored as lipids. Elevated levels of blood glucose and fats correlate with increased risks of cardiovascular diseases, diabetes, and specific types of cancers. Fatty acid (FA) deficiency is also linked to many disorders, including growth disturbance, infertility, and kidney failure (for review, see Kremmyda et al. 2011). Emerging evidence indicates that fat metabolism affects life span. Several C. elegans mutants with impaired fat metabolism have reduced life spans (Van Gilst et al. 2005; Taubert et al. 2006). Interestingly, some mutant worms with long life span display increased fat (Kimura et al. 1997; O'Rourke et al. 2009; Shi et al. 2013), whereas others are low in fat (Heestand et al. 2013). Thus, it remains unclear whether fat metabolism plays a causal role in the regulation of C. elegans life span.
Sterol regulatory element-binding proteins (SREBPs) are key transcription factors that govern fat metabolism. A high carbohydrate diet activates mammalian SREBPs and induces expression of lipogenic genes, including acetyl-CoA carboxylase (ACC), FA synthase (FAS), and stearoyl-CoA desaturase 1 (SCD1) (Horton et al. 1998; Hasty et al. 2000). These lipogenic genes are crucial for normal fat metabolism and organismal viability, as shown by mouse knockout studies (Abu-Elheiga et al. 2001; Miyazaki et al. 2001; Chirala et al. 2003). The mechanisms through which SREBPs regulate lipid metabolism appear to be conserved in C. elegans (McKay et al. 2003; Walker et al. 2010, 2011). The C. elegans SREBP homolog SBP-1 and the transcriptional coregulator mediator-15 (MDT-15) cooperatively control the expression of lipogenic genes, including those that encode SCDs (Taubert et al. 2006; Yang et al. 2006; Nomura et al. 2010; Walker et al. 2010). The SCDs catalyze the generation of unsaturated FAs (UFAs) by adding double bonds to the carbon chain in saturated FAs (SFAs). SREBP and its downstream enzymes appear to play key roles in various aspects of animal physiology, including reproduction and development (Yang et al. 2006; Brock et al. 2007).
The goal of this study was to identify genes that affect the ability of a glucose-rich diet to reduce life span. We performed a genome-wide RNAi screen on C. elegans that expressed a novel glucose reporter GFP. We identified >200 genes that potentially modulated the effects of dietary glucose on life span. We found that genetic inhibition of MDT-15 or SREBP further decreased the short life span of glucose-fed worms. Conversely, up-regulation of MDT-15 or SREBP restored normal life span in glucose-fed conditions. Dietary glucose increased the level of SREBP, which induced several key enzymes that convert SFAs to UFAs. In addition, SREBP/MDT-15 was necessary and sufficient for ameliorating the accumulation of SFAs during glucose feeding. Furthermore, the increased metabolic flow from glucose via SFA-to-UFA synthesis moderated the toxic effects of glucose on life span. These data indicate that reduction of SFAs by SREBP/MDT-15 prevents the accelerated aging caused by a high glucose diet.
Results
far-3p::GFP is a glucose-responsive, fluorescent C. elegans
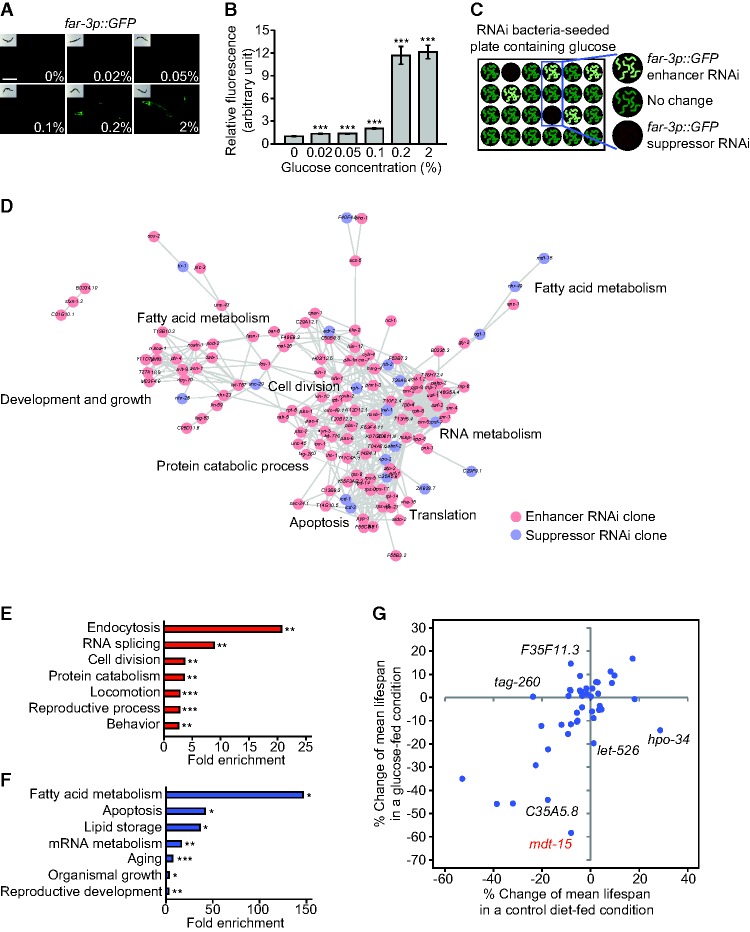
We sought to establish a glucose reporter that could be used in a genetic screen. We examined four available transgenic animals that expressed GFP under the control of glucose-responsive gene promoters (Supplemental Fig. S1A; Lee et al. 2009). Transgenic worms that expressed GFP under the control of the FA/retinol-binding protein 3 (far-3) promoter (far-3p::GFP) displayed prominent and consistent induction of fluorescence upon glucose feeding (Fig. 1A,B; Supplemental Fig. S1A). Expression of far-3p::GFP was also moderately induced in response to various sugars and sugar metabolites, including fructose, trehalose, and glycerol, which can be converted from and to glucose in vivo (Supplemental Fig. S1B,C). In contrast, far-3p::GFP was hardly induced by the following potential glucose-related stressors: elevated osmolarity by NaCl treatment, oxidative stress by paraquat treatment, endoplasmic reticulum (ER) stress by tunicamycin treatment, and increased temperature (Supplemental Fig. S1D–K). These results established far-3p::GFP as an in vivo glucose reporter.
Figure 1.
A genome-wide RNAi screen using a glucose-responsive GFP reporter C. elegans. (A) Addition of various concentrations of glucose significantly induced far-3p::GFP. Bar, 200 μm. (B) Quantification of the GFP intensity in A. Error bars represent standard error of the mean (SEM). (***) P < 0.001, two-tailed Student's t-tests. n ≥ 40 from two independent experiments. (C) Experimental design of the genome-wide RNAi screen. far-3p::GFP transgenic animals were cultured on dsRNA-expressing bacteria on 24-well solid NGM plates containing 2% glucose. Changes in fluorescence intensity for enhancer and suppressor RNAi clones were blindly and independently screened by three researchers. (D) A gene interaction network and gene ontology (GO) terms among enhancer and suppressor hits from our RNAi screen were analyzed by STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) (Szklarczyk et al. 2015) and DAVID (Database for Annotation, Visualization, and Integrated Discovery) (Huang et al. 2009). (E,F) GO terms annotated in the enhancer (E) and suppressor (F) RNAi clones, which increased and decreased the level of far-3p::GFP, respectively. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001, modified Fisher's exact tests (Huang et al. 2009). Because glucose and fat metabolism are closely related to each other, genes involved in FA metabolism may have been enriched in the suppressor RNAi clones. One noticeable caveat for this explanation is that far-3 is a FA metabolism-related gene, and this may have skewed our screen data for fat metabolism-related genes. (G) Percent mean life span changes by the suppressor RNAi clones from our screen with (Y-axis) or without (X-axis) additional glucose treatment compared with control RNAi. The life-shortening effect of mdt-15 RNAi (red) was much higher in glucose-enriched conditions than in control conditions. We also found that far-3 RNAi did not affect life span with or without additional glucose feeding (Supplemental Fig. S1L). See Supplemental Table S3 for statistical analysis of the life span data.
A genome-wide RNAi screen identifies genes that modulate the life-reducing effect of glucose
We performed a genome-wide screen to identify RNAi clones that influenced far-3p::GFP expression in response to glucose feeding (Fig. 1C). We found that 170 enhancer and 44 suppressor RNAi clones reproducibly increased and decreased, respectively, the expression of far-3p::GFP on a glucose-rich diet (Supplemental Tables S1, S2). Gene ontology (GO) analysis indicated that genes involved in endocytosis, RNA splicing, cell division, and protein catabolism were enriched in the enhancer RNAi clones (Fig. 1D,E). Genes involved in FA metabolism, apoptosis, lipid storage, mRNA metabolism, and aging were overrepresented in the suppressor RNAi clones (Fig. 1D,F). Next, we examined the effects of the suppressor RNAi clones on the life-shortening effects of glucose (Supplemental Table S1). RNAi targeting mdt-15, C35A5.8, let-526, hpo-34, tag-260, or F35F11.3 differentially affected life span in glucose-fed versus control conditions (Fig. 1G; Supplemental Fig. S2A–E and see the legend for details; Supplemental Table S1.). Thus, our genome-wide RNAi screen and life span assays identified genes that modulated the life-reducing effect of glucose.
MDT-15 and SBP-1 protect against life-reducing glucose toxicity
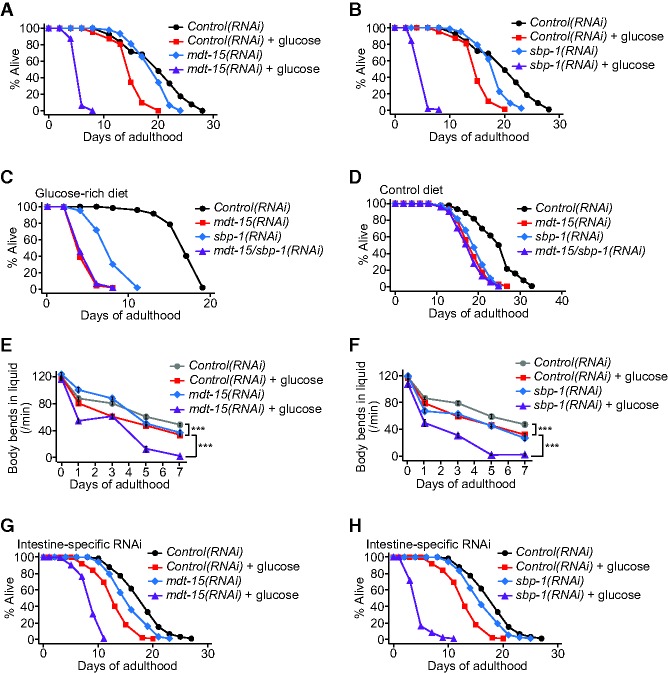
mdt-15 RNAi or mutation suppressed the induction of far-3 by glucose (Supplemental Fig. S3A–C) and further shortened the life span of animals in glucose-fed conditions (Figs. 1G, 2A; Supplemental Fig. S3D–F; Supplemental Table S1). MDT-15 functions as a transcriptional coregulator of fat metabolism and stress responses through interacting with transcription factors, including SREBP/SBP-1, nuclear hormone receptor-49 (NHR-49), and nuclear factor–erythroid-related factor 2 (Nrf2 [SKN-1]) (Taubert et al. 2006; Yang et al. 2006; Goh et al. 2014; Pang et al. 2014). sbp-1 RNAi or mutation substantially reduced life span in glucose-fed conditions, similar to that observed with mdt-15 RNAi (Fig. 2B; Supplemental Fig. S3G–I). In contrast, inhibition of NHR-49 or SKN-1 decreased life span independently of glucose feeding (Supplemental Fig. S3J,K). Consistent with the idea that SBP-1 and MDT-15 act together as a complex, genetic inhibition of sbp-1 did not further decrease the short life span of mdt-15(RNAi) worms on a glucose-rich diet (Fig. 2C,D). Knockdown of mdt-15 or sbp-1 also dramatically accelerated age-dependent declines in movement and feeding capacities upon glucose feeding (Fig. 2E,F; Supplemental Fig. S4A–J). These results suggest that MDT-15 and SBP-1 function together to protect worms from progeria in glucose-rich conditions.
Figure 2.
MDT-15 and SBP-1 are required for protecting worms from living too short upon glucose treatment. (A,B) Genetic inhibition of mdt-15 (A) or sbp-1 (B) by RNAi further shortened life span upon glucose treatment. (C,D) Shown are life span curves of worms treated with mdt-15/sbp-1 double RNAi on a glucose-rich diet (C) and a control diet (D). sbp-1 RNAi treatment did not further shorten the life span of mdt-15 RNAi-treated worms. Note that mdt-15(RNAi) worms may have lived too short upon glucose feeding to display further decreases in life span. (E,F) Knockdown of mdt-15 (E) or sbp-1 (F) enhanced age-dependent declines in motility (body bends in liquid) on a glucose-rich diet. (***) P < 0.001; P-values were calculated by using two-way ANOVA test. Error bars represent SEM. n = 15. Other motility assays—including measurements of body bends in solid medium and feeding rate—and additional repeats are shown in Supplemental Figure S4. See Supplemental Table S5 for statistical analysis of all of the measurements in E and F and Supplemental Figure S4. (G,H) mdt-15 RNAi (G) or sbp-1 RNAi (H) specifically in the intestine by using an nhx-2 promoter-driven rde-1 in an RNAi-defective rde-1(ne219) mutant background further decreased the short life span of glucose-fed worms. For the life span and motility assays, worms were treated with mdt-15 RNAi only during adulthood and were treated with sbp-1 RNAi for their whole lives. See the Supplemental Material for details. See Supplemental Tables S4 and S6 for additional repeats and statistical analysis of the life span data shown in this figure.
We examined tissue-specific effects of MDT-15 and SBP-1 on life span in response to a glucose-rich diet. As reported previously (McKay et al. 2003; Taubert et al. 2006; Walker et al. 2010), GFP expression driven by an mdt-15 promoter (mdt-15p::GFP) or an sbp-1 promoter (sbp-1p::GFP) was detected in multiple tissues (Supplemental Fig. S5A,B). RNAi against mdt-15 in the intestine, hypodermis, or neurons significantly shortened the life span of glucose-treated animals; mdt-15 RNAi in body wall muscle did not (Fig. 2G; Supplemental Fig. S5C–G; see the legends for the Supplemental Figures for further discussion). Intestine-specific sbp-1 RNAi further decreased the short life span of glucose-fed worms, whereas hypodermis-specific or body wall muscle-specific sbp-1 RNAi did not (Fig. 2H; Supplemental Fig. S5H–L). Thus, the intestine is a common tissue in which inhibition of mdt-15 or sbp-1 exacerbated the life span-shortening effects of glucose feeding.
Up-regulation of SBP-1/MDT-15 during glucose feeding protects animals from glucose toxicity
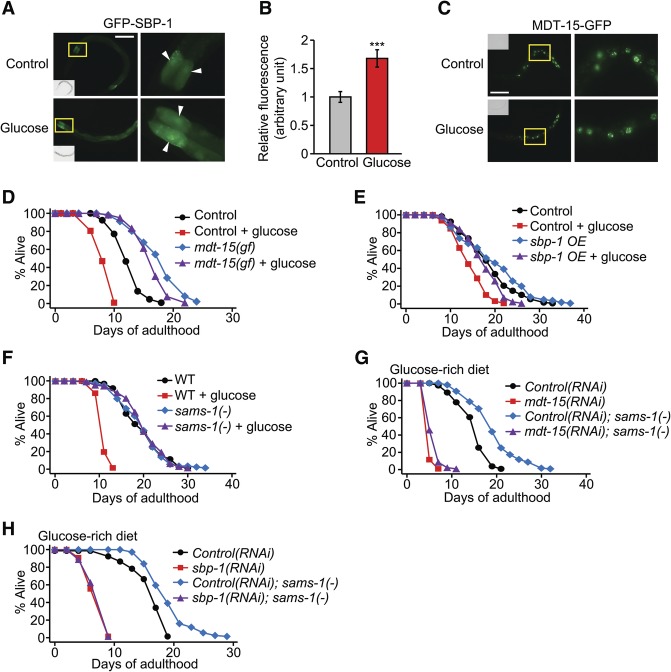
Next, we examined whether a glucose-rich diet influenced the activity of SBP-1 or MDT-15. Glucose-rich diet feeding increased overall levels of GFP-fused SBP-1 in intestinal cells (Fig. 3A,B; Supplemental Fig. S6A,B) while not increasing the mRNA levels of sbp-1 (Supplemental Fig. S6C). We also found that glucose treatment did not affect the level or expression pattern of MDT-15-GFP or mdt-15 mRNA (Fig. 3C; Supplemental Fig. S6D). Thus, dietary glucose appears to increase SBP-1 protein levels post-transcriptionally.
Figure 3.
SBP-1 and MDT-15 are sufficient for detoxifying the effects of glucose on life span. (A) Glucose-rich diet feeding increased the fluorescence intensity of GFP-SBP-1 proteins in the intestinal cells of IJ1208 sbp-1p::GFP::sbp-1 animals (L4 larvae). Arrowheads indicate the intestinal nuclei. Bar, 100 μm. Yellow boxes indicate the enlarged regions in A and C. We confirmed this result by using another GFP-SBP-1 transgenic strain, epEx141[sbp-1p::GFP::sbp-1; rol-6(su1006)] (Supplemental Fig. S6A,B; Walker et al. 2011). Fructose, another sugar, also increased GFP-SBP-1 levels and decreased life span (Supplemental Fig. S6E–G). (B) Quantification of GFP-SBP-1 levels in A. n ≥ 21 from three independent experiments. (C) Glucose feeding did not affect the expression pattern of MDT-15-GFP proteins in mdt-15p::mdt-15::GFP animals (L2 larvae). Bar, 50 μm. (D) The mdt-15(et14) gain-of-function (gf) mutation suppressed the shortened life span of worms fed with a glucose-rich diet. Please note that all animals that were used for the life span assay in D contained paqr-2(tm3410) mutations because the mdt-15(et14) mutation is very closely linked (0.33 cM) to paqr-2(tm3410) mutations (Svensk et al. 2013). (E) sbp-1 overexpression (sbp-1 OE; sbp-1p::GFP::sbp-1) largely restored normal life span in glucose-rich conditions. Injection marker (roller) transgenic worms (CF1290) were used as a control. (F) sams-1(ok3033) [sams-1(−)] mutation suppressed short life span caused by glucose-rich diet feeding. mdt-15(gf), sbp-1 overexpression, and sams-1(−) also suppressed the life span-shortening effect of 0.2% dietary glucose (Supplemental Fig. S7A–C). (G,H) Knockdown of mdt-15 (G) or sbp-1 (H) suppressed the restored life span by sams-1 mutation upon glucose-rich diet feeding. Note that inhibition of sams-1 increases life span (Hansen et al. 2005; Cabreiro et al. 2013; Ding et al. 2015), but we found that the sams-1(ok3033) mutation had little or no life span-extending effect in our experimental conditions. We speculate that different experimental conditions (e.g., bacterial food source) may have caused the different results. See Supplemental Table S7 for additional repeats and statistical analysis for the life span data shown in this figure.
We next asked whether up-regulation of SBP-1 or MDT-15 moderates the toxic effects of glucose on life span. An mdt-15 gain-of-function (gf) mutation, et14 (Svensk et al. 2013), restored normal life span in glucose-fed worms (Fig. 3D; Supplemental Fig. S7A). Similarly, the life span-reducing effect of glucose was significantly suppressed by sbp-1 overexpression (Fig. 3E; Supplemental Fig. S7B). Mutations in the S-adenosyl methionine synthetase gene (sams-1), which increase the activity of SBP-1 (Walker et al. 2011), conferred resistance to the life-shortening effects of glucose (Fig. 3F; Supplemental Fig. S7C). The life span-restoring effects of sams-1 mutations in high glucose conditions were largely dependent on mdt-15 and sbp-1 (Fig. 3G,H; Supplemental Fig. S7D,E). Thus, up-regulation of the SBP-1/MDT-15 complex appears to protect animals from the life-reducing effect of excessive dietary glucose.
Glucose feeding induces FA desaturases, transcriptional targets of SBP-1/MDT-15
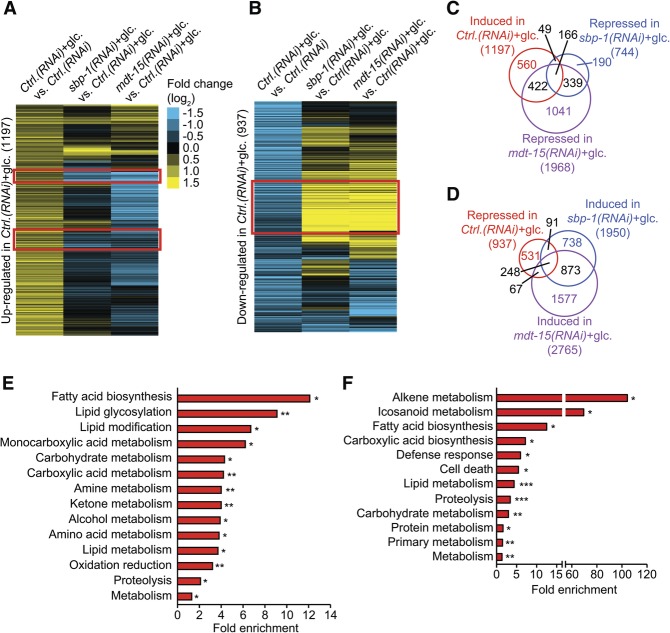
We performed RNA sequencing to measure the mRNA levels of genes that were differentially expressed by a glucose-rich diet and SBP-1/MDT-15 (P < 0.05, fold change >1.2) (Fig. 4A–D; Supplemental Fig. S8A–E). SBP-1 and MDT-15 shared a large number of common targets (Fig. 4C,D; Supplemental Fig. S8F–I), consistent with their roles as components of a transcription factor complex. Expression of a large subset of glucose-responsive genes was dependent on SBP-1 and MDT-15 (Fig. 4A–D). Various GO terms, including lipid metabolism and carbohydrate metabolism, were significantly enriched among the glucose-responsive genes whose expression was dependent on SBP-1 and MDT-15 (Fig. 4E,F). Overall, these data suggest that SBP-1 and MDT-15 regulate the expression of many common metabolic genes upon glucose-rich diet feeding.
Figure 4.
RNA sequencing identifies genes that are differentially regulated by glucose feeding and SBP-1/MDT-15. (A,B) Heat maps display clusters of the genes significantly induced (A) and repressed (B) by glucose-rich diet feeding (glc.) whose expression was reversed by sbp-1 RNAi or mdt-15 RNAi. Red boxes indicate SBP-1/MDT-15-dependent induction (A) and repression (B) of glucose-responsive genes. Ctrl.(RNAi) indicates control RNAi-treated wild-type (N2) worms. (C,D) Venn diagrams show that large fractions of glucose-induced genes (C) and glucose-repressed genes (D) are targets of SBP-1 and MDT-15. All of the overlaps of gene sets in C and D are significant. P < 10−50, hypergeometric probability test. (E) Overrepresented GO terms of the 166 overlapping genes in C. (F) Overrepresented GO terms of the 248 genes in D. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001, modified Fisher's exact tests (Huang et al. 2009).
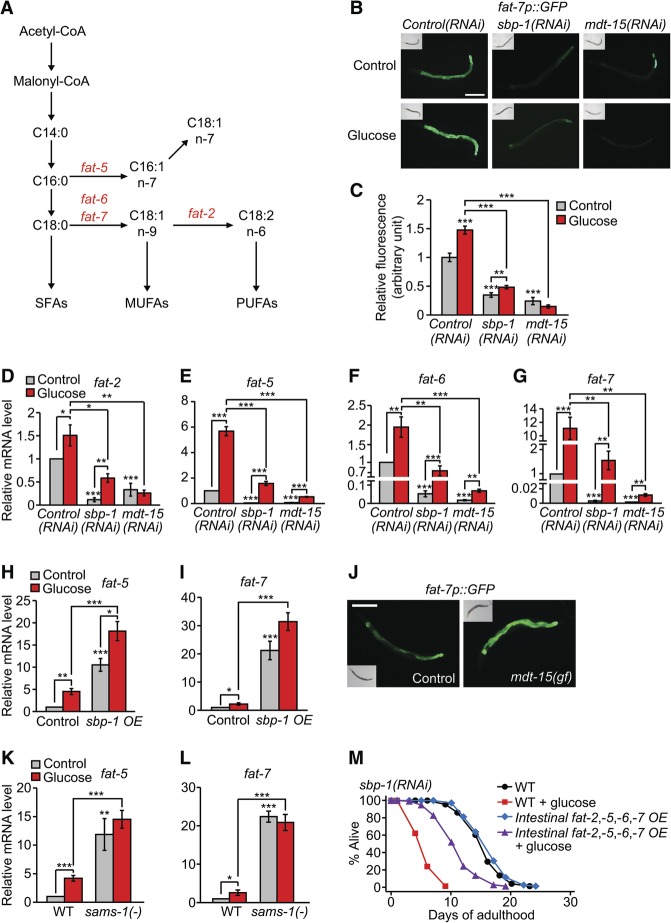
FA biosynthesis was significantly enriched among SBP-1/MDT-15-dependent glucose-induced GO terms (Fig. 4E). In particular, expression of the SCD1 homolog FAT-7 was substantially reduced in both sbp-1(RNAi) and mdt-15(RNAi) worms (Supplemental Table S8; Taubert et al. 2006; Yang et al. 2006; Nomura et al. 2010). FAT-7 is a key enzyme for FA biosynthesis and conversion of SFAs to monounsaturated FAs (MUFAs) and polyunsaturated FAs (PUFAs) (Fig. 5A). We confirmed the mRNA sequencing results of fat-7 by using GFP transgenic animals (Fig. 5B,C). We then expanded our expression analysis to include all seven C. elegans FA desaturases (fat-1 to fat-7). A glucose-rich diet induced fat-1, fat-2, fat-5, fat-6, and fat-7 (Fig. 5D–G; Supplemental Fig. S9A–G), although the expression of fat-2 and fat-6 was variable depending on the types of bacterial foods (Supplemental Fig. S9H–K; see the legends for the Supplemental Figures for discussion). RNAi targeting mdt-15 or sbp-1 reduced the expression of fat-2, fat-5, fat-6, and fat-7 (Fig. 5D–G; Supplemental Fig. S9D–G). Moreover, sbp-1 overexpression, the mdt-15 gain-of-function mutation, or the sams-1 mutation induced these FA desaturases (Fig. 5H–L; Supplemental Fig. S9H–K; Walker et al. 2011). Thus, glucose feeding appears to increase the expression of multiple FA desaturases, which are targets of the SBP-1/MDT-15 complex.
Figure 5.
FA desaturases are glucose-responsive targets of MDT-15 and SBP-1. (A) A simplified schematic of the FA synthesis pathway representing the steps that generate various SFAs, MUFAs, and PUFAs. Enzymatic steps that were catalyzed by the gene products of fat-5, fat-6, fat-7, and fat-2 are indicated. (B) Knockdown of sbp-1 or mdt-15 decreased the induction of fat-7p::GFP on a glucose-rich diet. Bar, 200 μm. (C) Quantification of the data shown in B. n ≥ 27 from three independent experiments. (D–G) Relative mRNA levels of fat-2 (D), fat-5 (E), fat-6 (F), and fat-7 (G) in sbp-1 RNAi-treated or mdt-15 RNAi-treated worms were measured by using quantitative RT–PCR (qRT–PCR). sbp-1 RNAi or mdt-15 RNAi significantly decreased the mRNA levels of fat-2, fat-5, fat-6, and fat-7 on a glucose-rich diet. n ≥ 4. (H,I) Relative mRNA levels of fat-5 (H) and fat-7 (I) were increased in sbp-1-overexpressing (sbp-1 OE) worms with or without glucose-enriched diet feeding. n = 4. Injection marker (roller) transgenic worms (CL2070) were used as a control. (J) The level of fat-7p::GFP was increased by mdt-15(et14) gain-of-function (gf) mutation. paqr-2(tm3410) mutant background was used for both control and mdt-15(gf) (Svensk et al. 2013). Bar, 200 μm. (K,L) The sams-1(ok3033) [sams-1(−)] mutation increased the expression of fat-5 (K) and fat-7 (L) regardless of glucose treatment. n ≥ 5. Error bars represent SEM. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001, two-tailed Student's t-test. (M) An intestinal promoter (ges-1p)-driven fat-2, fat-5, fat-6, and fat-7 overexpression (Intestinal fat-2,-5,-6,-7 OE) partly restored the very short life span of sbp-1(RNAi) worms upon glucose-rich diet feeding. See Supplemental Fig. S10A,B and the legends for the results regarding the effects of intestinal fat-2, fat-5, fat-6, and fat-7 overexpression on mdt-15(RNAi) and Control(RNAi) worms. We also found that genetic inhibition of multiple FA desaturases—fat-5, fat-6, and fat-7 or fat-2 and fat-5—did not have specific life span-shortening effects on a glucose-rich diet (Supplemental Fig. S10C,D). This negative result can be interpreted in many different ways, including redundancy of diverse branches in the FA synthesis pathway and of multiple FA desaturases (Fig. 5A).
We next determined the role of these multiple fat genes in the life-reducing effects of dietary glucose. We found that intestinal overexpression of FA desaturases (fat-2, fat-5, fat-6, and fat-7) significantly suppressed the short life span of sbp-1(RNAi) worms on a glucose-rich diet (Fig. 5M); the results with RNAi knockdown of these fat genes were inconclusive (Supplemental Fig. S10C,D and see the legend for details). These data suggest that FA desaturases can reduce the toxic effects of dietary glucose on life span.
SBP-1/MDT-15 converts SFAs to UFAs during glucose-rich diet feeding
We examined whether MDT-15 and SBP-1 affected life span by modulating fat metabolism in response to glucose feeding. We used various methods, including Oil-Red-O and Nile Red staining and coherent anti-Stokes Raman scattering (CARS) imaging, to measure internal fat levels. Consistent with previous reports (Schulz et al. 2007; Nomura et al. 2010; Pang et al. 2014), glucose treatment increased the overall fat levels (Fig. 6A,B; Supplemental Fig. S11A–D). The sbp-1(RNAi) animals displayed reduced levels of fat (Fig. 6A,B; Supplemental Fig. S11A–D), as previously shown (Ashrafi et al. 2003; McKay et al. 2003; Yang et al. 2006; Nomura et al. 2010). The effects of mdt-15 RNAi varied among the different assays (Fig 6A,B; Supplemental Fig. S11A–D), consistent with previous reports (Taubert et al. 2006; Arda et al. 2010). We found that a glucose-rich diet increased overall fat levels in sbp-1(RNAi) and mdt-15(RNAi) animals (Fig. 6A,B; Supplemental Fig. S11A–D), although the data show a substantial variability (Fig. 6B; Supplemental Fig. S11E). This result suggests that other factors that act in parallel to SBP-1/MDT-15 contribute to the accumulation of overall fat upon glucose feeding in addition to SBP-1 and MDT-15.
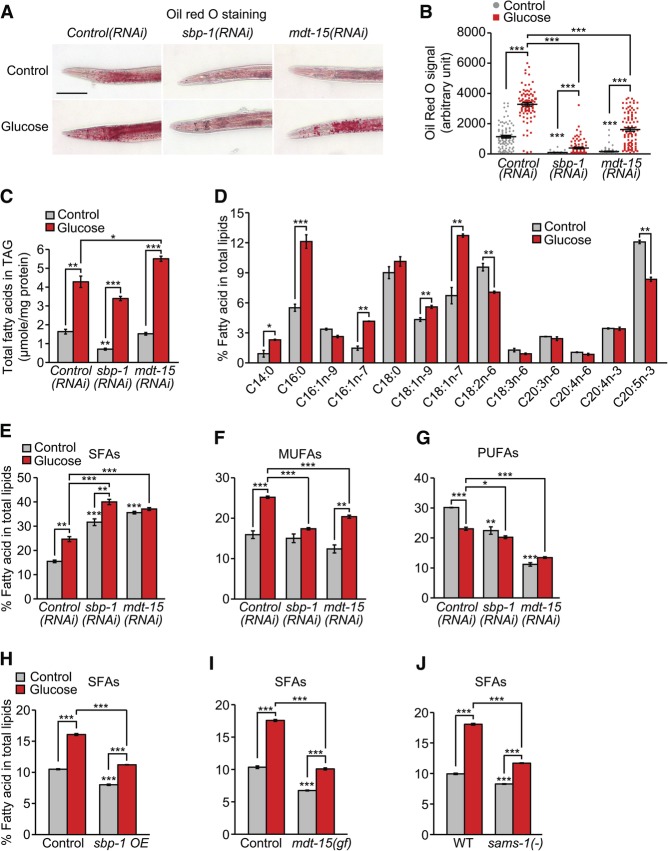
Figure 6.
Knockdown of mdt-15 or sbp-1 increases the levels of SFAs in glucose-fed worms. (A) Oil-Red-O staining of fixed worms, which were previously treated with control RNAi, mdt-15 RNAi, or sbp-1 RNAi on a control diet or a glucose-rich diet. Bar, 100 μm. (B) Quantification of the data shown in A. n ≥ 85 from five independent experiments. Note the variability of the signals, particularly among the glucose-treated mdt-15(RNAi) worms (Supplemental Fig. S11E). (C) The total amount of FAs in triacylglyceride (TAG) was analyzed by using gas chromatography/mass spectrometry (GC/MS). n = 3. (D) Total FA profiles of control RNAi-fed worms treated with or without additional glucose were analyzed by using GC/MS. n = 3. Fractions of each FA in the sum of the total FAs (mole/mole) were calculated. (E–G) Shown are the levels of SFAs (E), MUFAs (F), and PUFAs (G) in sbp-1(RNAi) or mdt-15(RNAi) worms with or without glucose-rich diet feeding. (H–J) The levels of SFAs in sbp-1-overexpressing (sbp-1 OE) (H), mdt-15(et14) gain-of-function (gf) (I), and sams-1(ok3033) [sams-1(−)] (J) animals. Injection marker transgenic worms (IJ1243) were used as a control for sbp-1-overexpressing animals, and paqr-2(tm3410) mutant background was used for both control and mdt-15(gf) (Svensk et al. 2013) animals. (WT) Wild type (N2). Total SFAs are the sum of C14:0, C16:0, and C18:0; total MUFAs are the sum of C16:1n-9, C16:1n-7, C18:1n-9, and C18:1n-7; total PUFAs are the sum of C18:2n-6, C18:3n-6, C20:3n-6, C20:4n-6, C20:4n-3, and C20:5n-3. Error bars represent SEM. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001, two-tailed Student's t-test.
We next performed gas chromatography/mass spectrometry (GC/MS) analysis to measure the composition of FAs in glucose-fed animals treated with sbp-1 RNAi or mdt-15 RNAi. The overall triacylglyceride (TAG) levels measured by GC/MS (Fig. 6C) were consistent with results obtained through other assays (Fig. 6A,B; Supplemental Fig. S11A–D). Glucose feeding increased the levels of SFAs (C14:0 and C16:0) and several types of MUFAs (C16:1n-7, C18:1n-9, and C18:1n-7) but did not increase the levels of PUFAs (Fig. 6D). Knockdown of sbp-1 or mdt-15 resulted in the accumulation of SFAs (C14:0, C16:0, and C18:0) (Fig. 6E; Supplemental Fig. S11F) but decreased the levels of MUFAs and PUFAs (in particular, C18:1n-9 and C18:2n-6) (Fig. 6F,G; Supplemental Fig. S11F; Taubert et al. 2006; Yang et al. 2006). Importantly, glucose feeding further increased the levels of SFAs (C14:0 and C16:0) in mdt-15(RNAi) and sbp-1(RNAi) animals (Supplemental Fig. S11F) while having a small effect on the levels of UFAs, particularly PUFAs (Fig. 6F,G; Supplemental Fig. S11F). Conversely, sbp-1 overexpression, the mdt-15(gf) mutation, or the sams-1 mutation consistently decreased overall SFA levels while having variable effects on UFA levels (Fig. 6H–J; Supplemental Fig. S12A–I). These results suggest that SBP-1 and MDT-15 counteract the accumulation of SFA-containing fat upon high glucose diet feeding.
The accumulation of SFAs and intermediate metabolites in glycolysis appears to reduce life span on a glucose-rich diet
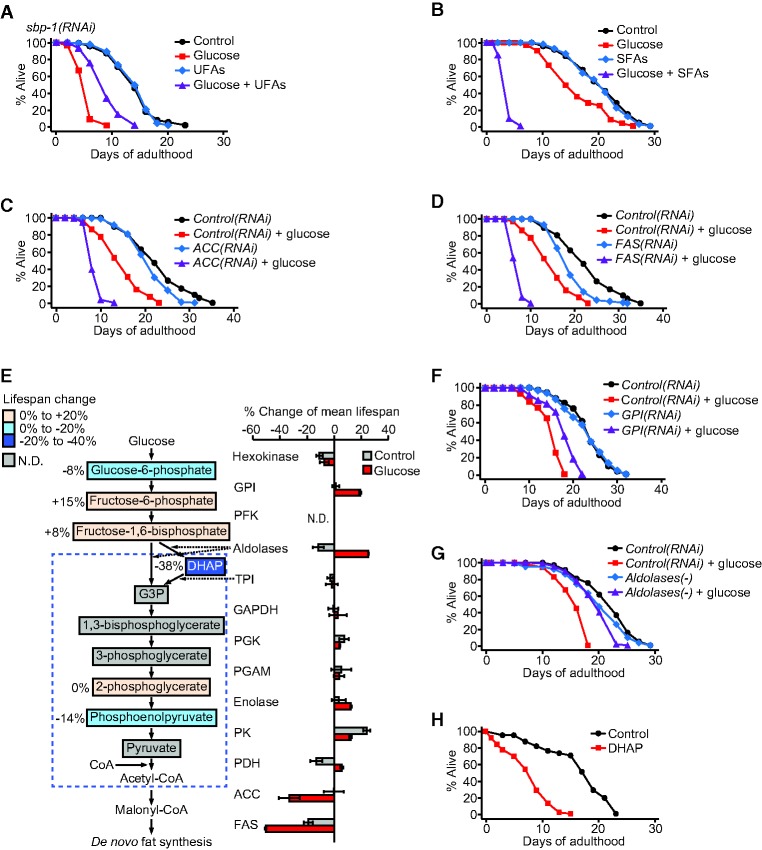
Are the changes in fat composition responsible for the shortened life span of glucose-fed mdt-15(−) and sbp-1(−) animals? We considered two possible causes: (1) decreased UFA levels and (2) increased SFA levels. To test these possibilities, we fed animals a glucose-rich diet with either UFAs or SFAs. Treatment with the UFA mixture (C18:1n-9, C18:1n-7, and C18:2n-6) partly restored the short life span of glucose-fed sbp-1(RNAi) worms, although the effects were variable (Fig. 7A; Supplemental Fig. S13C). Treatment with the UFA mixture or a major MUFA (C18:1n-9) had a small or no effect on the life span of glucose-treated mdt-15(RNAi) or mdt-15(−) worms (Supplemental Fig. S13A–E). In contrast, an SFA mixture (C14:0 and C16:0) or individual SFAs (C14:0 or C16:0) dramatically decreased the life span of animals fed a glucose-enriched diet while not reducing the life span of animals on a control diet (Fig. 7B; Supplemental Fig. S13F,G). Thus, accumulation of SFAs appears to be a major cause that reduces the life span of animals on a high glucose diet.
Figure 7.
Accumulation of intermediate metabolites from glucose to FA synthesis causes short life span. (A) UFA mixture (600 μM C18:1n-9, C18:1n-7, and C18:2n-6) feeding partly suppressed the short life span of the worms treated with sbp-1 RNAi on a glucose-enriched diet. (B) SFA mixture (600 μM C14:0 and C16:0) greatly shortened the life span of wild-type animals on a glucose-rich diet. NP-40 (0.1%) was added to all experimental conditions in A and B. (C,D) Knockdown of ACC (pod-2 RNAi) (C) or FAS (fasn-1 RNAi) (D) caused very short life span upon glucose-rich diet feeding. Worms were treated with pod-2 RNAi or fasn-1 RNAi only during adulthood. (E) Simplified glycolysis and FA synthesis pathways and percent changes of mean life span upon genetically inhibiting enzymes in the pathways. See Supplemental Table S9 for the genetic inhibition used for each enzymatic step. (DHAP) Dihydroxyacetonephosphate; (G3P) glyceraldehyde-3-phosphate; (GPI) glucose-6-phosphate isomerase; (PFK) phosphofructokinase; (TPI) triosephosphate isomerase; (GAPDH) glyceraldehyde-3-phosphate dehydrogenase; (PGK) phosphoglycerate kinase; (PGAM) phosphoglycerate mutase; (PK) pyruvate kinase; (PDH) pyruvate dehydrogenase. Error bars represent SEM of mean life spans from two independent experiments. Colored boxes for metabolites indicate percent changes in mean life span upon treatment with the respective metabolites. (N.D.) Not determined. See Supplemental Figure S15 and its legend for details. (F) Knockdown of GPI (gpi-1 RNAi) partly suppressed the short life span of glucose-treated animals. Note that previous reports showed that gpi-1 RNAi increased life span under normal feeding conditions (Hansen et al. 2005; Schulz et al. 2007), but it did not do so in our experimental conditions. We speculate that different experimental conditions (e.g., FUdR treatment) may have caused these different results. (G) Aldolases(−) [aldo-1(tm5782) mutation with aldo-2 RNAi treatment] largely suppressed the glucose toxicity on life span. Life span curves in C, D, F, and G are representative data of those shown in E. (H) DHAP treatment greatly reduced life span. See Supplemental Table S9 for additional repeats and statistical analysis for life span data shown in this figure.
SFAs may be toxic and synergistically enhance the life-shortening effects of glucose. Alternatively, the accumulation of SFAs may block a metabolic flow from glucose to SFA synthesis by inhibiting upstream enzymes and increases the level of toxic intermediates (Tong and Harwood 2006). We sought to reduce SFAs while increasing intermediate metabolites by genetically inhibiting key enzymes for de novo SFA synthesis, ACC (POD-2) and FAS (FASN-1) (Strable and Ntambi 2010). ACC(RNAi) and FAS(RNAi) animals exhibited very short life span phenotypes in glucose-enriched conditions (Fig. 7C–E), similar to mdt-15(−) or sbp-1(−) animals. In addition, knockdown of ACC or FAS did not further decrease the short life span of sbp-1(RNAi) or mdt-15(RNAi) worms on a glucose-rich diet (Supplemental Fig. S14C–J). These data suggest that ACC and FAS act in the same pathway with MDT-15/SBP-1 and that accumulation of intermediate metabolites between glucose and FAs causes the short life span.
To further specify the intermediate metabolites that reduced life span, we genetically inhibited each step in the glycolysis pathway (Fig. 7E and see also the legend for details). Genetic inhibition of glucose-6-phosphate isomerase (GPI) or fructose-1,6-bisphosphate aldolases (aldolases) significantly restored life span in animals on a glucose-rich diet (Fig. 7E–G). Thus, intermediate metabolites between the steps governed by aldolases and ACC appear to shorten life span (Fig. 7E). We next treated worms with available intermediate metabolites in glycolysis, and found that dihydroxyacetonephosphate (DHAP) significantly and consistently reduced life span (Fig. 7E,H; Supplemental Fig. S15A–J). Together, these data suggest that DHAP is the potential toxic molecule that causes short life span on a glucose-rich diet.
Discussion
SREBP and MDT-15 reduce the toxic effects of dietary glucose on life span by promoting the conversion of glucose to UFAs
Glucose is an essential energy source, but excessive amounts can shorten life span. In this study, we show that SREBP and MDT-15 reduce the toxic effects of glucose on life span by promoting fat conversion in C. elegans. Glucose feeding up-regulates the SBP-1/MDT-15 transcription factor complex, which induces expression of lipogenic genes, including FA desaturases. This prevents further accumulation of the SFAs and intermediate metabolites, including DHAP, which reduce life span. Thus, a normal metabolic flow from glucose to UFA synthesis is required to ameliorate the life-shortening effects of glucose. Several key data in our study support this model. First, both SBP-1 and MDT-15 were necessary and sufficient for reducing the life span-shortening effect of glucose. Second, SBP-1 and MDT-15 regulated the expression of many genes involved in fat metabolism in high glucose feeding conditions. Third, SBP-1/MDT-15 is necessary and sufficient to moderate the accumulation of SFAs in animals on high glucose diets. Fourth, SFAs further reduced the shortened life span of glucose-fed animals. Fifth, genetic inhibition of de novo FA synthesis enhanced the life span-shortening effect of glucose. These results indicate that an SBP-1/MDT-15-dependent metabolic flow from glucose via SFAs to UFAs moderates the life-shortening effect of glucose.
What toxic, life-shortening molecules could a glucose-rich diet produce? Excessive glucose directs a metabolic flow that favors FA synthesis. Increased levels of SFAs inhibit enzymes, such as ACC, by feedback regulation, which inhibits de novo fat synthesis (Tong and Harwood 2006). This may promote the accumulation of toxic intermediate metabolites in the glycolysis and FA synthesis pathways. Consistent with this model, we found that SFA feeding or inhibition of de novo FA synthesis enhanced the toxic effect of glucose on life span. Our data also indicate that intermediate metabolites, such as DHAP, have a life-shortening effect on a glucose-rich diet. Interestingly, DHAP is known to glycate proteins to generate advanced glycation end products (AGEs) (Hipkiss 2011). Consistently, a glucose-rich diet increases the levels of AGEs in C. elegans (Schlotterer et al. 2009). Thus, DHAP is likely to be accumulated in mdt-15(−) and sbp-1(−) animals in response to glucose feeding, which leads to a very short life span.
One of the limitations of our study results from the fact that full inactivation of the key lipogenic genes such as sbp-1, mdt-15, pod-2, or fasn-1 appears to cause lethality or growth defects. Because of this, we tested the physiological roles of these genes by using hypomorphic mutations, whole-life RNAi, or adult-only RNAi, which results in partial inhibition of the genes. Although these partial inhibition methods elicited glucose-specific life span phenotypes, our data regarding the effects of SBP-1 and MDT-15 inhibition on the expression of glucose-responsive genes or in overall fat synthesis were partial and should be interpreted with caution. It will be important to completely inhibit SBP-1 and MDT-15 in adulthood by exploiting new genetic tools, such as inducible knockout systems, in future studies.
Glucose-responsive lipid metabolism by SREBP and FOXO
The regulation of glucose-responsive lipid metabolism by SREBPs appears to be conserved from nematodes to mammals. Consistent with our data, glucose treatment up-regulates SREBPs in mammals and induces key lipogenic genes, such as ACC, FAS, and SCDs (Horton et al. 1998; Hasty et al. 2000). Glucose treatment increases ER stress, which activates SREBPs that are localized at the ER membrane (Wang et al. 2005). In addition, the nuclear liver X receptor (LXR) up-regulates SREBP-1 in response to glucose or insulin (Chen et al. 2004; Mitro et al. 2007). Glucose feeding decreases the activity of 5′-adenosine monophosphate-activated protein kinase (AMPK) in C. elegans (Schulz et al. 2007). Mammalian AMPK directly phosphorylates and inhibits SREBPs (Li et al. 2011). Thus, glucose treatment may decrease the activity of AMPK, relieving the inhibition of SBP-1 to promote lipogenesis.
Our previous study showed that glucose-rich diets shorten C. elegans life span by down-regulating the DAF-16/FOXO transcription factor and a glycerol channel (Lee et al. 2009). Interestingly, DAF-16/FOXO and glycerol channels are tightly associated with fat metabolism. Activation of DAF-16/FOXO increases fat levels and regulates the expression of key fat metabolism-related genes (Kimura et al. 1997; Murphy et al. 2003). Moreover, mutant mice lacking glycerol channel AQP7 have metabolic defects such as obesity or diabetes (Hibuse et al. 2005). This may be caused by defective glycerol usage, which can disturb proper fat metabolism, as glycerol is a crucial building block for triglyceride synthesis. Therefore, it will be interesting to determine whether DAF-16/AQP-1 and SBP-1/MDT-15 pathways interact with each other to regulate life span and fat metabolism on a glucose-rich diet.
Association of fat and glucose metabolism by SREBPs in pathology and aging models
Glucolipotoxicity underlies the deleterious effects of chronically high levels of glucose and FAs in pancreatic β cells (Poitout et al. 2010). Excessive amounts of glucose and FAs cause a functional decline and apoptosis of β cells, leading to metabolic disorders such as diabetes (Poitout et al. 2010). Our data are related to the glucolipotoxicity model at the organismal level because SFAs cause rapid death in animals fed a high glucose diet. In addition, our data regarding the beneficial effects of FA desaturase overexpression are consistent with the protective roles of mammalian SCD1 in SFA-induced pancreatic β-cell death (Busch et al. 2005). In contrast, mammalian SREBPs increase fat levels by promoting fat synthesis, which leads to glucolipotoxicity (Wang et al. 2003). This seems different from our current finding that SBP-1 moderated the toxic effects of glucose on C. elegans life span. Mammalian SREBPs regulate the expression of genes such as ACC and FAS, which are crucial for de novo FA synthesis (Horton et al. 1998; Hasty et al. 2000). Therefore, up-regulation of SREBPs increases the overall levels of FAs, including SFAs, which may enhance glucolipotoxicity. Our data indicate that C. elegans SBP-1 is crucial for the conversion of SFAs to UFAs (which reduces the life-decreasing effects of dietary glucose) as well as de novo fat synthesis. Thus, it will be crucial to determine detailed mechanistic roles of mammalian SREBPs in fat metabolic processes under glucose-rich conditions.
Activation of SREBPs is also linked to obesity, fatty liver diseases, and cardiovascular diseases (Walker and Näär 2012). Excessive SREBP activity can increase circulating cholesterol and lipids, which causes an accumulation of lipids in the liver and adipose tissues (Walker and Näär 2012). An adipose tissue-specific overexpression of SREBP-1c causes severe insulin resistance and high blood glucose (Shimomura et al. 1998). In contrast, SREBP-induced hepatic lipogenesis can reduce glucose toxicity by lowering blood glucose levels in mouse models (Becard et al. 2001; Takahashi et al. 2005). The C. elegans intestine is a major tissue for metabolism, similar to the mammalian liver (McGhee 2007). Thus, our data showing the beneficial effects of intestinal SBP-1 on glucose toxicity and life span are functionally relevant to data obtained in mouse models. It will be interesting to test the role of hepatic SREBPs in aging upon exposure to a high carbohydrate diet or in a diabetic model.
Materials and methods
Strains
All strains used in this study are described in the Supplemental Material.
Fluorescence imaging
Fluorescence imaging was performed as described previously with modifications (Gaglia et al. 2012). For glucose-rich diet feeding experiments, transgenic worms were fed with specific RNAi bacteria with or without 2% glucose from eggs to day 1 adults unless otherwise mentioned. sbp-1p::GFP::sbp-1 transgenic worms were treated with glucose from eggs to L4 larval stage. L4 larvae were chosen for sbp-1p::GFP::sbp-1 worm imaging because eggs in adult worms interfered with fluorescence signals from intestinal cells. mdt-15p::mdt-15::GFP worms were treated with glucose from eggs to L2 larval stage. Signals of the GFP-fused MDT-15 were strong at L2 stage but decreased during subsequent development. The transgenic worms were mounted on a 2% agarose pad and anesthetized by using 100 mM sodium azide (Daejung). Images of the worms were captured by using an AxioCam HRc (Zeiss Corporation) camera attached to a Zeiss Axioscope A.1 microscope (Zeiss Corporation). ImageJ (Schneider et al. 2012) was used to quantify the fluorescence intensity of whole body, and the background signals were subtracted. For measuring the intestinal expression of fat-6p::fat-6::GFP or fat-7p::GFP, the first anterior intestinal cells were quantified as described previously (Lemieux et al. 2011).
Genome-wide RNAi screen
The genome-wide RNAi screen was performed as described previously with modifications (Lamitina et al. 2006). See the Supplemental Material for details.
Life span assays
Life span assays were performed as described previously with some modifications (Lee et al. 2010). Statistical analysis of life span results was performed by using OASIS (Online Application of Survival Analysis, http://sbi.postech.ac.kr/oasis), and P-values were calculated by using a log rank (Mantel-Cox method) test (Yang et al. 2011). See the Supplemental Material for details.
Oil-Red-O staining
Oil-Red-O staining was performed as described previously with some modifications (O'Rourke et al. 2009). Worms were treated with specific RNAi with or without additional 2% glucose feeding from eggs to adulthood. Approximately 300 young adult (day 1) worms were then harvested with M9 buffer by washing three times and fixed with 50% isopropanol for 15 min. Oil-Red-O solution (0.5 g/100 mL in isopropanol; Sigma) was diluted in double distilled water (ddH2O) to 60% working solution, and precipitates were eliminated by filtering. Fixed worms were incubated in the working solution overnight at room temperature. Stained worms were washed with PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4), and PBST (PBS with 0.01% Triton X-100 [Daejung]) was used for placing the worms on a 2% agarose pad with micropipettes. DIC images were captured using an AxioCam HRc (Zeiss Corporation) camera attached to a Zeiss Axioscope A.1 microscope (Zeiss Corporation). ImageJ was used for the quantification of Oil-Red-O intensity (Schneider et al. 2012; Mehlem et al. 2013). Images were inverted, and thresholds were set as 90 for Oil-Red-O signals. Areas that had higher intensities than the threshold in anterior intestinal cells were quantified.
Locomotion assays
Locomotion was measured as described previously with modifications (Bansal et al. 2015). See the Supplemental Material for details.
mRNA sequencing and data analysis
Total RNA was isolated as described previously with modifications (Seo et al. 2015). Synchronized wild-type worms were treated with control RNAi (L4440), sbp-1 RNAi, or mdt-15 RNAi during development with or without additional 2% glucose treatment and harvested at day 1 adult stage. Total RNA of the worms was extracted by using RNAiso plus (Takara) and purified by using ethanol precipitation. Three independent biological repeats were used. The cDNA library was prepared by ChunLab (ChunLab, Inc.), and cDNA sequencing was performed by using a HiSeq 2500 platform. Quality-filtered reads were aligned to the reference genome sequence (Caenorhabditis elegans Bristol N2 [accession no. GCF_000002985.6]) by Bowtie2 (Langmead and Salzberg 2012), and differentially expressed genes were analyzed using the edgeR method (Robinson et al. 2010) by the CLRNASeq program (ChunLab, Inc.). Genes with significant changes in expression (P < 0.05, fold change >1.2) (Yao et al. 2015) were further analyzed, and genes whose FPKM (fragments per kilobase of transcript per million fragments) was zero were excluded. Scatter plot (Riedel et al. 2013) and heat map analyses (Ewald et al. 2015) were performed as previously described. Heat maps were generated by using Cluster 3.0 (Eisen et al. 1998) and Java Treeview (Saldanha 2004). Venn diagrams were generated by using Venn Diagram Plotter (http://omics.pnl.gov/software/venn-diagram-plotter). The significance of the overlaps among gene sets was calculated by using the hypergeometric probability test (https:// www.geneprof.org/GeneProf/tools/hypergeometric.jsp) (Halbritter et al. 2012).
Quantitative RT–PCR (qRT–PCR)
qRT–PCR was performed as described previously (Seo et al. 2013). Synchronized worms treated with sbp-1 RNAi or mdt-15 RNAi from eggs with or without 2% glucose treatment were harvested at young adult stage (day 1) with M9 buffer by washing three times. Synchronized worms were cultured on control RNAi with or without 2% glucose until L4 stage and then transferred onto pod-2 RNAi or fasn-1 RNAi plates. The pod-2 RNAi-treated or fasn-1 RNAi-treated worms were harvested after 24 h with M9 buffer by washing three times. RNA was isolated using RNAiso plus (Takara) and reverse-transcribed to cDNA using ImProm-II reverse transcriptase kit (Promega). Random primers (9-mers; Cosmogenetech) were used for reverse transcription except for the measurement of sbp-1 mRNA (Supplemental Fig. S6C), which was reverse-transcribed with oligo(dT) 15 primer (Promega). Quantitative real-time PCR was performed by using the StepOne real-time PCR system (Applied Biosystems), and relative quantity was analyzed by using comparative Ct methods described in the manufacturer's manual. The average of two technical repeats was used for each biological data set. mRNA levels of ama-1, which encodes an RNA polymerase II large subunit, were used for normalization. See the Supplemental Material for the sequence information of the primers used for the assays.
Lipid extraction and GC/MS assays
Lipid extraction was performed as described previously with some modifications (Yang et al. 2006). Day 1 adult worms treated with sbp-1 RNAi or mdt-15 RNAi from eggs with or without 2% glucose feeding were harvested and washed three times with ddH2O. Synchronized GFP::sbp-1-overexpressing animals and sams-1 mutants were cultured on OP50-seeded plates with or without 2% glucose treatment. paqr-2(tm3410) and mdt-15(et14) paqr-2(tm3410) worms were cultured on OP50-seeded plates without glucose addition until L4 stage and then transferred onto new OP50-seeded plates with or without 2% glucose. The worms were harvested after 24 h and washed three times with ddH2O. Approximately 400 μL of worm pellets was frozen in liquid nitrogen and stored at −80°C until use. The worm samples were homogenized by sonication on ice, and 10 μL of the worm lysates was used for quantifying total protein amount by using the BCA protein assay kit (Thermo). For each condition, 300 μL of worm lysate samples was used for the assay. Five milliliters of ice-cold chloroform/methanol (1:1) was added to each sample in a glass tube, followed by vortexing and incubation overnight at 4°C. After the incubation, 2 mL of Hajra's solution (0.2 M H3PO4, 1 M KCl) was added to the samples and mixed well. Chloroform layer and aqueous phase were separated by centrifugation (2000 rpm for 10 min), and the chloroform layer-containing lipid was extracted. Aqueous phase was re-extracted by the addition of 3 mL of chloroform followed by centrifugation. The combined chloroform layers were evaporated to 120 μL. Twenty microliters of the lipid extract was used for analyzing total lipid extract, and the remaining sample was separated by using thin-layer chromatography (TLC) to analyze TAG. To visualize lipids, 0.01% purimurin/80% acetone solution was used, and the lipid spots corresponding to TAG were scraped off. FA methyl esters were prepared by using sulfuric acid (2.5%)/methanol solution and analyzed by GC/MS (GCMS-QP2010, Shimadzu; HP-INOWAX capillary column, 30 m, 0.25 mm; Agilent).
Preparation of plates for FA feeding assays
FA feeding assays were performed as described previously with modifications (Yang et al. 2006). See the Supplemental Material for details.
Supplementary Material
Acknowledgments
We thank Dr. Cynthia Kenyon, Dr. Gary Ruvkun, and Dr. Marc Pilon for sharing strains, and Dr. Stefan Taubert, Dr. Yoontae Lee, and Dr. G-One Ahn for critical comments on the manuscript. We also thank anonymous reviewers for constructive comments, and all Lee laboratory members for help and discussion. This research was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (Ministry of Science, ICT, and Future Planning; NRF-2012R1A4A1028200 and NRF-2013R1A1A2014754), a grant of the Korean Health Technology R&D Project, Ministry of Health and Welfare (HI14C2337 to S.-J.V.L.), the Fostering Core Leaders of the Future Basic Science Program (NRF-2013H1A8A1003751 to D.-E.J. and S.-J.V.L.), the Global Frontier Project of the Advanced Biomass R&D Center funded by the Korean government (ABC-2015M3A6A2065746 to Y.L.), and the DGIST R&D Program funded by the Korean government (15-BD-06 to D.W.M.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.266304.115.
References
- Abu-Elheiga L, Matzuk MM, Abo-Hashema KA, Wakil SJ. 2001. Continuous fatty acid oxidation and reduced fat storage in mice lacking acetyl-CoA carboxylase 2. Science 291: 2613–2616. [DOI] [PubMed] [Google Scholar]
- Arda HE, Taubert S, MacNeil LT, Conine CC, Tsuda B, Van Gilst M, Sequerra R, Doucette-Stamm L, Yamamoto KR, Walhout AJ. 2010. Functional modularity of nuclear hormone receptors in a Caenorhabditis elegans metabolic gene regulatory network. Mol Syst Biol 6: 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi K, Chang FY, Watts JL, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. 2003. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 421: 268–272. [DOI] [PubMed] [Google Scholar]
- Bansal A, Zhu LJ, Yen K, Tissenbaum HA. 2015. Uncoupling lifespan and healthspan in Caenorhabditis elegans longevity mutants. Proc Natl Acad Sci 112: E277–E286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becard D, Hainault I, Azzout-Marniche D, Bertry-Coussot L, Ferre P, Foufelle F. 2001. Adenovirus-mediated overexpression of sterol regulatory element binding protein-1c mimics insulin effects on hepatic gene expression and glucose homeostasis in diabetic mice. Diabetes 50: 2425–2430. [DOI] [PubMed] [Google Scholar]
- Brock TJ, Browse J, Watts JL. 2007. Fatty acid desaturation and the regulation of adiposity in Caenorhabditis elegans. Genetics 176: 865–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch AK, Gurisik E, Cordery DV, Sudlow M, Denyer GS, Laybutt DR, Hughes WE, Biden TJ. 2005. Increased fatty acid desaturation and enhanced expression of stearoyl coenzyme A desaturase protects pancreatic β-cells from lipoapoptosis. Diabetes 54: 2917–2924. [DOI] [PubMed] [Google Scholar]
- Cabreiro F, Au C, Leung KY, Vergara-Irigaray N, Cocheme HM, Noori T, Weinkove D, Schuster E, Greene ND, Gems D. 2013. Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism. Cell 153: 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Liang G, Ou J, Goldstein JL, Brown MS. 2004. Central role for liver X receptor in insulin-mediated activation of Srebp-1c transcription and stimulation of fatty acid synthesis in liver. Proc Natl Acad Sci 101: 11245–11250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirala SS, Chang H, Matzuk M, Abu-Elheiga L, Mao J, Mahon K, Finegold M, Wakil SJ. 2003. Fatty acid synthesis is essential in embryonic development: fatty acid synthase null mutants and most of the heterozygotes die in utero. Proc Natl Acad Sci 100: 6358–6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding W, Smulan LJ, Hou NS, Taubert S, Watts JL, Walker AK. 2015. s-adenosylmethionine levels govern innate immunity through distinct methylation-dependent pathways. Cell Metab 22: 633–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. 1998. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci 95: 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald CY, Landis JN, Porter Abate J, Murphy CT, Blackwell TK. 2015. Dauer-independent insulin/IGF-1-signalling implicates collagen remodelling in longevity. Nature 519: 97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglia MM, Jeong DE, Ryu EA, Lee D, Kenyon C, Lee SJ. 2012. Genes that act downstream of sensory neurons to influence longevity, dauer formation, and pathogen responses in Caenorhabditis elegans. PLoS Genet 8: e1003133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh GY, Martelli KL, Parhar KS, Kwong AW, Wong MA, Mah A, Hou NS, Taubert S. 2014. The conserved Mediator subunit MDT-15 is required for oxidative stress responses in Caenorhabditis elegans. Aging Cell 13: 70–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbritter F, Vaidya HJ, Tomlinson SR. 2012. GeneProf: analysis of high-throughput sequencing experiments. Nat Methods 9: 7–8. [DOI] [PubMed] [Google Scholar]
- Hansen M, Hsu AL, Dillin A, Kenyon C. 2005. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet 1: 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasty AH, Shimano H, Yahagi N, Amemiya-Kudo M, Perrey S, Yoshikawa T, Osuga J, Okazaki H, Tamura Y, Iizuka Y, et al. 2000. Sterol regulatory element-binding protein-1 is regulated by glucose at the transcriptional level. J Biol Chem 275: 31069–31077. [DOI] [PubMed] [Google Scholar]
- Heestand BN, Shen Y, Liu W, Magner DB, Storm N, Meharg C, Habermann B, Antebi A. 2013. Dietary restriction induced longevity is mediated by nuclear receptor NHR-62 in Caenorhabditis elegans. PLoS Genet 9: e1003651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibuse T, Maeda N, Funahashi T, Yamamoto K, Nagasawa A, Mizunoya W, Kishida K, Inoue K, Kuriyama H, Nakamura T, et al. 2005. Aquaporin 7 deficiency is associated with development of obesity through activation of adipose glycerol kinase. Proc Natl Acad Sci 102: 10993–10998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipkiss AR. 2011. Energy metabolism and ageing regulation: metabolically driven deamidation of triosephosphate isomerase may contribute to proteostatic dysfunction. Ageing Res Rev 10: 498–502. [DOI] [PubMed] [Google Scholar]
- Horton JD, Bashmakov Y, Shimomura I, Shimano H. 1998. Regulation of sterol regulatory element binding proteins in livers of fasted and refed mice. Proc Natl Acad Sci 95: 5987–5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. 1997. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277: 942–946. [DOI] [PubMed] [Google Scholar]
- Kremmyda LS, Tvrzicka E, Stankova B, Zak A. 2011. Fatty acids as biocompounds: their role in human metabolism, health and disease: a review. part 2: fatty acid physiological roles and applications in human health and disease. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub 155: 195–218. [DOI] [PubMed] [Google Scholar]
- Lamitina T, Huang CG, Strange K. 2006. Genome-wide RNAi screening identifies protein damage as a regulator of osmoprotective gene expression. Proc Natl Acad Sci 103: 12173–12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-J, Murphy CT, Kenyon C. 2009. Glucose shortens the life span of C. elegans by downregulating DAF-16/FOXO activity and aquaporin gene expression. Cell Metab 10: 379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-J, Hwang AB, Kenyon C. 2010. Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Curr Biol 20: 2131–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux GA, Liu J, Mayer N, Bainton RJ, Ashrafi K, Werb Z. 2011. A whole-organism screen identifies new regulators of fat storage. Nat Chem Biol 7: 206–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Xu S, Mihaylova MM, Zheng B, Hou X, Jiang B, Park O, Luo Z, Lefai E, Shyy JY, et al. 2011. AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metab 13: 376–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee JD. 2007. The C. elegans intestine. In WormBook (ed. The C. elegans Research Community), WormBook, 10.1895/wormbook.1.133.1; http://www.wormbook.org. [DOI] [Google Scholar]
- McKay RM, McKay JP, Avery L, Graff JM. 2003. C elegans: a model for exploring the genetics of fat storage. Dev Cell 4: 131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlem A, Hagberg CE, Muhl L, Eriksson U, Falkevall A. 2013. Imaging of neutral lipids by oil red O for analyzing the metabolic status in health and disease. Nat Protoc 8: 1149–1154. [DOI] [PubMed] [Google Scholar]
- Mitro N, Mak PA, Vargas L, Godio C, Hampton E, Molteni V, Kreusch A, Saez E. 2007. The nuclear receptor LXR is a glucose sensor. Nature 445: 219–223. [DOI] [PubMed] [Google Scholar]
- Miyazaki M, Man WC, Ntambi JM. 2001. Targeted disruption of stearoyl-CoA desaturase1 gene in mice causes atrophy of sebaceous and meibomian glands and depletion of wax esters in the eyelid. J Nutr 131: 2260–2268. [DOI] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. 2003. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424: 277–283. [DOI] [PubMed] [Google Scholar]
- Nomura T, Horikawa M, Shimamura S, Hashimoto T, Sakamoto K. 2010. Fat accumulation in Caenorhabditis elegans is mediated by SREBP homolog SBP-1. Genes Nutr 5: 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke EJ, Soukas AA, Carr CE, Ruvkun G. 2009. C. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell Metab 10: 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang S, Lynn DA, Lo JY, Paek J, Curran SP. 2014. SKN-1 and Nrf2 couples proline catabolism with lipid metabolism during nutrient deprivation. Nat Commun 5: 5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poitout V, Amyot J, Semache M, Zarrouki B, Hagman D, Fontes G. 2010. Glucolipotoxicity of the pancreatic β cell. Biochim Biophys Acta 1801: 289–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel CG, Dowen RH, Lourenco GF, Kirienko NV, Heimbucher T, West JA, Bowman SK, Kingston RE, Dillin A, Asara JM, et al. 2013. DAF-16 employs the chromatin remodeller SWI/SNF to promote stress resistance and longevity. Nat Cell Biol 15: 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ. 2004. Java Treeview--extensible visualization of microarray data. Bioinformatics 20: 3246–3248. [DOI] [PubMed] [Google Scholar]
- Schlotterer A, Kukudov G, Bozorgmehr F, Hutter H, Du X, Oikonomou D, Ibrahim Y, Pfisterer F, Rabbani N, Thornalley P, et al. 2009. C. elegans as model for the study of high glucose-mediated life span reduction. Diabetes 58: 2450–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz TJ, Zarse K, Voigt A, Urban N, Birringer M, Ristow M. 2007. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab 6: 280–293. [DOI] [PubMed] [Google Scholar]
- Seo K, Choi E, Lee D, Jeong DE, Jang SK, Lee SJ. 2013. Heat shock factor 1 mediates the longevity conferred by inhibition of TOR and insulin/IGF-1 signaling pathways in C. elegans. Aging Cell 12: 1073–1081. [DOI] [PubMed] [Google Scholar]
- Seo M, Seo K, Hwang W, Koo HJ, Hahm JH, Yang JS, Han SK, Hwang D, Kim S, Jang SK, et al. 2015. RNA helicase HEL-1 promotes longevity by specifically activating DAF-16/FOXO transcription factor signaling in Caenorhabditis elegans. Proc Natl Acad Sci 112: E4246–E4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Li J, Zou X, Greggain J, Rodkaer SV, Faergeman NJ, Liang B, Watts JL. 2013. Regulation of lipid droplet size and phospholipid composition by stearoyl-CoA desaturase. J Lipid Res 54: 2504–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura I, Hammer RE, Richardson JA, Ikemoto S, Bashmakov Y, Goldstein JL, Brown MS. 1998. Insulin resistance and diabetes mellitus in transgenic mice expressing nuclear SREBP-1c in adipose tissue: model for congenital generalized lipodystrophy. Genes Dev 12: 3182–3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strable MS, Ntambi JM. 2010. Genetic control of de novo lipogenesis: role in diet-induced obesity. Crit Rev Biochem Mol Biol 45: 199–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensk E, Stahlman M, Andersson CH, Johansson M, Boren J, Pilon M. 2013. PAQR-2 regulates fatty acid desaturation during cold adaptation in C. elegans. PLoS Genet 9: e1003801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. 2015. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43: D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A, Shimano H, Nakagawa Y, Yamamoto T, Motomura K, Matsuzaka T, Sone H, Suzuki H, Toyoshima H, Yamada N. 2005. Transgenic mice overexpressing SREBP-1a under the control of the PEPCK promoter exhibit insulin resistance, but not diabetes. Biochim Biophys Acta 1740: 427–433. [DOI] [PubMed] [Google Scholar]
- Taubert S, Van Gilst MR, Hansen M, Yamamoto KR. 2006. A Mediator subunit, MDT-15, integrates regulation of fatty acid metabolism by NHR-49-dependent and -independent pathways in C. elegans. Genes Dev 20: 1137–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong L, Harwood HJ Jr. 2006. Acetyl-coenzyme A carboxylases: versatile targets for drug discovery. J Cell Biochem 99: 1476–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gilst MR, Hadjivassiliou H, Jolly A, Yamamoto KR. 2005. Nuclear hormone receptor NHR-49 controls fat consumption and fatty acid composition in C. elegans. PLoS Biol 3: e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AK, Näär AM. 2012. SREBPs: regulators of cholesterol/lipids as therapeutic targets in metabolic disorders, cancers and viral diseases. Clin Lipidol 7: 27–36. [Google Scholar]
- Walker AK, Yang F, Jiang K, Ji JY, Watts JL, Purushotham A, Boss O, Hirsch ML, Ribich S, Smith JJ, et al. 2010. Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes Dev 24: 1403–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AK, Jacobs RL, Watts JL, Rottiers V, Jiang K, Finnegan DM, Shioda T, Hansen M, Yang F, Niebergall LJ, et al. 2011. A conserved SREBP-1/phosphatidylcholine feedback circuit regulates lipogenesis in metazoans. Cell 147: 840–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Maechler P, Antinozzi PA, Herrero L, Hagenfeldt-Johansson KA, Bjorklund A, Wollheim CB. 2003. The transcription factor SREBP-1c is instrumental in the development of β-cell dysfunction. J Biol Chem 278: 16622–16629. [DOI] [PubMed] [Google Scholar]
- Wang H, Kouri G, Wollheim CB. 2005. ER stress and SREBP-1 activation are implicated in β-cell glucolipotoxicity. J Cell Sci 118: 3905–3915. [DOI] [PubMed] [Google Scholar]
- Yang F, Vought BW, Satterlee JS, Walker AK, Jim Sun ZY, Watts JL, DeBeaumont R, Saito RM, Hyberts SG, Yang S, et al. 2006. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature 442: 700–704. [DOI] [PubMed] [Google Scholar]
- Yang JS, Nam HJ, Seo M, Han SK, Choi Y, Nam HG, Lee SJ, Kim S. 2011. OASIS: online application for the survival analysis of lifespan assays performed in aging research. PLoS One 6: e23525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao L, Li Y, Knapp J, Smith P. 2015. Exploration of molecular pathways mediating electric field-directed Schwann cell migration by RNA-seq. J Cell Physiol 230: 1515–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.