Abstract
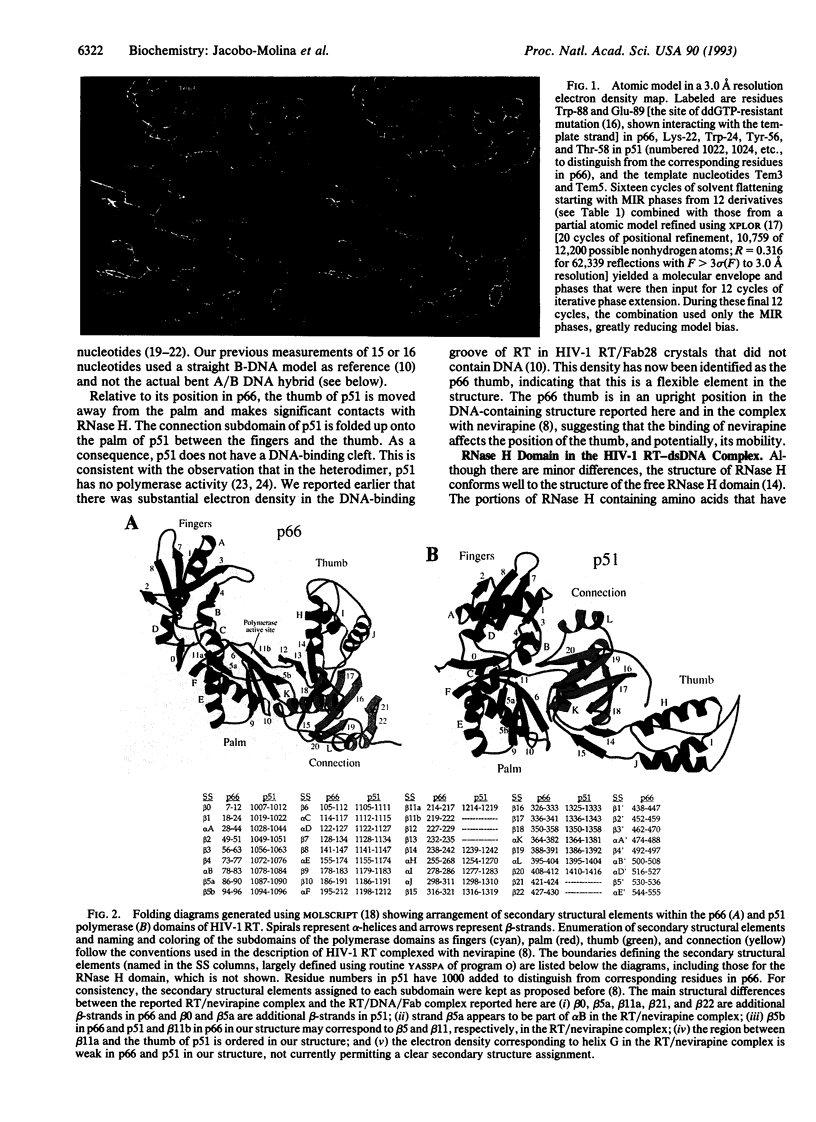
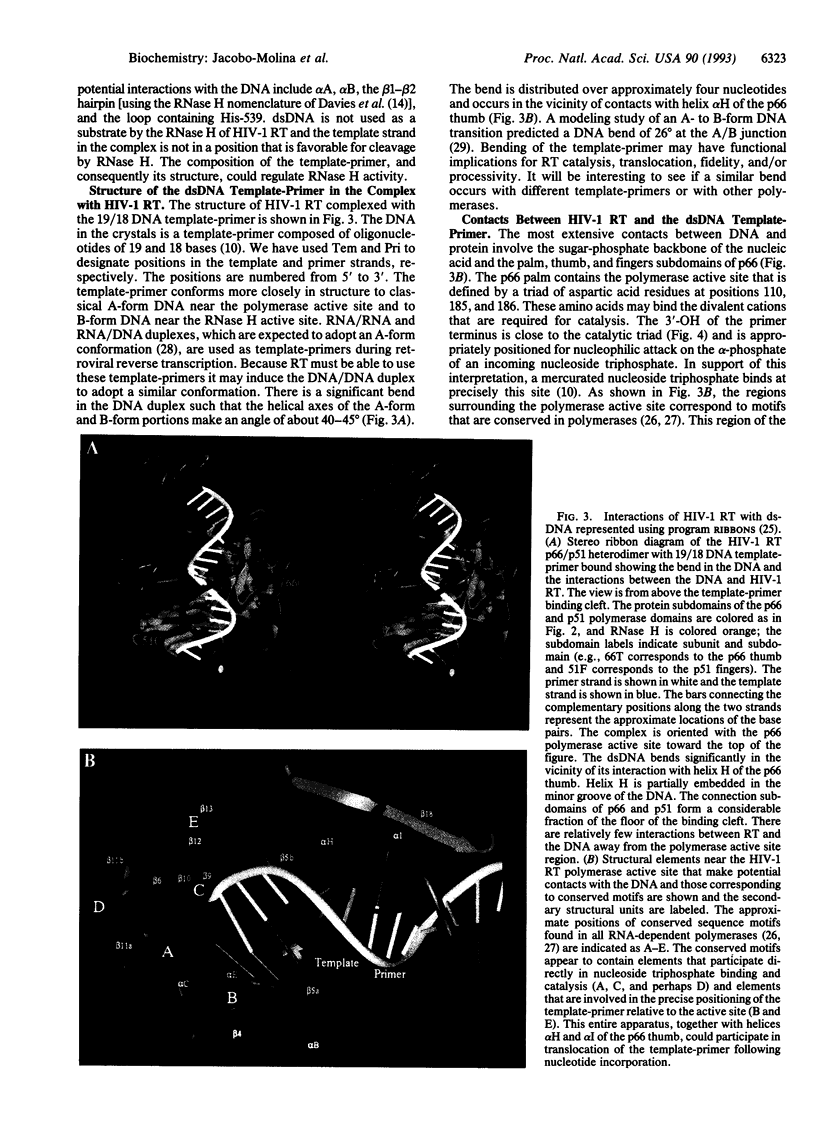
The crystal structure of a ternary complex of human immunodeficiency virus type 1 reverse transcriptase (HIV-1 RT) heterodimer (p66/p51), a 19-base/18-base double-stranded DNA template-primer, and a monoclonal antibody Fab fragment has been determined at 3.0 A resolution. The four individual subdomains of RT that make up the polymerase domains of p66 and p51 are named fingers, palm, thumb, and connection [Kohlstaedt, L. A., Wang, J., Friedman, J. M., Rice, P. A. & Steitz, T. A. (1992) Science 256, 1783-1790]. The overall folding of the subdomains is similar in p66 and p51 but the spatial arrangements of the subdomains are dramatically different. The template-primer has A-form and B-form regions separated by a significant bend (40-45 degrees). The most numerous nucleic acid interactions with protein occur primarily along the sugar-phosphate backbone of the DNA and involve amino acid residues of the palm, thumb, and fingers of p66. Highly conserved regions are located in the p66 palm near the polymerase active site. These structural elements, together with two alpha-helices of the thumb of p66, act as a clamp to position the template-primer relative to the polymerase active site. The 3'-hydroxyl of the primer terminus is close to the catalytically essential Asp-110, Asp-185, and Asp-186 residues at the active site and is in a position for nucleophilic attack on the alpha-phosphate of an incoming nucleoside triphosphate. The structure of the HIV-1 RT/DNA/Fab complex should aid our understanding of general mechanisms of nucleic acid polymerization. AIDS therapies may be enhanced by a fuller understanding of drug inhibition and resistance emerging from these studies.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnold E., Jacobo-Molina A., Nanni R. G., Williams R. L., Lu X., Ding J., Clark A. D., Jr, Zhang A., Ferris A. L., Clark P. Structure of HIV-1 reverse transcriptase/DNA complex at 7 A resolution showing active site locations. Nature. 1992 May 7;357(6373):85–89. doi: 10.1038/357085a0. [DOI] [PubMed] [Google Scholar]
- Davies J. F., 2nd, Hostomska Z., Hostomsky Z., Jordan S. R., Matthews D. A. Crystal structure of the ribonuclease H domain of HIV-1 reverse transcriptase. Science. 1991 Apr 5;252(5002):88–95. doi: 10.1126/science.1707186. [DOI] [PubMed] [Google Scholar]
- Delarue M., Poch O., Tordo N., Moras D., Argos P. An attempt to unify the structure of polymerases. Protein Eng. 1990 May;3(6):461–467. doi: 10.1093/protein/3.6.461. [DOI] [PubMed] [Google Scholar]
- Furfine E. S., Reardon J. E. Human immunodeficiency virus reverse transcriptase ribonuclease H: specificity of tRNA(Lys3)-primer excision. Biochemistry. 1991 Jul 23;30(29):7041–7046. doi: 10.1021/bi00243a001. [DOI] [PubMed] [Google Scholar]
- Goff S. P. Retroviral reverse transcriptase: synthesis, structure, and function. J Acquir Immune Defic Syndr. 1990;3(8):817–831. [PubMed] [Google Scholar]
- Gopalakrishnan V., Peliska J. A., Benkovic S. J. Human immunodeficiency virus type 1 reverse transcriptase: spatial and temporal relationship between the polymerase and RNase H activities. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10763–10767. doi: 10.1073/pnas.89.22.10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hostomsky Z., Hostomska Z., Fu T. B., Taylor J. Reverse transcriptase of human immunodeficiency virus type 1: functionality of subunits of the heterodimer in DNA synthesis. J Virol. 1992 May;66(5):3179–3182. doi: 10.1128/jvi.66.5.3179-3182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobo-Molina A., Arnold E. HIV reverse transcriptase structure-function relationships. Biochemistry. 1991 Jul 2;30(26):6351–6356. doi: 10.1021/bi00240a001. [DOI] [PubMed] [Google Scholar]
- Jacobo-Molina A., Clark A. D., Jr, Williams R. L., Nanni R. G., Clark P., Ferris A. L., Hughes S. H., Arnold E. Crystals of a ternary complex of human immunodeficiency virus type 1 reverse transcriptase with a monoclonal antibody Fab fragment and double-stranded DNA diffract x-rays to 3.5-A resolution. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10895–10899. doi: 10.1073/pnas.88.23.10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kohlstaedt L. A., Wang J., Friedman J. M., Rice P. A., Steitz T. A. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992 Jun 26;256(5065):1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- Larder B. A., Kemp S. D. Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (AZT). Science. 1989 Dec 1;246(4934):1155–1158. doi: 10.1126/science.2479983. [DOI] [PubMed] [Google Scholar]
- Le Grice S. F., Naas T., Wohlgensinger B., Schatz O. Subunit-selective mutagenesis indicates minimal polymerase activity in heterodimer-associated p51 HIV-1 reverse transcriptase. EMBO J. 1991 Dec;10(12):3905–3911. doi: 10.1002/j.1460-2075.1991.tb04960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merluzzi V. J., Hargrave K. D., Labadia M., Grozinger K., Skoog M., Wu J. C., Shih C. K., Eckner K., Hattox S., Adams J. Inhibition of HIV-1 replication by a nonnucleoside reverse transcriptase inhibitor. Science. 1990 Dec 7;250(4986):1411–1413. doi: 10.1126/science.1701568. [DOI] [PubMed] [Google Scholar]
- Nunberg J. H., Schleif W. A., Boots E. J., O'Brien J. A., Quintero J. C., Hoffman J. M., Emini E. A., Goldman M. E. Viral resistance to human immunodeficiency virus type 1-specific pyridinone reverse transcriptase inhibitors. J Virol. 1991 Sep;65(9):4887–4892. doi: 10.1128/jvi.65.9.4887-4892.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollis D. L., Brick P., Hamlin R., Xuong N. G., Steitz T. A. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. 1985 Feb 28-Mar 6Nature. 313(6005):762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- Pauwels R., Andries K., Desmyter J., Schols D., Kukla M. J., Breslin H. J., Raeymaeckers A., Van Gelder J., Woestenborghs R., Heykants J. Potent and selective inhibition of HIV-1 replication in vitro by a novel series of TIBO derivatives. Nature. 1990 Feb 1;343(6257):470–474. doi: 10.1038/343470a0. [DOI] [PubMed] [Google Scholar]
- Poch O., Sauvaget I., Delarue M., Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989 Dec 1;8(12):3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satow Y., Cohen G. H., Padlan E. A., Davies D. R. Phosphocholine binding immunoglobulin Fab McPC603. An X-ray diffraction study at 2.7 A. J Mol Biol. 1986 Aug 20;190(4):593–604. doi: 10.1016/0022-2836(86)90245-7. [DOI] [PubMed] [Google Scholar]
- Schatz O., Mous J., Le Grice S. F. HIV-1 RT-associated ribonuclease H displays both endonuclease and 3'----5' exonuclease activity. EMBO J. 1990 Apr;9(4):1171–1176. doi: 10.1002/j.1460-2075.1990.tb08224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selsing E., Wells R. D., Alden C. J., Arnott S. Bent DNA: visualization of a base-paired and stacked A-B conformational junction. J Biol Chem. 1979 Jun 25;254(12):5417–5422. [PubMed] [Google Scholar]
- Song Q., Yang G., Goff S. P., Prasad V. R. Mutagenesis of the Glu-89 residue in human immunodeficiency virus type 1 (HIV-1) and HIV-2 reverse transcriptases: effects on nucleoside analog resistance. J Virol. 1992 Dec;66(12):7568–7571. doi: 10.1128/jvi.66.12.7568-7571.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Clair M. H., Martin J. L., Tudor-Williams G., Bach M. C., Vavro C. L., King D. M., Kellam P., Kemp S. D., Larder B. A. Resistance to ddI and sensitivity to AZT induced by a mutation in HIV-1 reverse transcriptase. Science. 1991 Sep 27;253(5027):1557–1559. doi: 10.1126/science.1716788. [DOI] [PubMed] [Google Scholar]
- Wang B. C. Resolution of phase ambiguity in macromolecular crystallography. Methods Enzymol. 1985;115:90–112. doi: 10.1016/0076-6879(85)15009-3. [DOI] [PubMed] [Google Scholar]
- Wöhrl B. M., Moelling K. Interaction of HIV-1 ribonuclease H with polypurine tract containing RNA-DNA hybrids. Biochemistry. 1990 Nov 6;29(44):10141–10147. doi: 10.1021/bi00496a001. [DOI] [PubMed] [Google Scholar]






