Here we show that ClpB is a non-processive translocase that takes, at most, two steps on the polypeptide backbone before dissociation. These findings indicate that ClpB is not likely to translocate polypeptide through its axial channel as previously concluded.
Keywords: AAA+ motor proteins, chaperones, Förster resonance energy transfer (FRET), pre-steady-state kinetics, protein disaggregation
Abstract
Escherichia coli caseinolytic protease (Clp)B is a hexameric AAA+ [expanded superfamily of AAA (ATPase associated with various cellular activities)] enzyme that has the unique ability to catalyse protein disaggregation. Such enzymes are essential for proteome maintenance. Based on structural comparisons to homologous enzymes involved in ATP-dependent proteolysis and clever protein engineering strategies, it has been reported that ClpB translocates polypeptide through its axial channel. Using single-turnover fluorescence and anisotropy experiments we show that ClpB is a non-processive polypeptide translocase that catalyses disaggregation by taking one or two translocation steps followed by rapid dissociation. Using single-turnover FRET experiments we show that ClpB containing the IGL loop from ClpA does not translocate substrate through its axial channel and into ClpP for proteolytic degradation. Rather, ClpB containing the IGL loop dysregulates ClpP leading to non-specific proteolysis reminiscent of ADEP (acyldepsipeptide) dysregulation. Our results support a molecular mechanism where ClpB catalyses protein disaggregation by tugging and releasing exposed tails or loops.
INTRODUCTION
Escherichia coli caseinolytic protease (Clp)B is an ATP-dependent molecular chaperone belonging to the Clp/heat-shock protein (Hsp)100 family of chaperones and a member of the AAA+ [expanded superfamily of AAA (ATPases associated with diverse cellular activities)] superfamily. As a molecular chaperone, ClpB plays an important role in protein quality control by coupling the energy from ATP hydrolysis to protein disaggregation in collaboration with the DnaKJE chaperone system (DnaK, DnaJ and GrpE) [1,2].
The vital role ClpB plays in protein disaggregation has been demonstrated by the observation that clpB-null mutant strains of bacteria exhibit hypersensitivity to heat shock and other extreme stresses [3–5]. Many neurodegenerative diseases, such as Huntington's disease, Parkinson's disease, Alzheimer's disease, Kennedy's disease and others are increasingly found to involve protein aggregation and inclusion body formation [6,7]. Thus, improving our understanding of how ClpB and homologous enzymes can rescue proteins from aggregation could potentially lead to rational therapeutics [8,9].
E. coli ClpB is a multi-domain protein. From N- to C-terminus, monomeric ClpB is composed of an N-terminal domain, AAA+ domain 1 (D1) and AAA+ domain 2 (D2). Within D1 there is a unique coiled-coil M-domain (middle domain) that is absent from the homologous ClpA protein. Like ClpA and the yeast homologue of ClpB, Hsp104, ClpB contains two ATP binding and hydrolysis sites per monomer, one in each of the two AAA+ domains. Hsp100/Clp enzymes that contain two ATP-binding sites per monomer have been further classified as class 1 Hsp100/Clp enzymes [10]. This is in contrast with the class 2 enzymes that contain one ATP-binding site per monomer, ClpX being the best studied example ([11,12] and references therein).
Like other class 1 Hsp100/Clp enzymes, ClpB forms hexameric rings [13,14]. However, the molecular mechanism for how the hexameric rings of ClpB catalyse protein disaggregation remains obscure. In contrast, it has been well established that the hexameric rings of the homologous ClpA processively translocate a polypeptide substrate through the axial channel and into the proteolytic component, ClpP [15–17]. Existence of this knowledge is the pure consequence of the fact that the product of ClpA translocation is covalently modified by the protease ClpP. Because of this covalent modification, it is relatively straight forward to infer information about ClpA catalysed polypeptide translocation by simply examining the proteolytic degradation products produced by ClpP upon translocation catalysed by ClpA. In contrast, ClpB does not interact with any known protease. If hexamers of ClpB processively translocate a polypeptide substrate during protein disaggregation, then the product of this reaction is not covalently modified. Consequently, it has been difficult to determine if ClpB processively translocates or invokes some other mechanism to catalyse protein disaggregation.
To address the question of whether or not ClpB translocates a polypeptide substrate through its axial channel during protein disaggregation Weibezahn et al. [18] ‘forced’ ClpB to interact with the protease ClpP. This was accomplished by introducing the IGL loop from ClpA into ClpB. ClpA interacts with ClpP through the conserved IGL loop that exists at the base of AAA+ D2. This motif is missing in the primary structure of ClpB and upon introduction of this motif into ClpB [termed BAP (ClpB-ClpA-P-loop)] the enzyme formed a complex with ClpP. Using this construct, they reported the observation of proteolytic fragments of α-casein in the presence of BAP and ClpP that were not present in the absence of BAP or in the presence of wild-type ClpB and ClpP. Based on these observations, it was concluded that ClpB processively translocates polypeptide through its axial channel during protein disaggregation.
We developed a fluorescence stopped-flow method that allows us to examine ClpA catalysed polypeptide translocation in the absence of the proteolytic component, ClpP [19,20]. This was done, in part, to answer the question: does ClpA use the same mechanism to translocate polypeptide when ClpP is present compared with when ClpP is absent? Our emerging results are revealing that ClpP has a substantial impact on the translocation mechanism catalysed by ClpA [21,22]. This led us to the question: does introduction of the IGL loop into ClpB and subsequent interaction with ClpP up-regulate a translocation activity that is otherwise not present in wild-type ClpB? The idea that ClpP may up-regulate an activity in ClpB is a question that has been put forth by others [23].
Using single turnover, kinetic measurements that are sensitive to the molecular level events in polypeptide translocation in the absence of proteolysis, our results show that wild-type ClpB takes, at most, two translocation steps before rapid dissociation from the polypeptide substrate. This defines ClpB as a non-processive polypeptide translocase that probably binds to a protein aggregate, proceeds through one or two translocation steps (force generating steps), followed by rapid dissociation and rebinding. In this model, disaggregation occurs through repeating cycles of random binding, pulling and release. Further, using single-turnover FRET measurements that are sensitive to arrival of a translocated substrate into the central cavity of ClpP, we show that BAP does not actively translocate a polypeptide into the central cavity of ClpP for proteolytic degradation. Rather, the introduction of the IGL loop and presentation to ClpP probably induces opening of the axial pore in ClpP, thereby dysregulating ClpP and allowing for non-specific proteolysis reminiscent of the mechanism of ADEP (acyldepsipeptide) dysregulation of ClpP [24].
EXPERIMENTAL
Reagents and buffers
All chemicals were reagent grade. All buffers were prepared with distilled and deionized water produced from a Purelab Ultra Genetic system (Siemens Water Technology). Buffer H contains 25 mM HEPES, pH 7.5, at 25°C, 10 mM MgCl2, 2 mM 2-mercaptoethanol and 10% (v/v) glycerol. Buffer H is supplemented with 300, 200 or 50 mM NaCl indicated as buffer H300, H200 or H50, respectively. All experiments involving ClpA were performed in buffer H300 to compare with our previous reports. All experiments involving ClpB were performed in buffer H200 to compare with our previous binding studies [25]. All experiments involving BAP were performed in buffer H50 since BAP was not found to interact with ClpP at NaCl concentrations at or above 100 mM (result not shown). ADEP1 was kindly provided by Dr Joaquin Ortega (McMaster University, Hamilton, Canada).
Plasmid, protein and peptides
Wild-type E. coli ClpB, ClpA and ClpP were purified as described [21,26,27]. All protein concentrations throughout the manuscript are reported in monomer units. The recombinant plasmid containing BAP, pET24a(+)-BAP, was kindly provided by Dr Francis Tsai (Baylor College of Medicine, Houston, Texas) and verified by DNA sequencing. BAP was overexpressed in E. coli strain BL21(DE3) (Novagen). Cell paste was resuspended in Cell Cracking buffer [25 mM Tris, pH 8.0, at 4°C, 0.5 M NaCl, 10% (w/v) sucrose, 2 mM 2-mercaptoethanol] and disrupted with sonication on ice. Disrupted cells were centrifuged at 12000 g rpm in an SLA-3000 rotor at 4°C for 120 min. The supernatant containing BAP was filtered and loaded on to HisTrap™ FF column from GE Healthcare. After washing the HisTrap™ FF column with 20–30 column volumes of wash buffer [25 mM Tris, pH 8.0, at 25°C, 0.5 M NaCl, 20 mM imidazole, 20% (v/v) glycerol, 2 mM 2-mercaptoethanol], His-tagged BAP was eluted out with the buffer containing 0.5 M imidazole. The His6-tag on BAP was cleaved by His tagged Tobacco Etch Virus (His-TEV) (kindly provided by Dr Bingdong Sha, UAB) as a ratio of 30 mg his-tagged BAP/1 mg His-TEV. After cleavage, protein samples were adjusted to contain 1 M NaCl and loaded on a HisTrap™ FF column from GE Healthcare. The major amount of digested protein was eluted with buffer containing 15 mM imidazole. BAP was further purified through HiPrep™ 26/60 Sephacryl S-300 HR column from GE Healthcare with 20 mM Tris, pH 8.0, at 25°C, 1 M NaCl and 20% (v/v) glycerol. The purity of BAP was assessed to be greater than 95% by SDS/PAGE with Coomassie Brilliant Blue staining.
All the SsrA (small stable RNA A)-tagged polypeptides termed N-Cys-30-SsrA, N-Cys-40-SsrA and N-Cys-50-SsrA (Table 1) were synthesized by CPC Scientific. All peptides were certified >96% pure based on reverse phase HPLC and the mass was confirmed by MS. The recombinant plasmids for the 127- and 102-amino acid C-terminal truncations of αS1casein, termed αS1casein-127 and αS1casein-102, respectively were constructed and purified as described [28]. α-Casein that was used as a trap in stopped-flow experiments was purchased from Sigma. The plasmid for ClpPL139C was constructed using standard PCR methods with the QuikChange II XL Site-Directed Mutagenesis Kit from Stratagene. ClpPL139C was purified using the protocol for wild-type ClpP as described [21]. All fluorescent modifications to proteins or polypeptides were carried out with either Cy5-maleimide from GE Healthcare or fluorescein-5′-maleimide from Life Technologies as described [28]. The substrate and proteins are denoted with either Cy5- or Flu- for Cy5 and fluorescein modification, respectively.
Table 1. Polypeptide substrates.
| Name | Length (AA) | Sequence or source |
|---|---|---|
| N-Cys-30-SsrA | 30 | CTKSAANLKVKELRSKKKLAANDENYALAA |
| N-Cys-40-SsrA | 40 | CTGEVSFQAANTKSAANLKVKELRSKKKLAANDENYALAA |
| N-Cys-50-SsrA | 50 | CLILHNKQLGMTGEVSFQAANTKSAANLKVKELRSKKKLAANDENYALAA |
| αS1casein-102 | 102 | C-terminal 102 AA of αS1-casein |
| αS1casein-127 | 127 | C-terminal 127 AA of αS1-casein |
Single-turnover fluorescence/anisotropy stopped-flow experiments
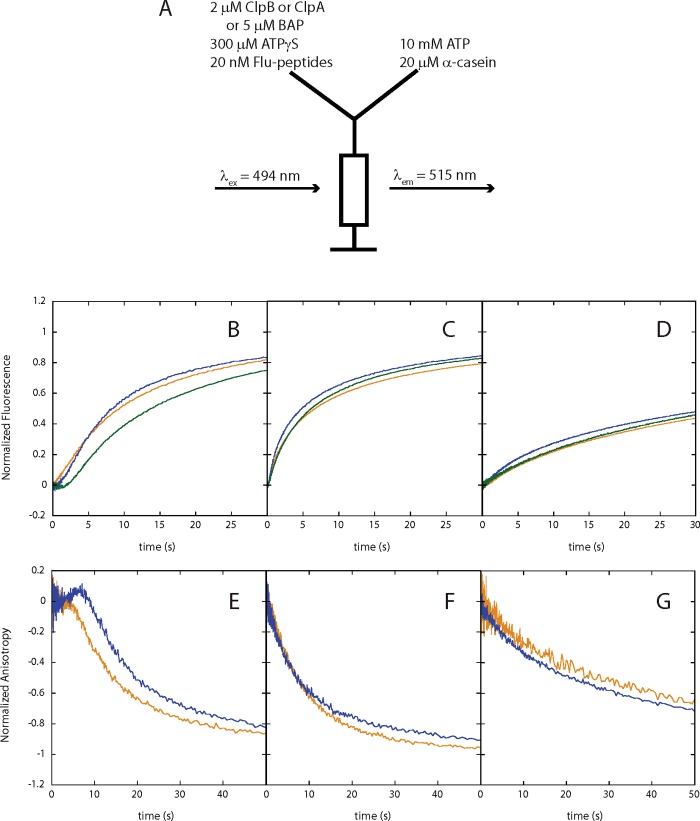
Single-turnover stopped-flow experiments for examining ClpA, ClpB or BAP translocation activity were performed using an SX.20 Applied Photophysics stopped-flow fluorometer (Leatherhead) with either fluorescence or anisotropy optics as illustrated in Figure 1(A) and as previously described [19,29].
Figure 1. ClpA catalyses processive polypeptide translocation, whereas ClpB and BAP do not.
Single-turnover fluorescence/anisotropy stopped-flow experiments for ClpA, ClpB and BAP in buffer H300, H200 and H50, respectively, on fluorescein labelled polypeptides (Table 1). (A) Experimental scheme illustrating the contents of each syringe that is rapidly mixed and exciting fluorescein on the polypeptide substrate at 494 nm, observing emission at 515 nm. Fluorescence time courses collected using Flu-N-Cys-30-SsrA (orange), Flu-N-Cys-40-SsrA (blue) and Flu-N-Cys-50-SsrA (green) for (B) ClpA, (C) ClpB and (D) BAP, and anisotropy time courses collected using Flu-αS1casein-102 (orange) and Flu-αS1casein-127 (blue) for (E) ClpA, (F) ClpB and (G) BAP.
The dissociation kinetic time courses were subjected to NLLS (non-linear least squares) analysis using a sum of two exponentials. The resultant observed rate constants, kobs,1 and kobs,2, were subjected to NLLS analysis using eqn (1),
| 1 |
where kmax,x is the maximum rate constant at saturating [ATP], Kapp,x is the concentration at half maximum kobs,x and kint is the ordinate intercept at zero [ATP].
Degradation tests by SDS/PAGE gel
Degradation tests were performed at 25°C by first mixing 10 μM BAP, 3 μM ClpA or 5 μM ADEP1, in the presence or absence of 5 mM nt (nucleotide) [ATP or ATPγS (adenosine 5′-[γ-thio]triphosphate)] with 3 μM ClpP. The sample was allowed to incubate for 5–10 min. The degradation reaction was initiated by the addition of α-casein (3 μM). At each time point, 15 μl of the reaction sample was mixed with 15 μl of 2× SDS loading buffer and heated at 100°C for 5 min. Degradation results were analysed by loading 20 μl of the denatured sample from each time point on a 15% SDS/PAGE gel followed by Coomassie Brilliant Blue staining.
Single-turnover FRET stopped-flow assays
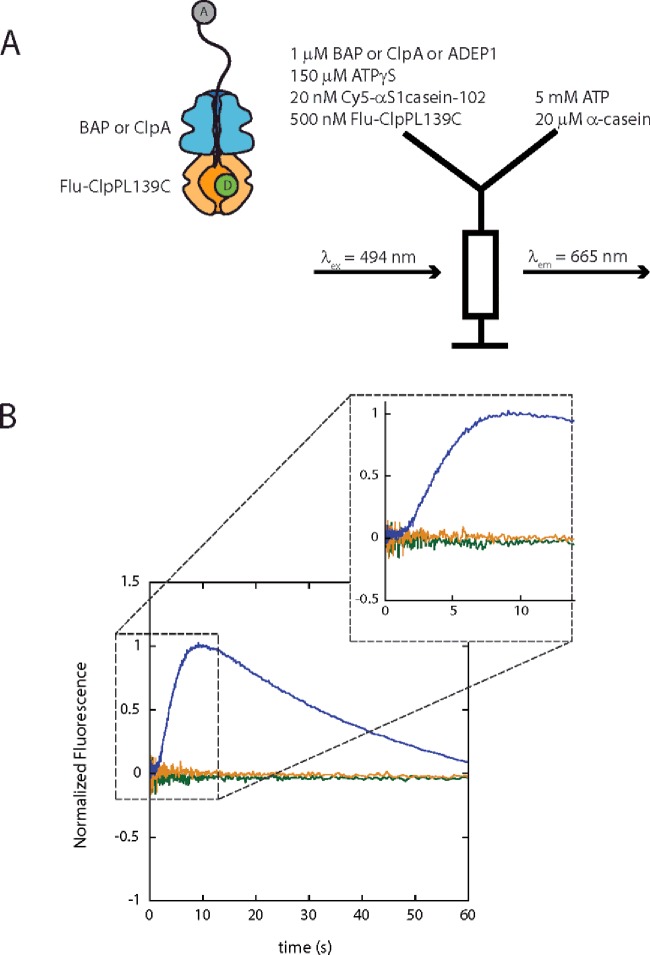
Single-turnover FRET stopped-flow experiments were performed using an SX.20 Applied Photophysics stopped-flow fluorometer (Leatherhead) as illustrated in Figure 3(A). One micromolar ClpA, BAP or 1 μM ADEP1 was pre-incubated with 500 nM Flu-ClpPL139C and 20 nM Cy5-αS1casein-102 in the presence of 150 μM ATPγS in either H300 (for ClpA and ADEP1) or H50 (for BAP) at 25°C. As shown in Figure 3(A), the pre-bound complex in the left syringe was then rapidly mixed with the contents in the right syringe, which are 5 mM ATP and 20 μM α-casein. Upon mixing, the final concentrations are 2-fold smaller. The fluorescein dye was excited at 494 nm. The kinetic time courses for sensitized emission of Cy5 were collected above 665 nm using a 665 nm long pass filter.
Figure 3. FRET experiments indicate that BAP does not translocate substrate into the central cavity of ClpP.
(A) Experimental scheme for single-turnover FRET experiments designed to examine translocation activity of BAP using ClpA as a positive control and ADEP1 as a negative control. (B) Normalized fluorescence emission monitored at 665 nm for BAP (orange), ClpA (blue) and ADEP1 (green). Experiments were performed in buffer H50 for BAP and buffer H300 for ClpA and ADEP1 separately.
Fluorescence emission spectra
Fluorescence emission spectra for different combinations of enzymes were collected over the range of 450–800 nm by exciting either the fluorescein dye at 494 nm or the Cy5 dye at 649 nm using a Fluorolog-3 spectrofluorometer (HORIBA Jobin Yvon).
RESULTS
Single-turnover examination of polypeptide translocation
To test the hypothesis that ClpB can processively translocate polypeptide substrates, we used the single-turnover fluorescence stopped-flow method that we developed to examine polypeptide translocation catalysed by ClpA in the absence of the proteolytic component [19,21,29]. This method is sensitive to polypeptide translocation in the absence of any covalent modification of the substrate being translocated. Since ClpB does not associate with any known peptidase, the single-turnover fluorescence stopped-flow method is an ideal approach to examine ClpB catalysed polypeptide translocation, if it occurs.
In our previous studies designed to examine ClpA catalysed polypeptide translocation we used a set of unstructured polypeptides ranging from 30 to 50 amino acids in length, all of which contain the SsrA sequence at the C-terminus (see Table 1 for sequence information) [19,28]. Our binding studies have shown that hexameric ClpB can associate with these substrates with a binding affinity of ∼ 4–12 nM [25]. Thus, we used this set of polypeptide substrates to test ClpB translocation activity. All three polypeptide substrates contain the SsrA sequence at the C-terminus and one cysteine residue at the N-terminus (Table 1). After labelling with fluorescein-5′-maleimide, each labelled polypeptide substrate contains a single fluorophore at the N-terminus.
To perform the single-turnover stopped-flow experiments, either 2 μM ClpB or ClpA was pre-incubated with 300 μM ATPγS and 20 nM fluorescently modified polypeptide in one syringe of the stopped-flow fluorometer. As schematized in Figure 1(A), 10 mM ATP and 20 μM α-casein were loaded into the other syringe. Upon rapid mixing of the contents of the two syringes, fluorescence emission was monitored at 515 nm. In the presence of ATPγS, both ClpB and ClpA will assemble into hexamers and bind to the fluorescently modified polypeptide substrates [25,28]. In this ‘pre-bound’ complex the fluorescence from fluorescein is quenched when either ClpA [19] binds or ClpB binds (Supplementary Figures S1A–S1D). Upon rapid mixing, ATP will exchange for ATPγS and the enzyme will either immediately dissociate or translocate the substrate followed by dissociation. In either case, when the enzyme dissociates the fluorescence will increase. The large excess of unlabelled α-casein is included to maintain single-turnover conditions by not allowing free ClpB to rebind the fluorescently-modified polypeptide. Supplementary Figure S2 shows that the α-casein is of sufficiently high concentration to maintain single turnover conditions. Thus, these experiments are single-turnover with respect to the fluorescently-modified polypeptide. Consequently, the ClpB that is pre-bound to the polypeptide substrate will either translocate and dissociate or immediately dissociate, but ClpB will not re-bind after rapid mixing with the trap for ClpB.
As previously reported for ClpA, the kinetic time courses from single-turnover fluorescence stopped-flow experiments performed as schematized in Figure 1(A) exhibit a lag followed by a signal enhancement (Figure 1B). This lag increases with increasing substrate length, e.g. the-50 amino acid substrate has the longest lag (solid green line) and the 30-amino acid substrate has the shortest lag (solid orange line). We have previously interpreted this lag to indicate that ClpA proceeds through multiple steps of translocation before dissociation from the polypeptide substrate [19,21,22,29].
To test whether ClpB can translocate a substrate that is known to be bound by ClpB, the stopped-flow experiment schematized in Figure 1(A) was performed with ClpB. Like ClpA, we also observed quenched fluorescence emission upon binding of ClpB to the polypeptide substrates examined in the present study (Supplementary Figure S1A–S1D). However, unlike the kinetic time courses collected using ClpA, the kinetic time courses collected using ClpB and the strategy schematized in Figure 1(A) do not exhibit a lag before fluorescence increase (Figure 1C). Rather, upon rapid mixing of polypeptide bound ClpB with ATP and trap for ClpB we observe immediate signal enhancement indicating rapid dissociation.
The stopped-flow experiment was performed with ClpB and three polypeptide substrates of length L=30, 40 and 50 amino acids. In all cases, the signal enhancements for each of the three substrates can be well described by a sum of two exponentials (discussed below). The observation of two phases suggests that at least two kinetic steps occur before ClpB dissociates from the substrate. However, unlike ClpA, there is no indication of a lag before the signal change occurs.
The absence of a lag phase alone does not indicate that ClpB does not translocate. For example, experiments designed to examine ssDNA translocation yield time courses that do not exhibit lag phases [30,31]. Translocation is revealed through the length dependence of the time courses collected with substrates of various lengths. By comparison, if processive translocation is occurring catalysed by ClpB one would expect to see a dependence of the kinetic time courses on polypeptide length. As seen in Figure 1(C), not only is a lag phase not observed, but no length dependence is observed for the 30, 40 and 50 amino acid substrates.
In ssDNA translocation experiments, signal is often the consequence of the motor approaching, interacting with and dissociating from the fluorophore and thereby affecting the fluorescence [30–33]. However, unlike ssDNA translocation, we have not observed a signal change as the motor, ClpA, approaches the fluorophore. This is probably the consequence of the fact that the environment is not changing as dramatically as the motor approaches the fluorophore in the polypeptide translocation case as it is for the nucleic acid motor. In the case of the polypeptide translocase, the fluorophore is attached to a protein and being approached by another protein. Consequently, the fluorophore is surrounded by amino acids and being approached by amino acids. In contrast, in the ssDNA translocase case, the fluorophore is on a highly charged poly-anion being approached by a protein of very different chemical composition.
ClpA binds specifically at the SsrA tag at the C-terminus and translocates a polypeptide substrate from the C- to N-terminus. However, the directionality of ClpB translocation, if it occurs, is not known. Consequently, one reason for not observing any length dependence in the time courses shown in Figure 1(C) is that ClpB translocates away from the fluorophore at the N-terminus. To test this, we used the identical substrate but moved the fluorophore to the C-terminus. Again, no lag or length-dependent signal change was observed (Supplementary Figure S3).
Another possible reason for not observing any indication of translocation by ClpB is that the 30–50 amino acid polypeptide substrates containing SsrA are natural substrates for ClpA, but may not be natural substrates for ClpB. We have shown that ClpB binds to these SsrA tagged substrates [25]. However, ClpB may dissociate upon mixing with ATP because it may be able to discriminate this substrate from other substrates. In contrast, α-casein is a natural substrate for ClpB [10] and was the substrate used in studies that concluded ClpB translocates substrate through its axial channel [18].
α-Casein is a mixture of αS1casein and αS2casein [34]. The translocation activity of ClpB was further examined using single-turnover anisotropy stopped-flow experiments and α-S1casein polypeptide substrates. Anisotropy was chosen because little observable fluorescence change was observed upon either ClpB or ClpA binding to the αS1casein substrates. When either ClpB or ClpA bind to these αS1casein substrates, we observe changes in fluorescence intensities that are difficult to interpret. In contrast, we observe a consistent and significant anisotropy increase when either ClpB or ClpA binds to these αS1casein substrates and this signal has been used to quantitatively examine polypeptide binding by us [25,28]. Thus, the pre-bound enzyme–polypeptide complex schematized in Figure 1(A) will exhibit an elevated anisotropy and, upon rapid mixing with ATP, the anisotropy is expected to decrease upon dissociation of the enzyme from the substrate. Moreover, anisotropy is not expected to be sensitive to the direction of translocation. Rather, the anisotropy signal will decrease when ClpB dissociates and should reflect the residence time of the enzyme on the polypeptide substrate. If translocation is occurring, ClpB should reside on the polypeptide substrate for a longer amount of time for each increase in substrate length.
In these experiments, we used two substrates with lengths of 102 and 127 amino acids derived from αS1casein. These constructs, which are shorter than full-length αS1casein were used because we have shown that multiple ClpB hexamers will associate with substrates longer than 127 amino acids [25]. We chose the shorter constructs because we wish to examine a single cycle of polypeptide translocation, if it occurs, catalysed by a single active complex of ClpB.
The single-turnover anisotropy stopped-flow experiments using two αS1casein substrates, αS1casein-102 and αS1casein-127 (Table 1), were performed as schematized in Figure 1(A). As expected, when the anisotropy stopped-flow experiments were carried out with ClpA, we observe a lag before the anisotropy decreases for experiments performed with αS1casein-102 (Figure 1E, orange solid line) and a longer lag for αS1casein-127 (Figure 1E, blue solid line). In contrast, the time courses collected from performing the same anisotropy stopped-flow experiments with ClpB neither exhibit a lag phase nor length dependence (Figure 1F). These results for both ClpA and ClpB are consistent with what we have observed from the single-turnover fluorescence stopped-flow experiments shown in Figures 1(B) and 1(C) for ClpA and ClpB, respectively.
It has been reported that ClpB exhibits enhanced disaggregation activity in a mixture of ATP and ATPγS with maximum activity observed with a 1:1 mixture [35]. In that study, it was concluded that the mixture of ATP and ATPγS serves the same role as the KJE chaperone system. Although not 1:1, it is important to note that the results exhibited in Figures 1(C), 1(D), 1(F) and 1(G) were carried out in a mixture of 150 μM ATPγS and 5 mM ATP. Control experiments carried out in the presence of 1 mM ATPγS and 1 mM ATP exhibit exponential time courses similar to those shown in Figure 1(C) for ClpB, although they appear to show slower dissociation kinetics (Supplementary Figure S4).
Based on the results of our single-turnover fluorescence/anisotropy stopped-flow experiments, we hypothesize that wild-type ClpB is a non-processive translocase. In the present study, we observe two phases in the kinetic time courses for dissociation of ClpB from the polypeptide substrate upon rapid mixing with ATP. These two phases must occur before ClpB dissociates. Thus, the two phases must represent events that ClpB is catalysing. These two phases may represent ClpB proceeding through one or two translocation steps followed by dissociation, where the suggestion of two steps is based on the observation of biphasic kinetic time courses.
Processivity, P, is the probability that the enzyme will step forward compared with dissociate and can be quantitatively defined by eqn (2):
| 2 |
where kT is the translocation rate constant, kd is the rate constant for dissociation from the polypeptide substrate, m is the step-size and N is the average distance translocated per binding event in units of amino acids [36]. Even though we do not have a measure of the step-size, m, for ClpB translocation, if ClpB takes one or two steps before dissociation then the average distance translocated per binding event would be N=1m or 2m. Thus, eqn (2) tells us that the processivity, P, would be P = 0.37 for one step or P = 0.61 for two steps. This represents a value that defines ClpB as an enzyme with low processivity or a non-processive translocase. This is in contrast with ClpA, which we have reported to have a processivity of P ∼ 0.88 at 300 μM ATP, indicating that ClpA translocates, on average, ∼100 amino acids per binding event at low [ATP; 19]. However, the processivity was too high to measure at [ATP] above 300 μM indicating that the processivity is much higher at physiological [ATP; 19].
Introducing the IGL loop into wild-type ClpB does not up-regulate ClpB translocation activity
Previous work used a variant of ClpB (BAP) to conclude ClpB translocates substrate through its axial channel. Those experiments were performed with the ClpB variant, BAP, ClpP and α-casein. Degradation of α-casein was observed both in the presence and absence of the KJE system. Those observations were used to conclude that ClpB translocates α-casein through its axial channel and out the other side of the hexameric ring [18]. Since our single-turnover experiments suggest wild-type ClpB may be a non-processive translocase, we asked the question: does the introduction of the IGL loop into ClpB (BAP) induce a translocation activity that is otherwise not present in wild-type ClpB? This is a question that has been posed by others [23]. To test this, we performed similar single-turnover fluorescence/anisotropy stopped-flow experiments as we did for ClpA and wild-type ClpB using the experimental scheme shown in Figure 1(A). The kinetic time courses for BAP in Figures 1(D) and 1(G), fluorescence and anisotropy, respectively, show similar length-independent exponentials as observed for ClpB in Figures 1(C) and 1(F). Since the kinetic time courses for BAP are similar to wild-type ClpB, our results suggest that introducing the IGL loop into ClpB does not induce a translocation activity of ClpB. Rather, the kinetic time courses indicate that both BAP and ClpB are unlikely to take more than one or two steps before dissociating from the polypeptide substrates.
BAP enlarges the axial pore of the ClpP tetradecamer
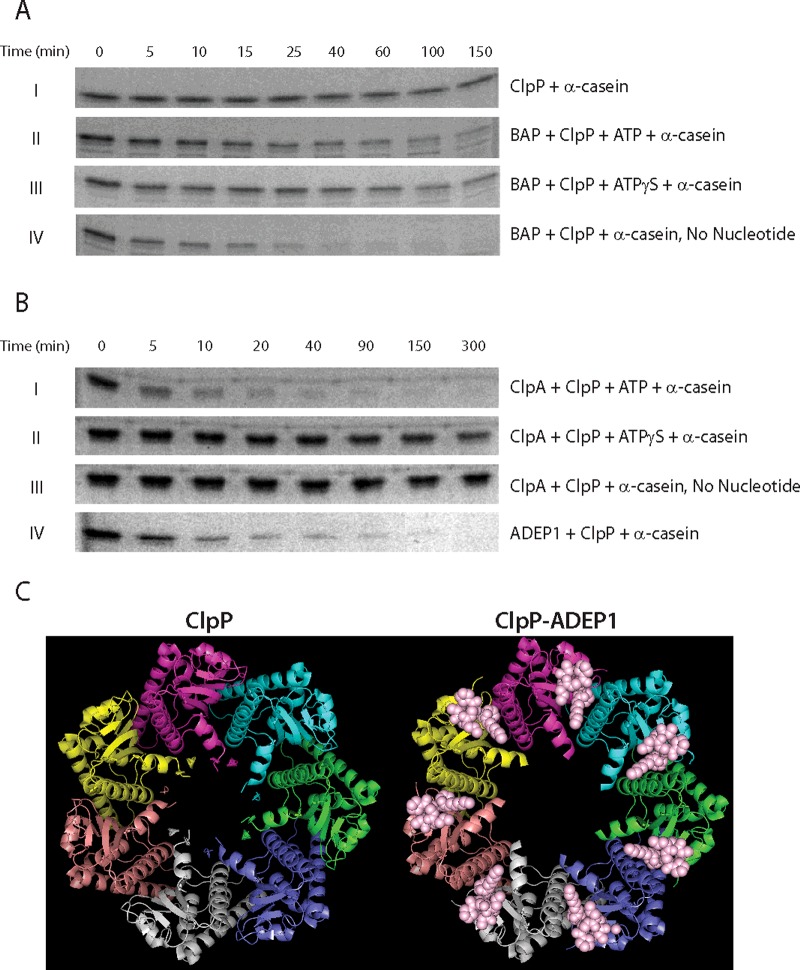
Our findings that neither ClpB nor BAP exhibit processive translocation from single-turnover experiments does not agree with the findings reported by Weibezahn et al. [18] where they reported that ClpB translocates substrate through its axial channel. To yield insight into this apparent discrepancy and to confirm activity of our preparation of BAP, we examined the proteolytic degradation products of α-casein produced by either BAP–ClpP or ClpAP (‘Experimental’). Here we used α-casein because it is the identical substrate used in the previous reported experiments that concluded that ClpB translocates substrate through its axial channel [18]. Moreover, we did not include the KJE system since degradation of α-casein was also observed in the absence of the KJE system. Different combinations of enzymes were incubated with α-casein in the presence or absence of nt as indicated in Figures 2(A) and 2(B). The density of the bands on the SDS/PAGE gel shown in Figures 2(A) and 2(B) represent the remaining amount of α-casein at defined incubation times indicated at the top of the gel images.
Figure 2. Introduction of the IGL loops into ClpB dysregulates ClpP and induces non-specific proteolysis.
(A) SDS/PAGE images for 3 μM α-casein in buffer H50 at 25°C in the presence of (I) 3 μM ClpP, (II) 10 μM BAP, 3 μM ClpP and 5 mM ATP, (III) 10 μM BAP, 3 μM ClpP and 5 mM ATPγS and (IV) 10 μM BAP and 3 μM ClpP. (B) SDS/PAGE images for 3 μM α-casein in buffer H300 at 25°C in the presence of (I) 3 μM ClpA, 3 μM ClpP and 5 mM ATP, (II) 3 μM ClpA, 3 μM ClpP and 5 mM ATPγS, (III) 3 μM ClpA and 3 μM ClpP and (IV) 5 μM ADEP1 and 3 μM ClpP. Degradation was examined at each indicated time point. (C) Top view of heptameric ClpP (left, PDB ID code 3KTG) and ClpP–ADEP1 complex (right, PDB ID code 3KTI). Each ClpP monomer is colour-coded. ADEP1 is represented in pink spheres in the ClpP–ADEP1 complex. Both images were prepared using PYMOL (the PyMOL molecular graphics system, version 1.5.0.4 Schrodinger, LLC).
Figure 2(AI) shows the results of incubating 10 μM α-casein with 3 μM ClpP in the absence of any other proteins or nt. The density of the α-casein band did not change significantly over 150 min. Although it has been shown that ClpP in the absence of any motor can degrade α-casein over a 24 h period, as expected, very little if any proteolysis is observed over 150 min [37]. When the same amount of α-casein was incubated with 10 μM BAP and 3 μM ClpP in the presence of 5 mM ATP, the density of the α-casein band appears to decrease slightly over 150 min (Figure 2AII). Interestingly, we also observed that α-casein was slowly degraded when incubating with 10 μM BAP and 3 μM ClpP in the presence of 5 mM ATPγS (Figure 2AIII). Since ATPγS is a poorly-hydrolysable ATP analogue, this result suggests that the BAP–ClpP complex could degrade polypeptides independent of ATP hydrolysis. To further test this, we incubated 10 μM α-casein with 10 μM BAP and 3 μM ClpP without any nt. Strikingly, the α-casein band shown in Figure 2(AIV), in the absence of any nt, rapidly diminished and disappeared within 1 h, which is even faster than in the presence of either ATP or ATPγS (Figures 2AII and 2AIII, respectively). This result indicates that α-casein is rapidly degraded by BAP–ClpP in the absence of any nt, which is not consistent with energy driven processive translocation of a substrate through the axial channel of BAP and into ClpP as previously concluded [18]. Since the mixing ratios of BAP and ClpP were not identical with those reported by Weibezahn et al. [18], all of the experiments shown in Figure 2(A) were reproduced in the conditions previously reported and with or without an ATP regenerating system. Our findings in all tested conditions are identical (Supplementary Figures S5A and S5B). Since degradation is observed both in the presence and in the absence of ATP, this observation leads us to conclude that this strategy cannot be used to conclude that BAP catalyses ATP-driven translocation of substrate through its axial channel.
As a positive control, we examined the proteolytic degradation of α-casein catalysed by ClpAP (Figure 2B). When incubating α-casein with ClpAP in the presence of ATP (Figure 2BI), α-casein was rapidly degraded. The observed rapid degradation of α-casein is consistent with ATP-driven translocation by ClpA of the substrate into the proteolytic chamber of ClpP. In contrast, no change in the density of the bands is observed up to 150 min when ClpA is incubated with ClpP, α-casein and ATPγS. However, a slight decrease in density may be present at 300 min, (Figure 2BII). Previous reports showed that polypeptides can be slowly degraded by ClpAP in the presence of ATPγS [38]. In stark contrast with what was observed with BAP, when ClpA, ClpP and α-casein were incubated in the absence of nt we did not observe any degradation of α-casein (Figure 2BIII).
We have previously reported that ClpA resides in a monomer—dimer–tetramer equilibrium in the absence of nt under our experimental conditions [27,39]. ClpA requires nt binding to form hexamers competent for binding to ClpP [40,41]. Consequently, ClpA doesn't associate with ClpP in the absence of nt because no hexamers are present to do so, which is probably the reason we do not observe degradation of α-casein by ClpAP in the absence of nt (Figure 2BIII). In contrast, ClpB and BAP do form hexamers in the absence of nt and therefore hexamers of BAP are present to associate with ClpP in the absence of nt [14,42–45] (Supplementary Figure S6). Degradation that is observed for BAP and ClpP in the absence of nt may be due to BAP interacting with ClpP and inducing enlargement of the axial pore of ClpP thereby dysregulating the proteolytic activity.
Several lines of evidence support BAP-induced dysregulation of ClpP. First, ClpP forms a tetradecamer with two stacked heptameric rings. The diameter of the two entrances to the proteolytic chamber located on opposite sides of its central cavity are ∼1.5 nm [46]. Previous reports showed that binding of ClpA to ClpP induces an opening of the entrances to the proteolytic chamber of the ClpP tetradecamer [47]. However, in the case of ClpA, hexameric ClpA remains associated with ClpP and probably shields the entrance, thereby precluding random diffusion of substrates into ClpP. Second, a previously discovered class of antibiotics, ADEPs, can also open up the entrances to the ClpP chamber [24,47–49]. In vivo, when the entrances are open, polypeptide substrates can diffuse into the chamber of ClpP and proteins are non-specifically degraded, leading to cell death.
ADEPs bind to tetradecameric ClpP on the apical and distal surfaces between adjacent ClpP monomers, where the IGL loop also binds [48]. Figure 2(C) shows the crystal structure of the surface of ClpP in the absence and presence of ADEPs [48]. As shown in Figure 2(C), the axial entrance of ClpP–ADEP1 complex in the right panel is larger than that of ClpP alone in the left panel. The diameter of the pore was reported to be 1.5 nm in the absence of ADEP and 3.0 nm when ADEP is bound. Moreover, Figure 2(BIV) shows the results from an experiment where ADEP1 was incubated with ClpP and α-casein. The image of the gel clearly shows that α-casein is rapidly degraded on a time scale similar to BAP, ClpP and α-casein in the absence of nt shown in Figure 2(AIV).
We hypothesize that since BAP can form hexamers in the absence of nt the enzyme is able to present the IGL loops to ClpP and induce enlargement of the axial pore. This, in turn, allows α-casein to randomly diffuse into the proteolytic core and undergo proteolytic degradation. This activity is probably slowed down in the presence of ATP or ATPγS because the hexameric rings of BAP are stabilized and/or the binding affinity to ClpP is stronger. Regardless, BAP may still catalyse some polypeptide translocation since proteolytic degradation is observed in the absence and presence of nt. In the presence of ATP, the observation of proteolytic degradation can be described by both processive translocation into ClpP and axial pore opening induced by binding of the chaperone. Thus, the question remains: in addition to inducing pore opening, does BAP also translocate some substrate into ClpP for proteolytic degradation?
Examination of entrance of polypeptide substrates into the ClpP cavity
We performed single-turnover FRET stopped-flow experiments to test whether BAP is able to processively translocate polypeptides into ClpP. We constructed a ClpP mutant, ClpPL139C, which has a cysteine substitution located in the central cavity of the ClpP tetradecamer and near the active site of degradation. ClpPL139C was labelled with fluorescein to serve as the donor in the FRET experiment resulting in Flu-ClpPL139C. The cysteine residue at the N-terminus of the polypeptide substrate, αS1casein-102 (Table 1), was labelled with Cy5-maleimide to serve as the acceptor in the FRET experiment resulting in Cy5-αS1casein-102. The emission spectrum of fluorescein has adequate overlap with the excitation spectrum of Cy5 over the range of 500–650 nm (Cy5 excitation spectrum not shown). With the exception of the choice of fluorophore an identical strategy was successfully employed by Reid et al. [17] to examine the directionality of ClpA catalysed polypeptide translocation into ClpP.
In order to preassemble the chaperone–peptidase machine bound to the polypeptide substrates, as shown in Figure 3(A), either 1 μM BAP or 1 μM ClpA was pre-incubated with 500 nM Flu-ClpPL139C and 20 nM Cy5-αS1casein-102 in the presence of 150 μM ATPγS. No sensitized emission of Cy5 was observed upon exciting the fluorescein in the pre-assembled ClpA-Flu-ClpPL139C bound to the polypeptide substrate (Supplementary Figure S7). Either single-turnover translocation catalysed by ClpA or BAP or dissociation of ClpA or BAP from the polypeptide substrate is triggered by rapidly mixing with 5 mM ATP and 20 μM trap for ClpB (α-casein). The FRET donor (fluorescein) inside the ClpP proteolytic cavity was excited at 494 nm. The sensitized emission of the FRET acceptor (Cy5) was monitored at 665 nm. In this single-turnover FRET stopped-flow experiment, the sensitized emission will occur only if the polypeptide substrate labelled with the acceptor is fully translocated into the ClpP central cavity and the acceptor is close to the donor.
The single-turnover FRET stopped-flow experiment performed with ClpA (Figure 3B solid blue line), exhibits a ∼2-s lag followed by a signal increase and then a decrease. The lag in the first ∼2 s indicates the average time that ClpA is translocating on the polypeptide substrate before the substrate enters the central cavity of the ClpP tetradecamer and sensitized emission from Cy5 is observed. The lag is consistent with processive polypeptide translocation catalysed by ClpA. Sensitized emission from Cy5 is due to arrival of the fluorophore in the central cavity of ClpP. The signal decrease indicates the release of degraded polypeptide substrates from ClpP. In contrast, the kinetic time course for BAP–ClpP (Figure 3B, solid orange line) did not show any FRET signal change under the same experimental setup. This result shows that the FRET acceptor labelled polypeptide was not processively translocated into the chamber of ClpP by BAP. Furthermore, this result suggests that ClpP does not up-regulate a translocation activity of ClpB. From this we conclude that both BAP and wild-type ClpB are non-processive polypeptide translocases with a processivity, P, between ∼0.3 and 0.6 based on eqn (2) and the assumption that one or two steps are taken, respectively.
The absence of the observation of FRET in this experiment presents an apparent discrepancy with the observed degradation on SDS gels in Figure 2. However, it is important to note that observation of disappearance of the α-casein band on the gel occurs with every cleavage event regardless of how the α-casein substrate arrived at the serine protease active site within the ClpP proteolytic chamber. In contrast, observation of the FRET signal change presented in Figure 3(B) for ClpAP requires complete and processive translocation into the central cavity of ClpP. A sustained build-up of substrate in the central cavity of ClpP will be required to observe a signal change. Diffusion of the α-casein substrate into and out of the cavity will occur with a similar rate and thus no net signal change will be observed. Consequently, the FRET experiment is not sensitive to random diffusion into ClpP.
To support the claim that the FRET signal is not sensitive to random diffusion into the central chamber of ClpP, we performed the same stopped-flow FRET experiment in the presence of ADEP1 with no ClpA or BAP. In this experiment, we know that ADEP1 dysregulates ClpP and proteolysis is the pure consequence of diffusion. As shown in Figure 3(B) (green trace), no FRET signal change is observed because the infrequent diffusion into and out of the cavity does not result in a sustained FRET signal change. Thus, we conclude that the FRET experiment is not sensitive to random diffusion into and out of ClpP, but is sensitive to processive translocation indicated by the FRET stopped flow experiment performed with ClpA.
ClpB is a non-processive polypeptide translocase
Fluorescence stopped-flow experiments performed with ClpB pre-bound to a polypeptide substrate followed by rapid mixing with ATP and trap for ClpB exhibit biphasic kinetic time courses. These biphasic kinetic time courses indicate that ClpB dissociates from the substrate in two kinetic steps (Figure 1C). This observation could indicate that ClpB takes one or two translocation steps followed by rapid dissociation. If the observed kinetic steps represent polypeptide translocation then the steps will be coupled to ATP-binding and hydrolysis. To test this we examined the kinetics of ClpB dissociating from the polypeptide substrate as a function of [ATP].
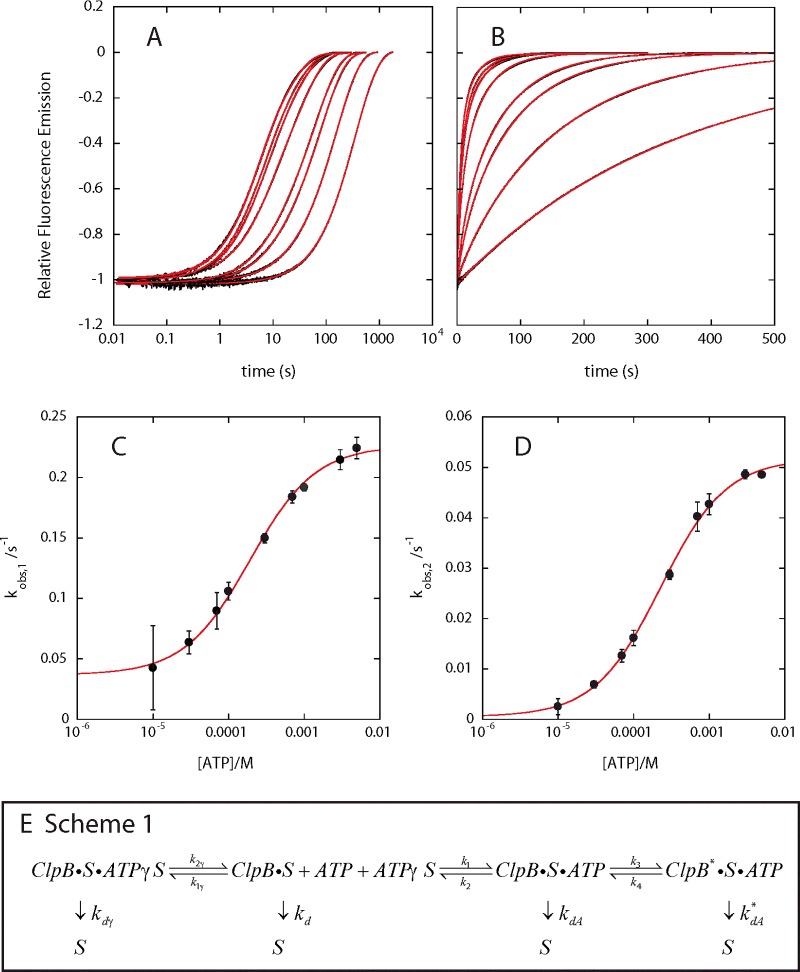
Fluorescence stopped-flow experiments were performed as schematized in Figure 1(A) using Flu-N-Cys-50-SsrA with the exception that the [ATP] in the second syringe was varied between 20 μM and 10 mM (10 μM–5 mM final mixing concentrations). Figures 4(A) and 4(B) shows the kinetic time-courses collected at each ATP concentration. In all cases, the fluorescence increases upon rapid mixing with ATP and trap indicating that ClpB is dissociating from the polypeptide substrate. Further, as the [ATP] is decreased, the dissociation kinetics clearly slow down. This observation indicates that dissociation from the polypeptide substrate is limited by the [ATP].
Figure 4. ClpB dissociates from polypeptide substrate in an ATP-dependent manner.
ATP-dependent single-turnover fluorescence stopped-flow experiments performed as schematized in Figure 1 with ClpB using Flu-N-Cys-50-SsrA as the polypeptide substrate with the final ATP concentration varying from 10 μM to 5 mM performed in buffer H200 at 25°C. Relative fluorescence emission time courses (black) were subjected to NLLS analysis using a summation of two exponentials. The result of the fit of each time course is shown in red displayed in both (A) logarithmic scale and (B) linear scale. The observed rate constants from the two exponential fits were plotted compared with [ATP] and subjected to NLLS analysis using eqn (1). The solid black circles in panel (C) are kobs,1 at each [ATP] and the solid line is the result of the fit to eqn (1) with kmax,1=(0.190±0.004) s−1, Kapp,1=(192±18) μM, kint=(0.037±0.004) s−1. Panel (D) is kobs,2 compared with [ATP] and the solid line is the result of the fit to eqn (1) with kmax,2=(0.0516±0.0006) s−1, Kapp,2=(226±16) μM and kint=0. (E) Potential kinetic scheme for ClpB catalysed protein disaggregation.
Each kinetic time course collected at a different [ATP] is well described by a sum of two-exponentials, where the red lines in Figures 4(A) and 4(B) indicate the fits and the blue lines indicate the experimental data. The two observed rate-constants from fitting of the kinetic time-courses are plotted in Figures 4(C) and 4(D). The uncertainty bars represent the S.D. from three independent measurements at each fixed [ATP]. Both observed rate-constants exhibit a hyperbolic dependence on [ATP]. This observation indicates that both kinetic steps are coupled to ATP binding [50].
The plots of kobs,x compared with [ATP] were subjected to NLLS analysis using eqn (1) [50]. The apparent dissociation equilibrium constants from this analysis were found to be Kapp,1=(192±18) μM and Kapp,2=(226±16) μM. The similarity of the two values could indicate that the two kinetic steps are coupled to the same ATP-binding step. The maximum observed rate constant at saturating [ATP] obtained from kobs,1 and kobs,2 are kmax,1=(0.190±0.004) s−1 and kmax,2=(0.0516±0.0006) s−1, respectively. Although it is clear that the plot of kobs,1 compared with ATP extrapolates to a positive value on the ordinate at zero [ATP], kobs,2 is equally well described if the curve intersects the origin or a small finite ordinate intercept. This result indicates that kobs,1 is a step that immediately follows ATP binding and this step is reversible with a reverse rate constant of kint=(0.037±0.004) s−1. In contrast, kobs,2 is a step that immediately follows ATP binding but is irreversible or the reverse rate constant is so small that it can be considered to be negligible [50].
One proposed elementary mechanism that can describe these observations is given by Scheme 1 in Figure 4(E). In Scheme 1 ClpB is prebound to a polypeptide substrate, S and ATPγS. In the presence of hydrolysable ATP the ClpB–polypeptide substrate complex can bind to ATP when ATPγS is released. The ClpB polypeptide and ATP complex can go through a conformational change that could represent a translocation step. In our experiments signal is observed when ClpB dissociates from the polypeptide substrate regardless of the nt ligation state or whether ClpB has translocated. Thus we can define a different dissociation rate constant for each state as kdγ, kd, kdA and kdA* to denote dissociation from the polypeptide substrate in the ATPγS bound state, the nt unligated state, the ATP bound state and the translocated ATP bound state, respectively.
If we assume that nt, regardless of type, binds with a diffusion controlled rate constant then k1γ=k1=∼1×108 M−1·s−1. Since the apparent equilibrium constant for ATP is ∼200 μM, this suggests that k2 is ∼2×104 s−1. The two steps that follow ATP binding are substantially slower than the ATP dissociation rate constant since kobs,1 and kobs,2 at saturating ATP concentration are kmax,1=(0.190±0.004) s−1 and kmax,2=(0.0516±0.0006) s−1. Therefore the ATP binding step being in a rapid equilibrium is a good assumption because k2 >> kmax,1 and kmax,2. This is consistent with the observed hyperbolic dependence of both kobs,1 and kobs,2 on [ATP].
Since the two observed rate constants are coupled to ATP binding, we propose that kobs,1 is a reflection of the reversible step with rate constants k3 and k4 (Scheme 1) and kobs,2 is a reflection of the irreversible dissociation after ATP binding, kdA. Thus, kmax,1=k3=(0.190±0.004) s−1 and the ordinate intercept is kint=k4=(0.037±0.004) s−1. Similarly, kobs,2 reflects the dissociation step and therefore kmax,2=kdA=(0.0516±0.0006) s−1. It is important to note that this represents a proposed minimal model that can describe the observations, but it will be the topic of substantial testing going forward.
DISCUSSION
Polypeptide translocases like ClpA, ClpX and HslU (heat-shock locus; ClpY) [51] processively translocate a polypeptide substrate through their axial channels and into the associated barrel shaped protease, ClpP or HslV (ClpQ), respectively. The homologous ClpB does not associate with any known protease. Appropriately, based largely on structural and functional comparisons with ClpA and ClpX, it has been proposed that ClpB translocates substrates through its axial channel and out the other side of the hexameric ring during protein disaggregation.
In addition to ClpB and yeast Hsp104, which both catalyse protein disaggregation, there are many AAA+ enzymes that only catalyse protein remodelling reactions and do not interact with proteases [52]. It has proven difficult to show whether such enzymes fully translocate a substrate through their axial channels, only pull on exposed tails [53] or take only a few translocation steps. The difficulty lies in the fact that the substrates in these reactions both enter and leave the reaction without covalent modification. Although the substrate may exhibit differences in structure before and after being remodelled by such an enzyme, this has proven difficult to monitor using transient state kinetic techniques. Having a method that allows us to examine the molecular level events in polypeptide translocation in the absence of proteolytic degradation has been lacking.
In recognition of the difficulty in acquiring supporting evidence for translocation by an enzyme that does not covalently modify its substrate, the Bukau group constructed a variant of ClpB that contained the IGL loop from ClpA [18]. This was done to force ClpB to interact with the protease ClpP. By forcing ClpB to interact with the proteolytic component, ClpP, they proposed that if proteolytic fragments are observed, ClpB must translocate a substrate through its axial channel and into ClpP. Here we have shown that the observation of proteolytic fragments can be described by two phenomena, both processive translocation and an ATP independent function that we propose is most probably dysregulation of ClpP.
To our knowledge, a control experiment with E. coli BAP, ClpP and no nt has not been reported. However, the Watanabe group reported observing no degradation catalysed by T. thermophilus TClpP (ClpP from T. thermophilus) and TBAP (BAP mutant of ClpB from T. thermophilus) in the absence of nt [54,55]. There does appear to be some degradation on their gel images between 60 and 90 min from the experiments performed in the absence of nt. Nevertheless, it is certainly clear that the proteolysis reaction is much faster when ATP is added. Those experiments were carried out at 55°C with 0.5 μM [55] or 2 μM [54] TClpP and 50 nM TBAP.
Both E. coli ClpB and E. coli BAP form hexamers in the absence of nt [14,42–45] (Supplementary Figure S6). In contrast, E. coli ClpA does not form hexamers in the absence of nt. Rather, ClpA resides in a monomer–dimer–tetramer equilibrium [27,39]. Here we have proposed that degradation is observed with BAP and ClpP in the absence of nt because hexameric BAP is present, able to interact with ClpP and open the axial channel. In contrast, ClpA in the absence of nt will not interact with ClpP because no hexameric ClpA is present to do so. The observation of no degradation in the absence of nt with TClpP and TBAP may be because TBAP does not form hexamers in the absence of nt. Furthermore, even if TBAP does form hexamers in the absence of nt there may not be a significant population of hexamers present in solution at the 50 nM protein concentration until nt shifts the equilibrium toward hexamers.
Up to two hexamers of ClpA bind the tetradecameric ClpP. The only reported affinity constant for hexameric ClpA interacting with tetradecameric ClpP is based on an α-casein degradation assay that concluded a value of 4 nM under the assumption that all of the ClpA is hexameric [40]. Even if the 4 nM binding constant for ClpA binding ClpP is correct, it seems unlikely that this would be the same for BAP interacting with ClpP.
To our knowledge, there is no reported affinity constant for hexameric BAP interacting with the two surfaces of tetradecameric ClpP. Bukau and colleagues [18] reported a size-exclusion chromatogram collected with 6 μM ClpB and 7 μM ClpP (both in monomer units, Axel Mogk personal communication) in the presence of 75 mM KCl. Although the elution profile is consistent with complex formation, it also spans an elution volume range consistent with BAP that is not bound by ClpP. In contrast, the degradation assays were carried out with 1 μM BAP (6 μM monomer) and 1.5 μM ClpP (21 μM monomer) and the KCl concentration is unclear. Even without an affinity constant for the BAP–ClpP interaction, we can still predict that there is probably less BAP–ClpP present in the degradation assay then there was in the size exclusion experiment.
We currently have no way to predict how much BAP–ClpP is present in solution to perform catalysis or the relative amounts of 1:1 (one BAP hexamer per ClpP tetradecamer) or 2:1 (two BAP hexamers per ClpP tetradecamer) complexes. This is important because a variety of protein concentrations and salt concentrations have been reported in studies using BAP. Thus it is unclear how much of each active form is present to catalyse the reaction in those experiments. In the present study, we report two conditions: 10 μM BAP monomer and 3 μM ClpP monomer and 6 μM BAP monomer and 21 μM ClpP monomer. In both cases, we see degradation when nt is left out. Thus, under these conditions, we conclude that proteolysis is also the result of an ATP-independent function leading us to conclude that this protein engineering method cannot be used to conclude energy driven translocation.
In the present study, we have shown that using rapid mixing kinetic techniques ClpB is most likely to take only one or two translocation steps before dissociation from a polypeptide substrate. Our observations are more consistent with the model that the Bukau and colleagues [56,57] previously proposed. In that work, they showed that ClpB binds protein aggregates and catalyses exposure of hydrophopic regions and this allows DnaKJE to bind and mediate dissociation and refolding of solubilized polypeptides into native proteins. In that model, disaggregation is accomplished through a mechanism that does not require complete translocation of a polypeptide chain.
Our observations have led us to the hypothesis that ClpB is a non-processive translocase, where a non-processive translocase is an enzyme that takes only one or a few steps before dissociating. This definition leads to the prediction that if ClpB is taking only one or two steps before dissociation then, as we observed, the dissociation kinetics would depend upon [ATP]. Since the signal in those experiments is coming from the polypeptide substrate, the observation that the reaction depends upon ATP indicates that the ATP dependent reactions occur before dissociation of the enzyme from the polypeptide.
Analysis of the stopped-flow time courses collected as a function of [ATP] has led us to propose the minimal mechanism given by Scheme 1 in Figure 4(E). However, since ClpB contains two ATP-binding and hydrolysis sites per monomer, it is entirely possible that the two kinetic steps we observe could be coupled to D1 and D2, respectively. Consequently, the dissociation kinetics remains to be examined using variants of ClpB that are ATP hydrolysis deficient at D1 and D2. Nevertheless, it is clear that ATP binding followed by a conformational change or hydrolysis occurs before ClpB dissociates from the polypeptide substrate [50,58]. This indicates that an ATP-driven process occurs before ClpB dissociates from the polypeptide substrate. This ATP-driven process could be a translocation step, ATP-driven motion of the M-domain or both. Regardless of the exact details of the elementary steps, these data still allow us to conclude that ClpB is taking, at most, one or two steps before dissociation, an observation that is not consistent with processive translocation of substrate through ClpB's axial channel. Therefore, the results presented here lead to the conclusion that ClpB is a non-processive translocase and probably only tugs on exposed tails and loops in protein aggregates.
Pulling on exposed loops has been previously proposed based on the observation that an unfolded luciferase molecule is proteolytically degraded by BAP–ClpP when it was flanked by folded YFP and CFP that were not degraded [59]. The observation that YFP and CFP were not unfolded in that construct indicates that they were not processively translocated by ClpB. Moreover, it was reported that the folded YFP was not unfolded during ClpB–KJE catalysed protein disaggregation in aggregates containing unfolded luciferase fused with folded YFP. This indicates that complete translocation of a substrate was not necessary for disaggregation.
A non-processive mechanism is consistent with the observation that hexamers of ClpB exhibit fast subunit exchange [53]. It has been reported that monomers of ClpB rapidly dissociate from the hexamer. The rapid disassembly of hexamers may play a role in the propensity of ClpB to dissociate before multiple translocation steps can occur.
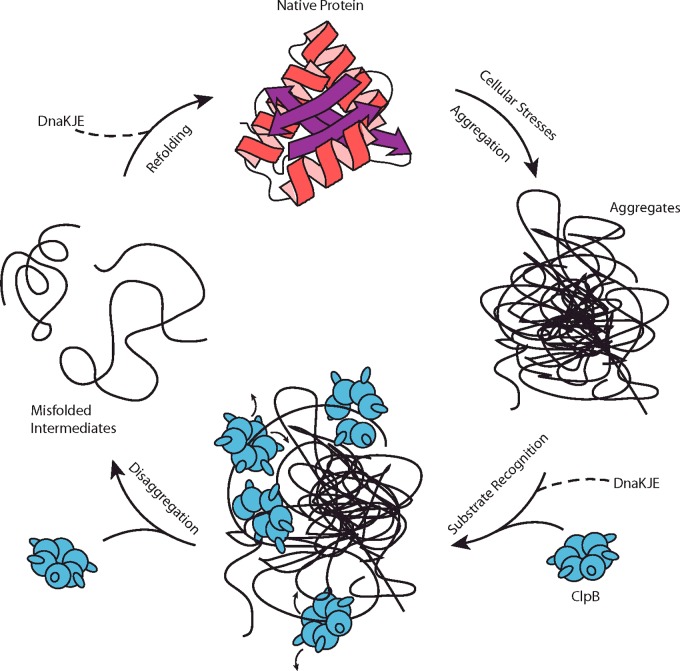
In summary, we propose the mechanistic model for ClpB catalysed protein disaggregation shown in Figure 5. When cells are under stress, native proteins can become misfolded or aggregated. With or without the assistance of the DnaK chaperone system, multiple ClpB hexamers will recognize and bind to the exposed unstructured regions of the protein aggregates [53]. We have recently reported that multiple ClpB hexamers can bind to a polypeptide chain longer than 127 amino acids [25]. Once bound, ClpB can mechanically pull on exposed tails or even loops [59] by taking one or two translocation steps to disrupt and loosen the structure of aggregates. ClpB then dissociates, probably as a consequence of rapid dissociation of the hexameric structure, followed by rebinding. The concerted effort of multiple ClpB hexamers binding to different parts of the aggregate tugging and releasing probably serves to fully resolve the protein aggregate. The unfolded or misfolded intermediates released from ClpB can refold back to the native state either spontaneously or with the assistance of the DnaKJE chaperone system as previously reported [56,57].
Figure 5. Schematic mechanism for ClpB catalysed protein disaggregation.
A native protein becomes unfolded or misfolded due to cellular stresses. In the crowded environment of the cell, such (partially) denatured proteins form aggregates. With or without DnaKJE, multiple ClpB hexamers can bind to exposed loops and tails of the aggregates. ClpB hexamers collectively disrupt the aggregate by the combined actions of individual ClpB hexamers tugging and pulling in one or two steps before dissociating. Released from the aggregate, the unfolded intermediates can refold to their native conformations spontaneously or with the assistance of DnaKJE.
Acknowledgments
We would like to thank James Shorter and Charles Lu for comments on this manuscript. We would also like to thank to J. Woody Robins for use of the fermenter core facility. The content discussed in the present study is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Biomedical Imaging and Bioengineering or the National Institutes of Health.
Abbreviations
- AAA
ATPase associated with various cellular activities
- AAA+
expanded superfamily of AAA
- ADEP
acyldepsipeptide
- ATPγS
adenosine 5′-[γ-thio]triphosphate
- BAP
ClpB–ClpA–P-loop
- Clp
caseinolytic protease
- D1
ATPase domain 1
- D2
ATPase domain 2
- Hsl
heat-shock locus
- Hsp
heat shock protein
- KJE
DnaK, DnaJ and GrpE
- M-domain
middle domain
- NLLS
non-linear least squares
- nt
nucleotide
- SsrA
small stable RNA A
- TBAP
BAP mutant of ClpB from T. thermophilus
- TClpP
ClpP from T. thermophilus
- TEV
Tobacco Etch Virus
AUTHOR CONTRIBUTION
Tao Li, Clarissa Weaver and Jiabei Lin performed experiments included in the manuscript and/or the supplemental data section. Elizabeth Duran constructed and isolated the ClpP L139C variant. Justin Miller isolated ClpP. Tao Li and Aaron Lucius designed experiments and wrote the manuscript.
FUNDING
This work was supported by the National Science Foundation [grant number MCB-1412624 (to A.L.L.)]; the National Institute of Biomedical Imaging and Bioengineering (NIBIB) [grant number T32EB004312 (to J.M.M.)].
References
- 1.Doyle S.M., Hoskins J.R., Wickner S. Collaboration between the ClpB AAA+ remodeling protein and the DnaK chaperone system. Proc. Natl. Acad. Sci. U.S.A. 2007;104:11138–11144. doi: 10.1073/pnas.0703980104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Motohashi K., Watanabe Y., Yohda M., Yoshida M. Heat-inactivated proteins are rescued by the DnaK.J-GrpE set and ClpB chaperones. Proc. Natl. Acad. Sci. U.S.A. 1999;96:7184–7189. doi: 10.1073/pnas.96.13.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Squires C.L., Pedersen S., Ross B.M., Squires C. ClpB is the Escherichia coli heat shock protein F84.1. J. Bacteriol. 1991;173:4254–4262. doi: 10.1128/jb.173.14.4254-4262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mogk A., Deuerling E., Vorderwulbecke S., Vierling E., Bukau B. Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation. Mol. Microbiol. 2003;50:585–595. doi: 10.1046/j.1365-2958.2003.03710.x. [DOI] [PubMed] [Google Scholar]
- 5.Mogk A., Schlieker C., Strub C., Rist W., Weibezahn J., Bukau B. Roles of individual domains and conserved motifs of the AAA+ chaperone ClpB in oligomerization, ATP hydrolysis, and chaperone activity. J. Biol. Chem. 2003;278:17615–17624. doi: 10.1074/jbc.M209686200. [DOI] [PubMed] [Google Scholar]
- 6.Ross C.A., Poirier M.A. Protein aggregation and neurodegenerative disease. Nat. Med. 2004;10(Suppl):S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 7.Wood J.D., Beaujeux T.P., Shaw P.J. Protein aggregation in motor neurone disorders. Neuropathol. Appl. Neurobiol. 2003;29:529–545. doi: 10.1046/j.0305-1846.2003.00518.x. [DOI] [PubMed] [Google Scholar]
- 8.DeSantis M.E., Leung E.H., Sweeny E.A., Jackrel M.E., Cushman-Nick M., Neuhaus-Follini A., Vashist S., Sochor M.A., Knight M.N., Shorter J. Operational plasticity enables hsp104 to disaggregate diverse amyloid and nonamyloid clients. Cell. 2012;151:778–793. doi: 10.1016/j.cell.2012.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shorter J. Hsp104: a weapon to combat diverse neurodegenerative disorders. Neurosignals. 2008;16:63–74. doi: 10.1159/000109760. [DOI] [PubMed] [Google Scholar]
- 10.Schirmer E.C., Glover J.R., Singer M.A., Lindquist S. HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem. Sci. 1996;21:289–296. doi: 10.1016/0968-0004(96)10038-4. [DOI] [PubMed] [Google Scholar]
- 11.Sauer R.T., Baker T.A. AAA+ proteases: ATP-fueled machines of protein destruction. Annu. Rev. Biochem. 2011;80:587–612. doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- 12.Baker T.A., Sauer R.T. ATP-dependent proteases of bacteria: recognition logic and operating principles. Trends Biochem. Sci. 2006;31:647–653. doi: 10.1016/j.tibs.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee S., Sowa M.E., Watanabe Y.H., Sigler P.B., Chiu W., Yoshida M., Tsai F.T. The structure of ClpB: a molecular chaperone that rescues proteins from an aggregated state. Cell. 2003;115:229–240. doi: 10.1016/S0092-8674(03)00807-9. [DOI] [PubMed] [Google Scholar]
- 14.del Castillo U., Alfonso C., Acebron S.P., Martos A., Moro F., Rivas G., Muga A. A quantitative analysis of the effect of nucleotides and the M domain on the association equilibrium of ClpB. Biochemistry. 2011;50:1991–2003. doi: 10.1021/bi101670s. [DOI] [PubMed] [Google Scholar]
- 15.Hoskins J.R., Kim S.Y., Wickner S. Substrate recognition by the ClpA chaperone component of ClpAP protease. J. Biol. Chem. 2000;275:35361–35367. doi: 10.1074/jbc.M006288200. [DOI] [PubMed] [Google Scholar]
- 16.Ortega J., Singh S.K., Ishikawa T., Maurizi M.R., Steven A.C. Visualization of substrate binding and translocation by the ATP-dependent protease, ClpXP. Mol. Cell. 2000;6:1515–1521. doi: 10.1016/S1097-2765(00)00148-9. [DOI] [PubMed] [Google Scholar]
- 17.Reid B.G., Fenton W.A., Horwich A.L., Weber-Ban E.U. ClpA mediates directional translocation of substrate proteins into the ClpP protease. Proc. Natl. Acad. Sci. U.S.A. 2001;98:3768–3772. doi: 10.1073/pnas.071043698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weibezahn J., Tessarz P., Schlieker C., Zahn R., Maglica Z., Lee S., Zentgraf H., Weber-Ban E.U., Dougan D.A., Tsai F.T., et al. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 2004;119:653–665. doi: 10.1016/j.cell.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 19.Rajendar B., Lucius A.L. Molecular mechanism of polypeptide translocation catalyzed by the Escherichia coli ClpA protein translocase. J. Mol Biol. 2010;399:665–679. doi: 10.1016/j.jmb.2010.03.061. [DOI] [PubMed] [Google Scholar]
- 20.Lohman T.M. Clipping along. J. Mol. Biol. 2010;399:663–664. doi: 10.1016/j.jmb.2010.04.063. [DOI] [PubMed] [Google Scholar]
- 21.Miller J.M., Lin J., Li T., Lucius A.L. E. coli ClpA catalyzed polypeptide translocation is allosterically controlled by the protease ClpP. J. Mol. Biol. 2013;425:2795–2812. doi: 10.1016/j.jmb.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller J.M., Lucius A.L. ATP-gamma-S competes with ATP for binding at domain 1 but not domain 2 during ClpA catalyzed polypeptide translocation. Biophys. Chem. 2014;185:58–69. doi: 10.1016/j.bpc.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shorter J., Lindquist S. Navigating the ClpB channel to solution. Nat. Struct. Mol. Biol. 2005;12:4–6. doi: 10.1038/nsmb0105-4. [DOI] [PubMed] [Google Scholar]
- 24.Brotz-Oesterhelt H., Beyer D., Kroll H.P., Endermann R., Ladel C., Schroeder W., Hinzen B., Raddatz S., Paulsen H., Henninger K., et al. Dysregulation of bacterial proteolytic machinery by a new class of antibiotics. Nat. Med. 2005;11:1082–1087. doi: 10.1038/nm1306. [DOI] [PubMed] [Google Scholar]
- 25.Li T., Lin J., Lucius A.L. Examination of polypeptide substrate specificity for Escherichia coli ClpB. Proteins. 2015;83:117–134. doi: 10.1002/prot.24710. [DOI] [PubMed] [Google Scholar]
- 26.Woo K.M., Kim K.I., Goldberg A.L., Ha D.B., Chung C.H. The heat-shock protein ClpB in Escherichia coli is a protein-activated ATPase. J Biol. Chem. 1992;267:20429–20434. [PubMed] [Google Scholar]
- 27.Veronese P.K., Stafford R.P., Lucius A.L. The Escherichia coli ClpA molecular chaperone self-assembles into tetramers. Biochemistry. 2009;48:9221–9233. doi: 10.1021/bi900935q. [DOI] [PubMed] [Google Scholar]
- 28.Li T., Lucius A.L. Examination of polypeptide substrate specificity for E. coli ClpA. Biochemistry. 2013;52:4941–4954. doi: 10.1021/bi400178q. [DOI] [PubMed] [Google Scholar]
- 29.Lucius A.L., Miller J.M., Rajendar B. Application of the sequential n-step kinetic mechanism to polypeptide translocases. Methods Enzymol. 2011;488:239–264. doi: 10.1016/B978-0-12-381268-1.00010-0. [DOI] [PubMed] [Google Scholar]
- 30.Fischer C.J., Lohman T.M. ATP-dependent translocation of proteins along single-stranded DNA: models and methods of analysis of pre-steady state kinetics. J. Mol. Biol. 2004;344:1265–1286. doi: 10.1016/j.jmb.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 31.Fischer C.J., Maluf N.K., Lohman T.M. Mechanism of ATP-dependent translocation of E.coli UvrD monomers along single-stranded DNA. J. Mol. Biol. 2004;344:1287–1309. doi: 10.1016/j.jmb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 32.Dillingham M.S., Wigley D.B., Webb M.R. Direct measurement of single-stranded DNA translocation by PcrA helicase using the fluorescent base analogue 2-aminopurine. Biochemistry. 2002;41:643–651. doi: 10.1021/bi011137k. [DOI] [PubMed] [Google Scholar]
- 33.Galletto R., Tomko E.J. Translocation of Saccharomyces cerevisiae Pif1 helicase monomers on single-stranded DNA. Nucleic Acids Res. 2013;41:4613–4627. doi: 10.1093/nar/gkt117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schulmeister U., Hochwallner H., Swoboda I., Focke-Tejkl M., Geller B., Nystrand M., Harlin A., Thalhamer J., Scheiblhofer S., Keller W., et al. Cloning, expression, and mapping of allergenic determinants of alphaS1-casein, a major cow's milk allergen. J. Immunol. 2009;182:7019–7029. doi: 10.4049/jimmunol.0712366. [DOI] [PubMed] [Google Scholar]
- 35.Doyle S.M., Shorter J., Zolkiewski M., Hoskins J.R., Lindquist S., Wickner S. Asymmetric deceleration of ClpB or Hsp104 ATPase activity unleashes protein-remodeling activity. Nat. Struct. Mol. Biol. 2007;14:114–122. doi: 10.1038/nsmb1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lucius A.L., Maluf N.K., Fischer C.J., Lohman T.M. General methods for analysis of sequential “n-step” kinetic mechanisms: application to single turnover kinetics of helicase-catalyzed DNA unwinding. Biophys. J. 2003;85:2224–2239. doi: 10.1016/S0006-3495(03)74648-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jennings L.D., Lun D.S., Medard M., Licht S. ClpP hydrolyzes a protein substrate processively in the absence of the ClpA ATPase: mechanistic studies of ATP-independent proteolysis. Biochemistry. 2008;47:11536–11546. doi: 10.1021/bi801101p. [DOI] [PubMed] [Google Scholar]
- 38.Li D.H., Chung Y.S., Gloyd M., Joseph E., Ghirlando R., Wright G.D., Cheng Y.Q., Maurizi M.R., Guarne A., Ortega J. Acyldepsipeptide antibiotics induce the formation of a structured axial channel in ClpP: A model for the ClpX/ClpA-bound state of ClpP. Chem. Biol. 2010;17:959–969. doi: 10.1016/j.chembiol.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Veronese P.K., Lucius A.L. Effect of temperature on the self-assembly of the Escherichia coli ClpA molecular chaperone. Biochemistry. 2010;49:9820–9829. doi: 10.1021/bi101136d. [DOI] [PubMed] [Google Scholar]
- 40.Maurizi M.R., Singh S.K., Thompson M.W., Kessel M., Ginsburg A. Molecular properties of ClpAP protease of Escherichia coli: ATP-dependent association of ClpA and clpP. Biochemistry. 1998;37:7778–7786. doi: 10.1021/bi973093e. [DOI] [PubMed] [Google Scholar]
- 41.Veronese P.K., Rajendar B., Lucius A.L. Activity of Escherichia coli ClpA bound by nucleoside Di- and triphosphates. J. Mol. Biol. 2011;409:333–347. doi: 10.1016/j.jmb.2011.02.018. [DOI] [PubMed] [Google Scholar]
- 42.Zolkiewski M., Kessel M., Ginsburg A., Maurizi M.R. Nucleotide-dependent oligomerization of ClpB from Escherichia coli. Protein Sci. 1999;8:1899–1903. doi: 10.1110/ps.8.9.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim K.I., Cheong G.W., Park S.C., Ha J.S., Woo K.M., Choi S.J., Chung C.H. Heptameric ring structure of the heat-shock protein ClpB, a protein-activated ATPase in Escherichia coli. J. Mol. Biol. 2000;303:655–666. doi: 10.1006/jmbi.2000.4165. [DOI] [PubMed] [Google Scholar]
- 44.Akoev V., Gogol E.P., Barnett M.E., Zolkiewski M. Nucleotide-induced switch in oligomerization of the AAA+ ATPase ClpB. Protein Sci. 2004;13:567–574. doi: 10.1110/ps.03422604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee S., Choi J.M., Tsai F.T. Visualizing the ATPase cycle in a protein disaggregating machine: structural basis for substrate binding by ClpB. Mol. Cell. 2007;25:261–271. doi: 10.1016/j.molcel.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J., Hartling J.A., Flanagan J.M. The structure of ClpP at 2.3 A resolution suggests a model for ATP-dependent proteolysis. Cell. 1997;91:447–456. doi: 10.1016/S0092-8674(00)80431-6. [DOI] [PubMed] [Google Scholar]
- 47.Effantin G., Maurizi M.R., Steven A.C. Binding of the ClpA unfoldase opens the axial gate of ClpP peptidase. J. Biol. Chem. 2010;285:14834–14840. doi: 10.1074/jbc.M109.090498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee B.G., Park E.Y., Lee K.E., Jeon H., Sung K.H., Paulsen H., Rubsamen-Schaeff H., Brotz-Oesterhelt H., Song H.K. Structures of ClpP in complex with acyldepsipeptide antibiotics reveal its activation mechanism. Nat. Struct. Mol. Biol. 2010;17:471–478. doi: 10.1038/nsmb.1787. [DOI] [PubMed] [Google Scholar]
- 49.Alexopoulos J.A., Guarne A., Ortega J. ClpP: a structurally dynamic protease regulated by AAA+ proteins. J. Struct. Biol. 2012;179:202–210. doi: 10.1016/j.jsb.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 50.Johnson K.A. 1 Transient-state kinetic analysis of enzyme reaction pathways. In: David S.S., editor. In The Enzymes. Academic Press; 1992. pp. 1–61. [Google Scholar]
- 51.Missiakas D., Schwager F., Betton J.M., Georgopoulos C., Raina S. Identification and characterization of HsIV HsIU (ClpQ ClpY) proteins involved in overall proteolysis of misfolded proteins in Escherichia coli. EMBO J. 1996;15:6899–6909. [PMC free article] [PubMed] [Google Scholar]
- 52.Neuwald A.F., Aravind L., Spouge J.L., Koonin E.V. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 53.Werbeck N.D., Schlee S., Reinstein J. Coupling and dynamics of subunits in the hexameric AAA+ chaperone ClpB. J. Mol. Biol. 2008;378:178–190. doi: 10.1016/j.jmb.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 54.Nakazaki Y., Watanabe Y.H. ClpB chaperone passively threads soluble denatured proteins through its central pore. Genes Cells. 2014;19:891–900. doi: 10.1111/gtc.12188. [DOI] [PubMed] [Google Scholar]
- 55.Watanabe Y.H., Nakazaki Y., Suno R., Yoshida M. Stability of the two wings of the coiled-coil domain of ClpB chaperone is critical for its disaggregation activity. Biochem. J. 2009;421:71–77. doi: 10.1042/BJ20082238. [DOI] [PubMed] [Google Scholar]
- 56.Goloubinoff P., Mogk A., Zvi A.P., Tomoyasu T., Bukau B. Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc. Natl. Acad. Sci. U.S.A. 1999;96:13732–13737. doi: 10.1073/pnas.96.24.13732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Diamant S., Ben-Zvi A.P., Bukau B., Goloubinoff P. Size-dependent disaggregation of stable protein aggregates by the DnaK chaperone machinery. J. Biol. Chem. 2000;275:21107–21113. doi: 10.1074/jbc.M001293200. [DOI] [PubMed] [Google Scholar]
- 58.Lucius A.L., Lohman T.M. Effects of temperature and ATP on the kinetic mechanism and kinetic step-size for E. coli RecBCD helicase-catalyzed DNA unwinding. J. Mol. Biol. 2004;339:751–771. doi: 10.1016/j.jmb.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 59.Haslberger T., Zdanowicz A., Brand I., Kirstein J., Turgay K., Mogk A., Bukau B. Protein disaggregation by the AAA+ chaperone ClpB involves partial threading of looped polypeptide segments. Nat. Struct. Mol. Biol. 2008;15:641–650. doi: 10.1038/nsmb.1425. [DOI] [PubMed] [Google Scholar]