Abstract
In HIV-1-infected patients, variation at the HLA class I locus is associated with disease progression, but few studies have assessed the influence of HLA alleles on HIV-1 CRF01_AE infection, which is dominant in Thailand. We hypothesized that alleles predicted to confer more effective immune responses, such as HLA-B*46, would protect against disease progression. HLA typing was performed on HIV-1 incident cases surviving until 1998–1999 and HIV-1-negative matched controls from Thai army cohorts enrolled between 1991 and 1995. We assessed associations between class I alleles and disease progression subsequent to HLA typing. Ninety-nine HIV-1-incident cases were followed for a median of 3.7 years after HLA typing; during this time, 58 participants died. Two alleles were associated with mortality: HLA B*51 was protective (3-year survival B*51pos vs. B*51neg: 75% vs. 52%; p = 0.034) whereas Cw*04 was deleterious (3-year survival Cw*04pos vs. Cw*04neg: 39% vs. 60%; p = 0.027). HLA-B*46 was not associated with disease progression. Alleles present at different frequencies in HIV-1-incident compared with HIV-1-negative men included HLA-A*02:03, B*35, B*15, and C*08. 1. In conclusion in this Thai army cohort, HLA-B*51 was associated with lower mortality, confirming that this allele, which is protective in clade B HIV-1 infection, has a similar effect on HIV CRF01_AE infection. The deleterious effect of HLA-Cw*04 must be interpreted with caution because it may be in linkage disequilibrium with disease-susceptible HLA-B alleles. We did not find that HLA-B*46 was protective. These findings may inform vaccine development for areas of the world in which HIV-1 CRF01_AE infection is prevalent.
Introduction
Immunologic control of HIV-1 infection is associated with effective CD8+ T cell responses against the virus. This link is supported by the fact that specific class I MHC alleles—which mediate the CD8+ T cell immune response—have been associated with lower plasma HIV-1 RNA levels. In a genome-wide association study of HIV-1 controllers, amino acids in the HLA class I binding groove were found to be the major genetic determinant of HIV-1 control.1 However, many previous studies have focused on clade B or C HIV-1 infection,2 and relatively little is known about the association between different HLA class I alleles and disease progression in patients infected with HIV-1 clades found predominantly in South and Southeast Asia, such as HIV-1 CRF01_AE. Such information may be helpful in identifying strategies to develop an effective global HIV-1 vaccine.
We hypothesized that alleles predicted to confer more effective immune responses, such as HLA-B*46, would protect against disease progression in patients infected with HIV-1 CRF01_AE. This hypothesis was based on in vitro work showing selective down-regulation of different HLA alleles by HIV-1 Nef and predicting more effective immune responses in individuals with HLA-B*46,3 which may have arisen from recombination between an HLA-B and -C allele. Because this particular allele is common in Thailand, we assessed whether HLA-B*46 had a protective effect against HIV-1 in this population.
To identify HLA alleles associated with disease progression in patients who acquire HIV-1 CRF01_AE, we studied a subset of men enrolled in a longitudinal cohort of Royal Thai army recruits between 1991 and 1995. HLA was typed on subjects surviving until 1998–1999 (samples were not available from those who died before then). In this cohort of HIV-1 survivors, we assessed associations between HLA class alleles and disease progression, CD4 cell count, and HIV-1 RNA. We also compared the frequency of HLA class I alleles in HIV-1-positive men to HIV-1-negative men enrolled in the same cohort.
Materials and Methods
Participants for this study were drawn from a cohort of Thai military recruits. Starting in 1991, a total of 21,701 HIV-1-seronegative male army recruits were followed with blood tests every 6 months until their discharge from the military, generally after serving 2 years.4–6 Because military service is compulsory, these men were believed to be representative of the population of men of that age in Thailand. During 1998–1999, the first follow-up survey among these seroconverters was performed.7 Members of the HIV-1-positive cohort who had seroconverted while in the Royal Thai Army were recontacted.
Follow-up interviews and blood tests were conducted on the surviving members (HIV-1-positive survivors) of the original group. At the time of the first follow-up, 99 had blood samples collected and available to be evaluated for HLA typing. Their blood was also tested for CD4 cell count and HIV-1 RNA. In 2005–2006, those seroconverters who were alive during the first follow-up survey in 1998–1999 were recontacted for the second follow-up survey.8 This second follow-up serves as the end date for the survival analyses. To estimate time to mortality from all causes, the primary endpoint in this analysis was death. HIV-1 seroconverters who received antiretroviral therapy (ART) were censored at the time treatment was started.
In addition to the HIV-1-incident cases, a control group of HIV-1-seronegative men at the time of their discharge from the military was matched by age and geographic district of residence to the seropositive group. Blood was obtained from these control subjects and HLA typed.
HLA typing was performed by polymerase chain reaction with sequence-specific primers (PCR-SSP)9 on blood samples obtained during the first follow-up survey in 1998–1999. Associations between HLA alleles with >10% prevalence and subsequent mortality were analyzed (Kaplan–Meier, exact log-rank tests). CD4 cell counts and HIV-1 RNA levels were measured during 1998–1999, the time of the first follow-up survey when the blood samples for HLA testing were collected. CD4 cell counts and HIV-1 RNA levels were compared between HLA groups (Wilcoxon rank-sum tests). Fisher's exact test evaluated associations between HLA alleles and HIV status. Analyses were exploratory (not adjusted for multiple comparisons).
The study was approved by institutional review boards in Thailand, the Walter Reed Army Institute of Research, and the Johns Hopkins School of Public Health.
Results
Between 1991 and 1995, 228 men became HIV-1 seropositive, and these incident cases form the basis of the cohort in this study. The date of seroconversion in these 228 men is known within a 6-month window. HLA typing by PCR-SSP was performed on 99 HIV-1-incident cases who were alive in 1998–1999; samples were not available from those who died before this time. HLA typing was also performed on 178 HIV-1-negative age- and region-matched controls from the same military cohorts (male recruits) enrolled between 1991 and 1995. Among the HIV-1-positive seroconvertors, there were no significant differences in age, region, and year of seroconversion between the 99 participants whose blood samples were available for HLA typing and those whose blood samples were not available for testing. Among the HIV-1-negative controls, those who had HLA typing performed were more likely to be from the Upper-Northern region of Thailand (86.5%) than those who did not have testing done (62.3%) (p < 0.001); age was comparable between both groups.
All infected patients had HIV CRF01-AE. None received ART prior to 2001. Of the 99 HIV-1 seroconverters, 19 received antiretroviral therapy after 2001 and were censored at the time treatment was started.
At the time of HLA typing, the median age (Q1, Q3) of the HIV-1-positive group was 28 years (28, 29), the median CD4 cell count was 322 (137, 413) cells/mm3, and the median HIV-1 RNA was 4.7 (4.2, 5.1) log10 copies/ml.
The HLA-A, B, and C allele frequencies are summarized in Table 1. For HLA-B, four-digit allele typing was not examined because three patients had nonspecific determinations (1501/25, 1502/13, 1520/25).
Table 1.
Frequency of HLA Alleles in the Study Population
| Allele | N (percentage with allele) |
|---|---|
| A*02 | 41 (41%) |
| A*0203 | 10 (10%) |
| A*0207 | 10 (10%) |
| A*03 | 1 (1%) |
| A*11 | 63 (64%) |
| A*24 | 32 (32%) |
| A*26 | 3 (3%) |
| A*31 | 5 (5%) |
| A*33 | 20 (20%) |
| A*34 | 1 (1%) |
| A*68 | 3 (3%) |
| A*74 | 2 (2%) |
| B*07 | 6 (6%) |
| B*08 | 1 (1%) |
| B*13 | 21 (21%) |
| B*15 | 30 (30%) |
| B*18 | 7 (7%) |
| B*27 | 7 (7%) |
| B*35 | 3 (3%) |
| B*38 | 5 (5%) |
| B*40 | 23 (23%) |
| B*44 | 3 (3%) |
| B*46 | 30 (30%) |
| B*48 | 3 (3%) |
| B*51 | 20 (20%) |
| B*52 | 5 (5%) |
| B*55 | 7 (7%) |
| B*56 | 3 (3%) |
| B*57 | 2 (2%) |
| B*58 | 18 (18%) |
| C*01 | 35 (35%) |
| C*03 | 38 (38%) |
| C*04 | 18 (18%) |
| C*06 | 3 (3%) |
| C*07 | 29 (29%) |
| C*08 | 24 (24%) |
| C*12 | 7 (7%) |
| C*14 | 13 (13%) |
| C*15 | 12 (12%) |
Class I HLA alleles and HIV disease progression
The 99 HIV-1-positive patients were followed for a median (Q1, Q3) of 3.7 (1.5, 6.5) years after HLA typing; during this time, 58 patients died. The most common causes of death, based on review of death certificates and medical records, were AIDS-related etiologies (74%), tuberculosis (5.2%), sepsis (5.2%), suicide (3.4%), and unspecified (3.4%) (for each of the other causes of death, only one participant was affected). We examined the association between mortality and class I MHC for those HLA alleles that were present in 10% or more of the cohort (Table 2).
Table 2.
HLA Allele Expression and Mortality
| Allele | N deaths among those expressing the allele | p-value | Effect |
|---|---|---|---|
| A*02 | 26 | 0.42 | |
| A*0203 | 7 | 0.76 | |
| A*0207 | 6 | 0.91 | |
| A*11 | 35 | 0.24 | |
| A*24 | 18 | 0.99 | |
| A*33 | 12 | 0.81 | |
| B*13 | 14 | 0.65 | |
| B*15 | 20 | 0.49 | |
| B*40 | 12 | 0.39 | |
| B*46 | 19 | 0.35 | |
| B*51 | 7 | 0.034 | Protective |
| B*58 | 13 | 0.13 | |
| C*01 | 23 | 0.24 | |
| C*03 | 26 | 0.23 | |
| C*04 | 15 | 0.027 | Deleterious |
| C*07 | 13 | 0.063 | |
| C*08 | 15 | 1.0 | |
| C*14 | 5 | 0.19 | |
| C*15 | 5 | 0.24 |
This analysis was limited to those alleles for which there was ≥10% expression in the study population. Exact log rank p-values are presented.
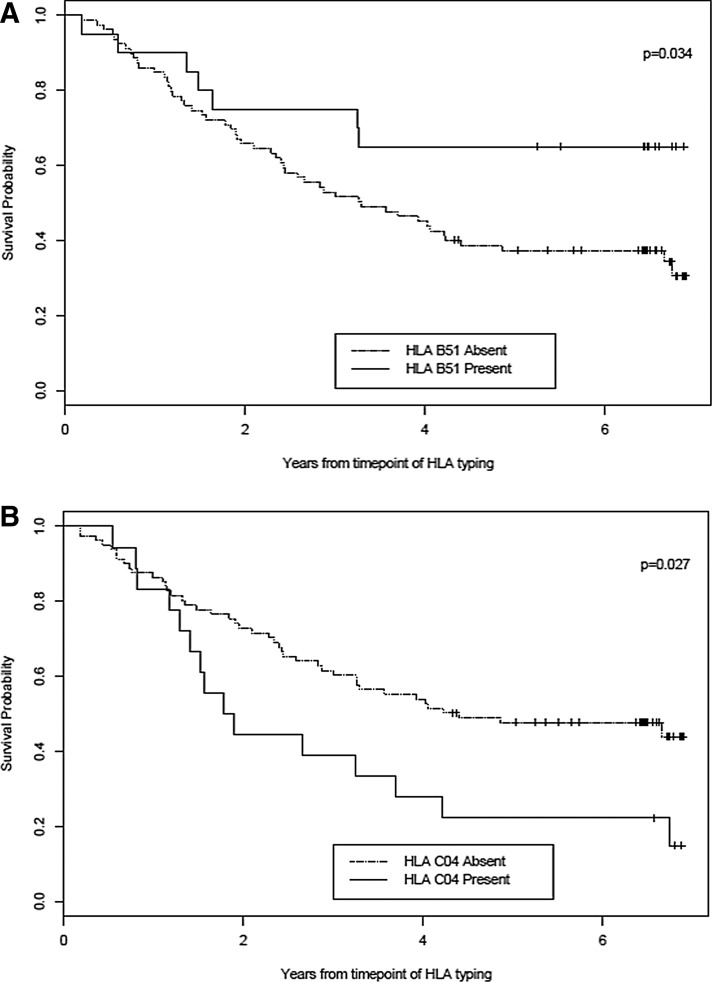
Two HLA alleles were associated with mortality: HLA-B*51 was protective and HLA-Cw*04 was deleterious (Table 2). The 3-year survival of patients who were HLA-B*51 positive was significantly greater than those who were HLA-B*51 negative (75% vs. 52%, p = 0.034) (Fig. 1A). HLA-B*51 was not associated with CD4 cell count (Table 3). By contrast, the 3-year survival of patients who were HLA- C*04 positive was significantly lower than those who were HLA-C*04 negative (39% vs. 60%; p = 0.027) (Fig. 1B). Patients who were HLA-C*04 positive had lower CD4 cell counts than those who were C*04 negative (median 167 vs. 333/mm3, p = 0.047) (Table 3). However, neither HLA-B*51 nor HLA-C*04 was associated with HIV-1 RNA level.
FIG. 1.
Kaplan–Meier curves showing the relationship between survival probability and HLA-B*51 status (A) and HLA-Cw*04 status (B).
Table 3.
HLA Allele Expression Versus CD4 Cell Counts and HIV-1 RNA Level
| Allele | N | CD4 cell count (median) | HIV-1 RNA (median, log10) |
|---|---|---|---|
| B*51 | |||
| Absent | 79 | 325 | 4.7 |
| Present | 20 | 321 | 4.4 |
| p-value | 0.77 | 0.21 | |
| C*04 | |||
| Absent | 81 | 333 | 4.6 |
| Present | 18 | 167 | 5.0 |
| p-value | 0.047 | 0.25 | |
| C*07 | |||
| Absent | 70 | 273 | 4.7 |
| Present | 29 | 358 | 4.6 |
| p-value | 0.035 | 0.72 | |
CD4 cell counts and HIV-1 RNA levels were measured during 1998–1999, the time of the first follow-up survey when the blood samples for HLA testing were collected. Wilcoxon rank-sum p-values are presented.
To determine whether the association of HLA-Cw*04 with HIV-1 disease progression might be mediated by deleterious HLA-B alleles, we assessed for potential linkage disequilibrium (LD) in the overall study population of HIV-infected survivors and HIV-uninfected controls. (There were only three HIV-positive survivors who were B*35 positive so we could not assess the impact of this HLA type on HIV progression in this study.) We observed that HLA-B*35 commonly occurred with HLA-Cw*04 (17 HLA B*35 and Cw*04 vs. three HLA-B*35 only, p < 0.0001, Fisher's exact test). Linkage disequilibrium between Cw*04 and HLA-B*35 or with other alleles may explain our findings; some HLA-B*35 alleles have been associated with rapid HIV-1 disease progression.10,11
There was a trend for an association between mortality and C*07 positivity (protective, p = 0.063) (Table 2); this allele was also associated with higher CD4 cell counts (C*07pos: 358 vs. C*07neg: 273/mm3, p = 0.035) (Table 3). HLA-C*07 was not associated with HIV-1 RNA level.
Although we hypothesized that HLA-B*46—an unusual allele found mainly in Thailand—would be protective against HIV disease progression, we did not see an effect of this allele on mortality or an association with CD4 cell count or HIV-1 RNA level.
Class I HLA alleles in HIV-1-infected men and HIV-1-negative controls
To assess whether particular HLA alleles were overrepresented in participants who did not acquire HIV-1 infection, we examined the frequency of HLA alleles in HIV-1-infected men vs. HIV-1 negative controls. The control group consisted of 178 HIV-1-seronegative men at the time of their discharge from the military who were matched by age and geographic district of their residence to the seropositive group at the time that the original cohort was defined in 1991–1995. HLA frequencies in the HIV-1-negative group were similar to that of the general Thai population (the latter available on the HLA Allele Frequency Net12). HIV-1-infected men were less likely than HIV-1-negative controls to express HLA-A*02:03 (proportion with allele, 0.10 vs. 0.21, p = 0.02, Fisher's exact test) and HLA-B*35 (0.03 vs. 0.10, p = 0.052). HIV-1-positive men were more likely than HIV-negative controls to express HLA-B*15 (0.30 vs. 0.19, p = 0.036) and C*08 (0.24 vs. 0.10, p = 0.003).
Discussion
In this Thai army cohort of men infected with HIV-1 CRF01_AE, we found that two HLA class I alleles were associated with mortality: HLA-B*51 positivity was associated with lower mortality whereas HLA-Cw*04 was associated with higher mortality. We did not find evidence for a protective effect of HLA-B*46, which we had hypothesized might be associated with more effective immune responses against the virus.
The finding in our study that HLA-B*51 positivity was associated with lower mortality confirms that this allele, which has been previously reported to be protective in clade B HIV-1 infection, has a similar effect on HIV-1 CRF01_AE infection. Among genes that have been associated with HIV-1 disease progression, HLA-B alleles play a dominant role; for example, HLA-B*57:01 is associated with HIV control whereas other HLA-B alleles are associated with disease susceptibility (reviewed by Goulder et al.13). Importantly, HLA-B*51:01 has been associated with both disease protection14–16 and disease susceptibility, depending on the HIV clade involved and when during an epidemic the association was assessed. In a cohort of patients infected with clade B HIV-1 through contaminated blood, Zhang et al. found that control of HIV-1 levels and higher CD4 cell counts were associated with T cell responses to unmutated HLA-B*51-restricted epitopes.17 In an important study of the impact of HLA type on HIV-1 evolution, however, Kawashima et al. found evidence for accumulation over time of viral mutations that led to escape from HLA-B*51-restricted CD8+ cytotoxic T lymphocytes.18 This adaption of the virus to HLA type at the population level may explain why HLA-B*51 appeared to be protective early in the epidemic but not protective later on.
In our study population, the participants were infected with HIV-1 CRF01_AE between 1991 and 1995, which was relatively early in the epidemic. Future research should examine whether the accumulation of escape mutations over time ameliorates the protective effect of this allele. Perhaps the relatively similar HIV-1 RNA levels between the HLA-B*51-positive and HLA-B*51-negative participants reflect the development of escape mutations, but viral sequencing studies within infected participants are needed to determine if this is the explanation for the finding. In addition, studies of the presence or absence of key B*51 epitopes in the CRF01_AE consensus sequence may be informative.19
The deleterious effect of HLA-Cw*04 must be interpreted with caution because this allele may be in linkage disequilibrium with disease-susceptible HLA-B alleles. Indeed, an early study in five U.S. cohorts, in which patients were presumably infected with clade B HIV-1 infection, concluded that the HLA-B*35/Cw*04 haplotype was associated with rapid disease progression.20 Subsequent work, however, revealed that the association with HLA-Cw*04 was due to linkage disequilibrium between this allele and HLA-B*35x alleles, and that it was, in fact, the latter that were linked to disease susceptibility.10 In our study population, we found evidence that HLA-Cw*04 is in linkage disequilibrium with HLA-B*35, suggesting the possibility that the effect may be related to disease-susceptible B*35 alleles or perhaps other deleterious alleles. Larger studies are needed to assess this possibility, and these investigations may reveal other genes associated with disease progression.
We did not confirm our prediction that HLA-B*46 would be associated with slower progression to AIDS.3 HLA-B*46, because it arose from a recombination between HLA-Cw*1 and HLA-B*15, has a cytoplasmic tail that matches HLA-B alleles and, therefore, should be down-regulated by HIV-1. Because HLA-B*46, by virtue of its key HLA-Cw01 sequences, should inhibit NK cells expressing NK inhibitory receptor 2 (NKIR-2) from killing HIV-infected cells, down-regulation of HLA-B*46 would expose the infected cells to attack by NK cells unless the infected person also expressed other HLA-C alleles that inhibit NKIR-2. Definitively testing the original hypothesis, however, would require an analysis of whether persons who are HLA-B46 positive and negative for particular HLA-C alleles—such as HLA-Cw*1, 3, 7, and 8—that are recognized by NKIR-2 have slower HIV-1 disease progression (there were no participants in our study who were HLA-B*46 positive but negative for all of those HLA-C alleles). Our sample size was not large enough to test his hypothesis definitively and larger studies are necessary to exclude an association. Indeed, more recent studies have suggested that HLA-B*46, which is a common allele in Asians,19,21 is actually associated with increased susceptibility to HIV-1 infection.22 Examining associations between HLA-B*46 and specific HLA-C alleles with disease acquisition and progression in these larger studies may provide additional insights.
How do our results compare with other studies of HLA class I alleles and disease progression in HIV-1 CRF01_AE infection? In a recently published study by Mori et al. of 557 Thais with HIV-1 CRF01_AE infection in a hospital-based cohort, HLA-B*3505 was found to be a protective allele23; this allele is one of the B*35 PY alleles, which are not associated with rapid HIV progression, and may have a unique peptide binding groove that might enhance HIV control. We were not able to examine this allele in our study because B*35 positivity was rare and our sample size was not large enough to assess its contribution to disease progression. In the study by Mori et al.23 HLA-B*51 was not associated with HIV RNA level. Similarly, in our study, HLA-B*51 was not associated with HIV-1 RNA but was associated with mortality. Future work should examine whether HLA-B*51 is associated with mortality in larger cohorts, although these studies will be difficult now that effective antiretroviral therapy is in widespread use.
We also found that several class I HLA alleles were overrepresented or underrepresented among HIV-1-infected men as compared with age- and region-matched HIV-1-negative controls, although these associations may be affected by survivor bias. HLA frequencies in the HIV-1-negative controls were similar to the general Thai population but an apparent association between HIV-1 acquisition and HLA type might be confounded by underrepresentation in the survivors of alleles associated with rapid disease progression, such as some of the deleterious HLA-B*35 alleles; indeed, HIV-1-infected survivors were less likely than HIV-1-negative controls to express HLA-B*35.
Our study has several limitations. First, the small sample size makes it impossible to evaluate the effect of rare HLA types on HIV disease progression; in fact, we assessed only alleles with a gene frequency of 10% or more. Second, because blood was not available on all seroconvertors, we assessed mortality from the time of HLA typing forward; if patients with particular HLA types were more likely to die or to be lost to follow-up between the time of seroconversion (1991–1995) and the time of HLA typing (1998–1999), then this survivor bias could confound our analysis. We attempted to address this possibility by focusing on survival going forward in time from the date of HLA typing in a cohort of patients all of whom had the testing at approximately the same time. Third, this exploratory analysis was not adjusted for multiple comparisons. We limited the number of comparisons by analyzing only HLA alleles with >10% prevalence, but interpretation needs to consider the potential for false-positive findings.
Another limitation of our study is that we did not have data on high-resolution HLA typing on the participants in the study. For the alleles with significant associations—and for other alleles in this study—there is a low frequency of polymorphisms within the Thai population: for example, HLA B*51 is predominantly HLA B*51:01 and HLA B*46 is commonly HLA B*46:01.24 The latter is part of a common Thai haplotype consisting of A*02:07-Cw*01-B*46:01-DRB1*09-DQB1*03:03. It is, therefore, unlikely that higher resolution typing to detect these infrequent polymorphisms with the sample size in this study would significantly confound these results, but additional studies with high-resolution typing should be pursued.
Our study also has several strengths. First, we were able to assess the impact of common HLA alleles on mortality, a clinical outcome of substantial interest. Second, we had excellent follow-up and complete mortality data on the 99 participants who underwent HLA typing. Third, as mentioned, because the study was performed at a time before widespread antiretroviral therapy was available, we were able to assess the association between HLA type and mortality. Future work on the impact of HLA alleles on HIV-1 CRF_01 acquisition and disease progression as well as studies of virus sequence evolution in response to immune pressure are needed to inform development of a global HIV-1 vaccine.
Acknowledgments
This work was supported by a cooperative agreement (W81XWH-07-2-0067) between the Henry M. Jackson Foundation for the Advancement of Military Medicine and the U.S. Department of Defense, and by the National Institutes of Health-funded Harvard University Center for AIDS Research (NIAID 5P30AI060354-08). The opinions or assertions contained herein are the private views of the authors, and are not to be construed as official, or as reflecting true views or positions of the United States Army or the Department of Defense. We would like to thank Neha Patel for her assistance in preparing this manuscript. We would like to express our appreciation for reviewer comments that strengthened the manuscript.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.International HIVCS, Pereyra F, Jia X, et al. : The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 2010;330(6010):1551–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leslie A, Matthews PC, Listgarten J, et al. : Additive contribution of HLA class I alleles in the immune control of HIV-1 infection. J Virol 2010;84(19):9879–9888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen GB, Gandhi RT, Davis DM, et al. : The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity 1999;10(6):661–671 [DOI] [PubMed] [Google Scholar]
- 4.Celentano DD, Nelson KE, Lyles CM, et al. : Decreasing incidence of HIV and sexually transmitted diseases in young Thai men: Evidence for success of the HIV/AIDS control and prevention program. AIDS 1998;12(5):F29–36 [DOI] [PubMed] [Google Scholar]
- 5.Carr JK, Sirisopana N, Torugsa K, et al. : Incidence of HIV-1 infection among young men in Thailand. J Acquir Immune Defic Syndr 1994;7(12):1270–1275 [PubMed] [Google Scholar]
- 6.Celentano DD, Nelson KE, Suprasert S, et al. : Risk factors for HIV-1 seroconversion among young men in northern Thailand. JAMA 1996;275(2):122–127 [PubMed] [Google Scholar]
- 7.Rangsin R, Chiu J, Khamboonruang C, et al. : The natural history of HIV-1 infection in young Thai men after seroconversion. J Acquir Immune Defic Syndr 2004;36(1):622–629 [DOI] [PubMed] [Google Scholar]
- 8.Rangsin R, Piyaraj P, Sirisanthana T, et al. : The natural history of HIV-1 subtype E infection in young men in Thailand with up to 14 years of follow-up. AIDS 2007;21(Suppl 6):S39–46 [DOI] [PubMed] [Google Scholar]
- 9.Bunce M, O'Neill CM, Barnardo MC, et al. : Phototyping: Comprehensive DNA typing for HLA-A, B, C, DRB1, DRB3, DRB4, DRB5 & DQB1 by PCR with 144 primer mixes utilizing sequence-specific primers (PCR-SSP). Tissue Antigens 1995;46(5):355–367 [DOI] [PubMed] [Google Scholar]
- 10.Gao X, Nelson GW, Karacki P, et al. : Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N Engl J Med 2001;344(22):1668–1675 [DOI] [PubMed] [Google Scholar]
- 11.Gao X, O'Brien TR, Welzel TM, et al. : HLA-B alleles associate consistently with HIV heterosexual transmission, viral load, and progression to AIDS, but not susceptibility to infection. AIDS 2010;24(12):1835–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzalez-Galarza FF, Takeshita LY, Santos EJ, et al. : Allele frequency net 2015 update: New features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucl Acids Res 2015;43(Database issue):D784–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goulder PJ, Brander C, Tang Y, et al. : Evolution and transmission of stable CTL escape mutations in HIV infection. Nature 2001;412(6844):334–338 [DOI] [PubMed] [Google Scholar]
- 14.Kaslow RA, Carrington M, Apple R, et al. : Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat Med 1996;2(4):405–411 [DOI] [PubMed] [Google Scholar]
- 15.O'Brien SJ, Gao X, and Carrington M: HLA and AIDS: A cautionary tale. Trends Mol Med 2001;7(9):379–381 [DOI] [PubMed] [Google Scholar]
- 16.Kawashima Y, Kuse N, Gatanaga H, et al. : Long-term control of HIV-1 in hemophiliacs carrying slow-progressing allele HLA-B*5101. J Virol 2010;84(14):7151–7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Peng Y, Yan H, et al. : Multilayered defense in HLA-B51-associated HIV viral control. J Immunol 2011;187(2):684–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawashima Y, Pfafferott K, Frater J, et al. : Adaptation of HIV-1 to human leukocyte antigen class I. Nature 2009;458(7238):641–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lazaro E, Tram LT, Bellecave P, et al. : Molecular characterization of HIV-1 CRF01_AE in Mekong Delta, Vietnam, and impact of T-cell epitope mutations on HLA recognition (ANRS 12159). PloS One 2011;6(10):e26244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carrington M, Nelson GW, Martin MP, et al. : HLA and HIV-1: Heterozygote advantage and B*35-Cw*04 disadvantage. Science 1999;283(5408):1748–1752 [DOI] [PubMed] [Google Scholar]
- 21.Prentice HA, Ehrenberg PK, Baldwin KM, et al. : HLA class I, KIR, and genome-wide SNP diversity in the RV144 Thai phase 3 HIV vaccine clinical trial. Immunogenetics 2014;66(5):299–310 [DOI] [PubMed] [Google Scholar]
- 22.Huang X, Ling H, Mao W, et al. : Association of HLA-A, B, DRB1 alleles and haplotypes with HIV-1 infection in Chongqing, China. BMC Infect Dis 2009;9:201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mori M, Wichukchinda N, Miyahara R, et al. : HLA-B*35: 05 is a protective allele with a unique structure among HIV-1 CRF01_AE-infected Thais, in whom the B*57 frequency is low. AIDS 2014;28(7):959–967 [DOI] [PubMed] [Google Scholar]
- 24.Puangpetch A, Koomdee N, Chamnanphol M, et al. : HLA-B allele and haplotype diversity among Thai patients identified by PCR-SSOP: Evidence for high risk of drug-induced hypersensitivity. Front Genet 2014;5:478. [DOI] [PMC free article] [PubMed] [Google Scholar]