To the editor:
Wnt signaling in hematopoietic cells and the bone marrow microenvironment plays a critical role in maintaining the pool of hematopoietic stem cells (HSCs) and in regulating differentiation.1,2 Wnt signaling is tightly regulated by the interplay of multiple cytoplasmic components, with Wnt activity being highest in HSCs and lower in more mature myeloid cells.1 Moreover, graded Wnt signaling has differential effects, with high activation leading to bone marrow failure and less profound activation leading to HSC expansion.1,3-5 Wnt activation has also been implicated in self-renewal of leukemia stem cells and is associated with a poorer outcome in acute myeloid leukemia (AML).6
However, our understanding of Wnt signaling in hematopoietic cells is incomplete; a holistic view is needed to understand how alterations to Wnt pathway components affect disease processes. This was most recently illustrated in a paper in Blood by Kühnl et al,7 who examined the role of the putative tumor suppressor CXXC5, a CXXC-type Zn-finger protein that interacts with disheveled (DVL) and impairs Wnt signaling in leukemia cell lines. Contrary to expectations, in AMLs, downregulation of CXXC5 expression via epigenetic silencing was associated with upregulation of cell cycling genes, coordinated with downregulation of genes implicated in leukemogenesis (eg, WT1, GATA2, KMT2A/MLL, DNMT3B, and RUNX1), and a better prognosis. The latter observation likely reflects the prevalence of Core Binding Factor AMLs in this series, which have low expression of CXXC5.
In previous studies, we and others have drawn attention to the role of loss of several genes on 5q, APC (5q22),8 and CSNK1A1 (5q32),4 encoding negative regulators of the Wnt pathway, in the pathogenesis of therapy-related myeloid neoplasms (t-MN) or high-risk myelodysplastic syndromes (MDS)/AML with a del(5q), as well as MDS with an isolated del(5q).9 The interstitial deletions of 5q are typically large, as virtually all patients have loss of 5q14-33, and confer haploinsufficient expression of many genes in the deleted interval. CXXC5, which maps within the deleted interval (5q31.2) and is downregulated in myeloid disorders with a del(5q),10 can be added to this growing list.
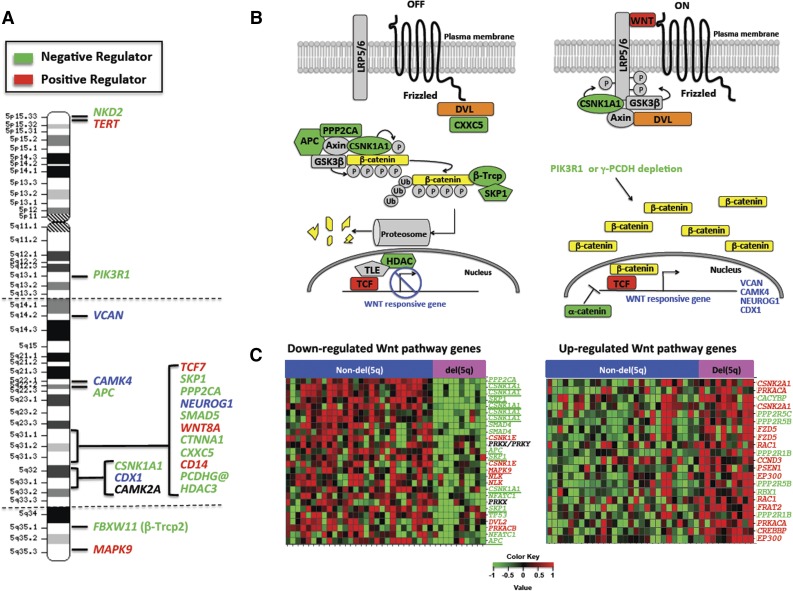
We posited that deregulation of the Wnt pathway extends beyond haploinsufficiency for these 3 genes on 5q. We used the Molecular Signature Database to investigate protein-coding genes that mapped to chromosome 5, and found that 17 genes (APC, CAMK2A, CAMK4, CD14, CDX1, CSNK1A1, FBXW11, MAPK9, NEUROG1, NKD2, PIK3R1, PPP2CA, SKP1, TCF7, TERT, VCAN, and WNT8A) are statistically enriched in the Pathway Interaction Database β-catenin nuclear pathway, Kyoto Encyclopedia of Genes and Genomes Wnt signaling pathway, and/or BIOCARTA GSK3 pathway (false discovery rate < 0.002). Moreover, CTNNA1, HDAC3, SMAD5, and the PCDHG@ cluster genes on 5q have also been implicated as regulators of the Wnt signaling pathway. All of these genes are expressed in AMLs (SRA061655), and most map within the deleted segment of 5q (Figure 1A) and either encode proteins involved in regulating the CTNNB1 destruction complex or are CTNNB1 transcriptional targets (Figure 1B).
Figure 1.
The long (q) arm of chromosome 5 is highly enriched in Wnt signaling genes. (A) The location of genes on chromosome 5 encoding proteins within the Wnt signaling pathway. Dashed horizontal lines indicate the segment typically deleted in myeloid neoplasms with a del(5q). Green and red text identifies genes encoding known negative and positive regulators of Wnt/CTNNB1 activity, respectively. Blue text identifies CTNNB1 target genes. (B) Wnt signaling pathway illustrating that del(5q) genes encode multiple negative (green) or positive (red) key regulators of Wnt signaling. CTNNB1 target genes on 5q are identified in blue text. DVL (orange) maps to 17p13.1, a region commonly lost in del(5q) t-MN. (C) Color-coded heat maps of significantly (FDR < 0.2) downregulated (green) or upregulated (red) WNT signaling pathway genes (Kyoto Encyclopedia of Genes and Genomes) in t-MN patients with a del(5q) (n = 10) vs non-del(5q) (n = 28) (GSE39991). In both groups, one-third of patients had t-MDS, and two-thirds had t-AML. TP53 deletion and/or mutations were detected in 9/10 (90%) del(5q) and 3/25 (12%) non-del(5q) patients. Abnormalities of chromosome 7 were detected in 5/10 (50%) del(5q) and 9/28 (32%) non-del(5q) patients. The non-del(5q) group included patients with +8 (2/28; 7%), KMT2A/MLL (4/28; 14%), or RUNX1 (2/28, 7%) translocations, and other complex karyotypes (8/29; 28%). Three patients had a normal karyotype. The downregulation of genes encoding negative (green) and upregulation of positive (red) Wnt pathway regulators in del(5q) patients is consistent with an active Wnt signature. Genes on chromosome 5 are underlined.
Database for Annotation, Visualization and Integrated Discovery pathway analysis of significantly deregulated probe sets (limma with false discovery rate < 0.2) between del(5q) and non-del(5q) t-MN cases (GSE39991) revealed that there is a significant downregulation of several genes encoding negative regulators (APC, CSNK1A1, SKP1, SMAD4, NFAT, PPP2CA, and TP53), and upregulation of genes encoding positive regulators (CCND1, CSNK2A1, CREBBP, EP300, FRAT2, FZD2, PRKACA, RAC1) of Wnt signaling, consistent with activation of this pathway (Figure 1C). Downregulation of several of the Wnt signaling genes (APC, CSNK1A1, PPP2CA, and SKP1) is likely a result of hemizygous deletion of 5q. In addition, DVL2 and TP53, 2 additional pathway components, were downregulated, likely because of their location on 17p13.1, commonly deleted in del(5q) patients.
The mechanism by which haploinsufficiency of 5q genes leads to clonal dominance is poorly understood. Homozygous loss of multiple negative regulators of Wnt signaling would likely lead to profound activation and apoptosis of HSCs.1 In contrast, we propose that haploinsufficient loss of multiple Wnt regulators as a result of a del(5q) may work in concert to finely tune Wnt activity, promoting expansion of HSCs, ultimately giving rise to myeloid neoplasms and contributing to clonal expansion. Adding further complexity to Wnt signaling regulation, the positive and negative regulators of Wnt signaling may have epistatic effects. Wnt signaling is also deregulated in MDS with an isolated del(5q), suggesting that additional genetic abnormalities influence the disease phenotype.11 It will be important for future studies to quantitate the level of Wnt/CTNNB1 activation in all myeloid neoplasms with a del(5q) and to validate the Wnt pathway as a candidate therapeutic target.
Authorship
Acknowledgments: This work was supported by Public Health Service, National Institutes of Health, National Cancer Institute grants CA40046 (M.M.L.B. and J.R.D) and CA190372 (M.M.L.B.), and by a grant from the Edward P. Evans Foundation (M.M.L.B.).
Contribution: A.S. analyzed data and cowrote the manuscript; J.N. performed experiments and analyzed data; S.-C.C. analyzed data; J.R.D. supervised research and analyzed data; and M.M.L.B. supervised research, analyzed data, and cowrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Angela Stoddart, Section of Hematology/Oncology, University of Chicago, 900 E. 57th St, KCBD 7th floor, Chicago, IL 60637; e-mail: astoddar@bsd.uchicago.edu.
References
- 1.Luis TC, Naber BA, Roozen PP, et al. Canonical wnt signaling regulates hematopoiesis in a dosage-dependent fashion. Cell Stem Cell. 2011;9(4):345–356. doi: 10.1016/j.stem.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 2.Malhotra S, Kincade PW. Wnt-related molecules and signaling pathway equilibrium in hematopoiesis. Cell Stem Cell. 2009;4(1):27–36. doi: 10.1016/j.stem.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qian Z, Chen L, Fernald AA, Williams BO, Le Beau MM. A critical role for Apc in hematopoietic stem and progenitor cell survival. J Exp Med. 2008;205(9):2163–2175. doi: 10.1084/jem.20080578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schneider RK, Ademà V, Heckl D, et al. Role of casein kinase 1A1 in the biology and targeted therapy of del(5q) MDS. Cancer Cell. 2014;26(4):509–520. doi: 10.1016/j.ccr.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trowbridge JJ, Xenocostas A, Moon RT, Bhatia M. Glycogen synthase kinase-3 is an in vivo regulator of hematopoietic stem cell repopulation. Nat Med. 2006;12(1):89–98. doi: 10.1038/nm1339. [DOI] [PubMed] [Google Scholar]
- 6.Ysebaert L, Chicanne G, Demur C, et al. Expression of beta-catenin by acute myeloid leukemia cells predicts enhanced clonogenic capacities and poor prognosis. Leukemia. 2006;20(7):1211–1216. doi: 10.1038/sj.leu.2404239. [DOI] [PubMed] [Google Scholar]
- 7.Kühnl A, Valk PJ, Sanders MA, et al. Downregulation of the Wnt inhibitor CXXC5 predicts a better prognosis in acute myeloid leukemia. Blood. 2015;125(19):2985–2994. doi: 10.1182/blood-2014-12-613703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stoddart A, Fernald AA, Wang J, et al. Haploinsufficiency of del(5q) genes, Egr1 and Apc, cooperate with Tp53 loss to induce acute myeloid leukemia in mice. Blood. 2014;123(7):1069–1078. doi: 10.1182/blood-2013-07-517953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith SM, Le Beau MM, Huo D, et al. Clinical-cytogenetic associations in 306 patients with therapy-related myelodysplasia and myeloid leukemia: the University of Chicago series. Blood. 2003;102(1):43–52. doi: 10.1182/blood-2002-11-3343. [DOI] [PubMed] [Google Scholar]
- 10.Treppendahl MB, Möllgård L, Hellström-Lindberg E, Cloos P, Grønbaek K. Downregulation but lack of promoter hypermethylation or somatic mutations of the potential tumor suppressor CXXC5 in MDS and AML with deletion 5q. Eur J Haematol. 2013;90(3):259–260. doi: 10.1111/ejh.12045. [DOI] [PubMed] [Google Scholar]
- 11.Boultwood J, Pellagatti A, Cattan H, et al. Gene expression profiling of CD34+ cells in patients with the 5q- syndrome. Br J Haematol. 2007;139(4):578–589. doi: 10.1111/j.1365-2141.2007.06833.x. [DOI] [PubMed] [Google Scholar]