Abstract
In vertebrate species, the innate immune system down-regulates protein translation in response to viral infection through the action of the double-stranded RNA (dsRNA)-activated protein kinase (PKR). In some teleost species another protein kinase, Z-DNA-dependent protein kinase (PKZ), plays a similar role but instead of dsRNA binding domains, PKZ has Zα domains. These domains recognize the left-handed conformer of dsDNA and dsRNA known as Z-DNA/Z-RNA. Cyprinid herpesvirus 3 infects common and koi carp, which have PKZ, and encodes the ORF112 protein that itself bears a Zα domain, a putative competitive inhibitor of PKZ. Here we present the crystal structure of ORF112-Zα in complex with an 18-bp CpG DNA repeat, at 1.5 Å. We demonstrate that the bound DNA is in the left-handed conformation and identify key interactions for the specificity of ORF112. Localization of ORF112 protein in stress granules induced in Cyprinid herpesvirus 3-infected fish cells suggests a functional behavior similar to that of Zα domains of the interferon-regulated, nucleic acid surveillance proteins ADAR1 and DAI.
Keywords: DNA viruses, interferon, protein-nucleic acid interaction, stress granule, X-ray crystallography, Zalpha domain
Introduction
The nucleic acid double-helix can adopt three main conformations termed A, B, and Z. Among these conformations, the Z-DNA or Z-RNA is the only one that has a left-handed configuration and is named for its characteristic zig-zag backbone shape (1). Because the Z conformation is energetically unfavorable under physiological conditions, for long it was not clear whether it plays a role in biological processes (2). This changed with the discovery of Zα domains, which belong to the winged helix-turn-helix fold family and bind with high specificity and affinity to both Z-DNA and Z-RNA (reviewed in Ref. 3). Until now, these domains have only been found in proteins involved in the antiviral interferon response pathway and act either as part of host foreign nucleic acid surveillance proteins or as their viral inhibitors (3).
The unique ability of Zα domains to stabilize the left-handed conformation under physiological conditions of both DNA and RNA has led to extensive structural studies aiming at understanding the underlying mechanism. Crystal structures of DNA complexes with Zα domains from the RNA editing enzyme ADAR1 (adenosine deaminase acting on RNA), the DNA sensor DAI (DNA activator of interferon regulatory factors), the Z-DNA dependent protein kinase PKZ,3 and the poxviral interferon response inhibitor E3L have revealed a highly conserved set of interactions (4–7). Previously, we solved the first structure of such a domain (Zα from ADAR1) in complex with double-stranded RNA (dsRNA), which shows very similar interactions (8), suggesting that Zα domains interact in vivo with either dsDNA, dsRNA, or their hybrids.
Despite extensive in vitro studies, little is known about the context of Zα targeting in vivo or other functional roles of the domain. Recent studies showed that Zα domains localize in stress granules (SGs): ribonucleoprotein particles formed by stalled ribosomes upon translational arrest during different forms of cellular stress (9, 10). The same work showed that nucleic acid binding is a requirement for this localization and that a Zα domain fusion can target other proteins to arsenite-induced SGs. These findings suggest that Zα domains can also bind endogenous nucleic acid and that viral infection is not the only condition under which they are functional.
Genomes of several bony fish species (Cyprinoformes and Salmoniformes such as Danio rerio, Cyprinus carpio, Carassius auratus, and Salmo salar) encode a paralogue of the RNA-dependent protein kinase PKR named PKZ (11). PKR is a well studied cytoplasmic sensor of dsRNA that, upon recognition of dsRNA by its dsRNA-binding domains, is activated by autophosphorylation (12). The activated PKR then phosphorylates the eukaryotic initiation factor 2α, leading to a shutdown of protein translation. PKZ, which instead of dsRNA-binding domains has two Zα domains (11), was shown to phosphorylate eukaryotic initiation factor 2α similarly to PKR in response to CpG DNA, but not to poly(IC), a classic dsRNA activator of PKR (13). Previously, we and others solved the crystal structures of the PKZ-Zα domain (7, 14) and clarified the details of the interaction of PKZ-Zα with Z-DNA.
Cyprinid herpesvirus 3 (CyHV-3; species Cyprinid herpesvirus 3, genus Cyprinivirus, family Alloherpesviridae, order Herpesvirales) is an emerging fish pathogen that has a major negative impact in common and koi carp populations both in the wild and in aquaculture (15). Interestingly, its large genome that consists of a dsDNA molecule of 295 kb (16) encodes some genes with close relatives in viral families such as Poxviridae and Iridoviridae but are absent in Herpesviridae. Recently, we demonstrated that CyHV-3 ORF112 encodes a Zα-like domain (17), leading us to hypothesize that this protein could represent an inhibitor of PKZ. This parallels poxviruses that use E3L, another Zα-containing protein, to block PKR and other elements of the interferon response (18). Recombinant vaccinia virus with deleted the E3L-Zα domain was shown to lose pathogenicity in mice (19), and the E3L-Zα was demonstrated to be required for the efficient blocking of the PKR pathway in mouse embryonic fibroblasts (20).
We have previously shown that the structure of the apo-protein of CyHV-3 ORF112 adopts the Zα fold, whereas our in vitro experiments suggested that ORF112 interacts with CpG DNA in the left-handed conformation (17). Here, we aimed at understanding the specificity and mechanism of binding of ORF112 by determining the crystal structure of its complex with dsDNA. We find that ORF112-Zα forms dimeric structures on DNA that have a role in stabilizing the protein·DNA complex and shows interactions that extend beyond the minimal (CG)3 binding site. Furthermore, we use this structural information to understand its relationship with PKZ and the poxvirus inhibitor of interferon response, E3L. Finally, we show evidence that ORF112 localizes in stress granules, cytoplasmic ribonucleoproteins involved in antiviral responses, as do Zα domains of ADAR1 and DAI (9).
Experimental Procedures
Cloning, Expression, and Purification
The Zα domain of ORF112 (BAF48926.1 residues 187–278) was cloned in a pET28a vector with NheI/XhoI restriction enzymes as a His tag N-terminal fusion protein. The construct was expressed in Escherichia coli strain BL21 (DE3). Cell cultures with 0.6–0.9 OD were induced with 0.7 mm IPTG. After 3 h, cells were harvested by centrifugation (5000 × g) at 4 °C. Chemical cell lysis was performed with Bugbuster (Novagen) in the presence of 1 mm PMSF, mixture of proteinase inhibitors (Complete Mini, EDTA-free; Roche), and Benzonase (Novagen) for 1 h at 4 °C. The protein extract was loaded on a HiTrap IMAC-Sepharose FF column (GE Healthcare). The column was then washed with 30 mm imidazole, and the protein was eluted using a gradient of 30–250 mm imidazole. The His tag was cleaved with 10 units of thrombin during an overnight dialysis at 4 °C against MonoS buffer A (10 mm HEPES, pH 6.9, 20 mm NaCl) supplemented with 5 mm EDTA. The cleaved protein was loaded on a Mono S 4.6/100 PE (GE Healthcare). The column was washed with a gradient of 20–120 mm NaCl. The protein was then eluted with 120–500 mm NaCl gradient, and the fraction content was evaluated by gel electrophoresis. Buffer exchange and concentration was performed with Amicon-Ultra centrifugal filters (Merck Millipore). Finally, the protein was concentrated at 35 mg/ml in 10 mm HEPES, pH 7.4, 20 mm NaCl and used in crystallization trials.
Complex Crystallization
A T(CG)9 DNA oligonucleotide was purchased from Integrated DNA Technologies and dissolved in MilliQ water. The oligonucleotides were annealed overnight in a PCR machine using a temperature gradient from 80 °C to room temperature, decreasing 1 °C every 12 min. Protein and oligonucleotide concentration estimations were based on absorbance measurements using a NanoDrop device. For crystallization a complex mix of ORF112-Zα (1.2 mm) with T(CG)9 (0.3 mm) was screened against solutions of three-dimensional structure screen (Molecular Dimensions). Initial small hexagonal crystals were obtained in 1.05 m lithium sulfate, 0.1 m HEPES, pH 7.5. The crystals were optimized, and the best quality ones were obtained in 0.9 m lithium sulfate, 0.1 m HEPES, pH 7.6. Such crystals were harvested and cryoprotected either in 20% glycerol, 20% PEG200, or Paratone-N and flash frozen in liquid nitrogen.
Data Collection, Structure Determination, and Phylogenetic Analysis
X-ray diffraction data of crystals frozen in liquid nitrogen were collected at Beamline ID29 of European Synchrotron Radiation Facility/Grenoble synchrotron at 100 K using 0.976 Å x-ray wavelength. The best quality data were obtained from crystals cryoprotected in 20% PEG200. The XDS package was used to process the data (21). The complex crystallized in the P3221 space group with unit cell dimensions and angles a = 44.82 Å, b = 44.82 Å, c = 140.08 Å, α = β = 90.00°, γ = 120.00°. Initial phases were obtained by molecular replacement using a composite model of a truncated chain (48 residues of 69) of Zα ORF112 (PDB code 4HOB) with 5 bases of a Z-DNA strand from the ADAR1-Zα DNA complex (PDB code 1QBJ). The composite model was constructed after superposition of the two structures in PyMOL. Our starting model was then refined in Phenix (22) followed by manual rebuilding in Coot (23) to a final R/Rfree of 0.18/0.21. The final model has phi-psi angles for all protein residues within the favored region of the Ramachandran plot.
The asymmetric unit of the crystals contains two Zα domains and two Z-DNA chains of 6 bases (one third of the T(CG)9). The electron density for the DNA shows continuity between asymmetric units and neighboring unit cells forming infinite helixes spanning the crystal lattice in three directions. The overhanging T is disordered and not visible in the electron density. A form of crystal disorder in which the missing terminal phosphates of the DNA backbone in the continuous helices are not aligned among them explains the disappearance of the DNA ends and leads to an apparent asymmetric unit that contains only a third of the physical DNA molecule. Similar cases have been observed in crystal structures of Z-DNA when the helical axis coincides with a crystallographic axis (7, 24, 25). In the final model, the density of two solvent exposed residues (Lys-233A and Gln-270A) was very weak, and thus these side chains were not modeled. In addition the N-terminal 36 residues (6 originating from the expression vector), as well as the last C-terminal residue, are not seen in the electron density and thus are not modeled. Arg-258B also did not show strong density but guided by the interaction of the same residue in chain A, we were able to model this residue in two alternative conformational states. The final model contains 120 waters as well as 4 sulfate ions bound to the highly charged ORF112 surfaces. Details about data collection and refinement statistics are listed in Table 1. The PISA (26) software was used to obtain information about protein-DNA and protein-protein interfaces and assemblies. Structure-guided alignments were performed in UCSF Chimera (27) and phylogenetic analysis in PhyML (28) as implemented on the publicly available server Phylogeny.fr (29). Representations of the structure and structural alignments were generated in PyMOL (30). The RMSDs from the structural alignments in Fig. 1b refer to chain A from each structure with the exception of drPKZ, in which chain B was used because chain A has a disordered region. The structure and structure factors have been deposited to the RCSB database (PDB code 4WCG).
TABLE 1.
Data collection and refinement statistics
| Data collection | |
| Space group | P3221 |
| Cell dimensions | |
| a, b, c (Å) | 44.82, 44.82, 140.08 |
| α, β, γ (°) | 90.0, 90.0, 120.0 |
| Resolution range (Å) | 46.69–1.5 (1.58–1.5)a |
| Rmergeb | 0.037 (0.68) |
| Mean I/σ(I) | 22.34 (2.96) |
| Completeness (%) | 99.76 (99.85) |
| Multiplicity | 7.12 (7.34) |
| Refinement | |
| Resolution (Å) | 46.69–1.5 (1.56–1.5) |
| Number of reflections | 26960 (2923) |
| Rworkc | 0.18 (0.24) |
| Rfree (5%) | 0.21 (0.29) |
| Number of atoms | |
| Protein | 1021 |
| DNA | 246 |
| Solvent | 140 |
| Average B-factors | |
| Protein | 29.40 |
| DNA | 24.48 |
| Solvent | 36.93 |
| RMSDs | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.05 |
a Statistics for the highest resolution shell are shown in parentheses.
b Rmerge = ΣhklΣi|Ii(hkl) − <I(hkl)>|/ΣhklΣiIi(hkl).
c Rwork/Rfree = Σ||Fobs| − |Fcalc||/Σ|Fobs|.
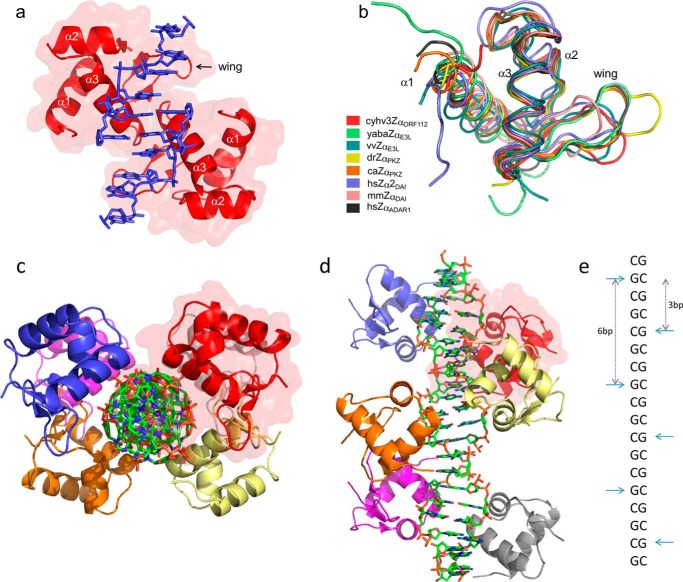
FIGURE 1.
Overall structure of Cyprinid herpesvirus 3 Zα domain in complex with Z-DNA and its fold similarity to other members of Zα family. a, representation of the asymmetric unit of CyHV-3 ORF112-Zα: two protein domains (red with semi-transparent surface) and two Z-DNA strands (in blue). b, structural alignments (Cα trace) of Zα family members: Cyprinid herpesvirus 3 ORF112-Zα (cyhv3ZαORF112, PDB code 4WCG), Yaba-like E3L-Zα (yabaZαE3L, PDB code 1SFU), vaccinia virus E3L-Zα (vvZαE3L, PDB code 1OYI), D. rerio PKZ-Zα (drZαPKZ, PDB code 4LB5), C. auratus PKZ-Zα (caZαPKZ, PDB code 4KMF), Homo sapiens DAI-Zα2 (hsZα2DAI, PDB code 3EYI), Mus musculus DAI-Zα (mmZαDAI, PDB code 1J75), and Homo sapiens ADAR1-Zα (hsZαADAR1, PDB code 1QBJ). c and d, representation of reconstructed biological assembly of T(CG)9 oligonucleotide. The view is perpendicular to (c) or along (d) the DNA axis. DNA duplex is represented as a stick model. Proteins are depicted as the ribbons; six monomers decorate T(CG)9 oligonucleotide. Red transparent surface marking one of the monomers serves as a reference. e, schematic representation of the arrangement of the ORF112-Zα domains along the Z-DNA helix. The CH-π interaction between the guanosines in the syn conformation and Tyr-257 is used as the reference and is indicated by horizontal cyan arrows. Vertical arrows indicate binding site spacing (number of base pairs) between adjacent monomers.
Cell Culture, Virus, and Treatment
Common carp brain (CCB) cells (31) were cultured in minimum essential medium (Sigma) containing 4.5 g/liter glucose (d-glucose monohydrate; Merck) and 10% FCS as described previously (32). The CyHV-3 FL strain was isolated from the kidney of a fish that died from CyHV-3 infection (32). To induce stress granule formation, CCB cells were incubated at 25 °C for 30 min in media supplemented with 1 mm sodium arsenite (Sigma) and then were washed twice with complete medium and allowed to recover for 15 min before further processing. Purification of CyHV-3 virions (American strain: accession code ABG42939.1) and mass spectrometry analyses by two-dimensional LC MS/MS were performed as described previously (33).
Immunofluorescent Staining and Confocal Microscopy
CCB cells were fixed in PBS containing 4% (w/v) paraformaldehyde for 15 min at 4 °C and then 10 min at 20 °C. After washing with PBS, samples were permeabilized in PBS containing 0.2% (w/v) Triton X-100 at 20 °C for 10 min. Immunofluorescent staining (incubation and washes) was performed in PBS containing 10% FCS (v/v). CCB cells were incubated at 37 °C for 60 min with mouse polyclonal sera raised against CyHV-3 ORF112 protein and rabbit polyclonal antibodies raised against CyHV-3 purified virions or rabbit polyclonal antibodies raised against HuR/ELAVL1 protein (Proteintech). After three washes, samples were incubated at 37 °C for 30 min with Alexa Fluor 488-conjugated goat anti-mouse IgG (H+L) (Life Technologies) and with Alexa Fluor 568-conjugated goat anti-rabbit IgG (H+L) (Life Technologies) as the secondary antibodies. After washing, cells were mounted using Prolong Gold antifade reagent with DAPI (Invitrogen). Confocal microscopy analyses were performed as described previously (34).
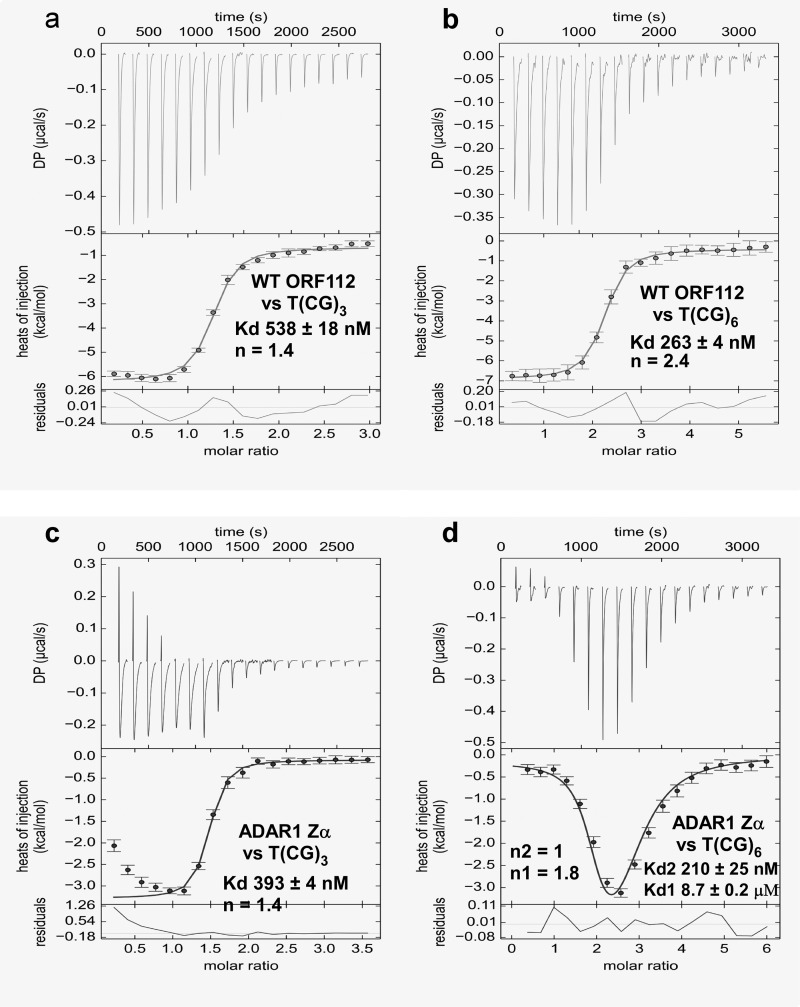
Isothermal Titration Calorimetry
Binding heat was measured on ITC200 instrument (GE Healthcare) at 25 °C and 1000 rpm. Oligonucleotides T(CG)3 and T(CG)6 were purchased from Integrated DNA Technologies and annealed. Protein and DNA storage solutions were exchanged against 10 mm HEPES, pH 6.5, 50 mm NaCl with Amicon ultracentrifugal filters (Merck Millipore). Briefly, experiments consisted of 18 injections of 2 μl of protein to oligonucleotide (concentrations used were optimized for optimal curve fit and are indicated in the corresponding figure legends). After each injection, the system was allowed to equilibrate for 3 min. Raw data were integrated using NITPIC software (35), and fitting was carried out with SEDPHAT (one-site models: Levenberg-Marquardt algorithm) (36) or in CHASM (two-site models) (37). Plots were created with GUSSI (evoked in SEDPHAT).
Results
Overview of ORF112-Zα·DNA Complex Structure
Our recently obtained crystal structure of PKZ-Zα (7) suggested that, for at least some Zα domains, the binding site might extend beyond the minimal Zα binding site of a 6-bp duplex. For ORF112-Zα, we performed several trials with different oligonucleotide lengths and were able to obtain co-crystals with a T(CG)9 dsDNA (attempts to crystallize CyHV-3 ORF112-Zα with smaller DNA fragments proved unsuccessful). The crystals of the complex belong to P3221 space group. We determined the structure of the complex at 1.5 Å using molecular replacement. The asymmetric unit consists of two Zα monomers and two strands of 6-bp DNA with the DNA forming apparent pseudocontinuous helices along the crystal lattice. Thus, we chose the bases in the asymmetric unit in a way that captures the maximal number of interactions without the need to apply crystal symmetry operators (Fig. 1a). Symmetry operations can then recreate the pseudocontinuous helix, which is spanning the entire crystal. Structural alignment shows that the two monomers in the asymmetric unit are very similar (0.257 Å RMSD). In agreement with our findings from the structure of the apo-protein (17), the ORF112-Zα represents a typical winged helix-turn-helix domain fold. The free and DNA-bound forms are very similar (0.253–0.361 Å RMSD depending on the monomer choice). Structural alignment with other members of the Zα family, despite low sequence identity, reveals extensive structural conservation, with the most prominent differences found in the wing region (Fig. 1b).
The arrangement of ORF112-Zα domains along the DNA helix was defined by taking as reference the critical and conserved CH-π interaction between Tyr-257 and guanosine 4 (G4) in syn conformation. ORF112-Zα binding sites are separated by 6 bp on the same strand. Binding to the other strand is shifted by 3 bases (Fig. 1, c–e). This arrangement was previously observed only in the tetragonal crystal form of PKZ-Zα with T(CG)6 (7), which is the only other Zα co-crystal structure with CpG repeats longer than (CG)3 and also showing a perfectly continuous DNA helix along the crystal. Importantly, in this arrangement, the two ORF112-Zα domains come within protein-protein contact distance, an interaction that involves the end of helix 3 and the wing of each monomer in a symmetric fashion that we describe in more detail below.
The ORF112-Zα DNA Binding Induces a Novel Protein-Protein Interface
The crystal structure of the prototypic Zα domain (from the RNA editing enzyme ADAR1) showed two monomers that reside on opposite sides of the DNA helix with no interactions between them and each monomer forming hydrogen bonds with a single DNA strand (4). Following structures of other Zα domains bound to (CG)3 dsDNA showed a similar organization, suggesting that Zα domains bind to each DNA strand independently. However, our recently described structure of free ORF112-Zα revealed extensive protein-protein interactions between Zα domains leading to a domain swapped dimer (17), suggesting a role for dimer formation in DNA binding.
In the protein·DNA complex, the dimer interface is mostly stabilized by van der Waals interactions and exhibits a symmetrical interaction between chains A and B (Fig. 2, a and b). The two interacting monomers are related by a noncrystallographic, 2-fold symmetry. The C-terminal part of the recognition helix α3 of monomer A tightly packs against strand S2 of the wing and helix α3 of monomer B. This interaction provides structural support for the wing and fixes its orientation. In total, 7 residues of each monomer are involved in the interaction with a contact area of 370 Å2 (∼10% of the total surface area). Two backbone hydrogen bonds are formed between the N-H of Lys-267 (chain A) with the backbone carbonyl group of Glu-261 (chain B) and its symmetric Lys-267 (chain B) with Glu-261 (chain A) (Fig. 2b). This protein-protein interface, although extensive, only forms on protein bound to DNA, because we have shown that in vitro the protein, in the absence of sulfate ions, is monomeric (17).
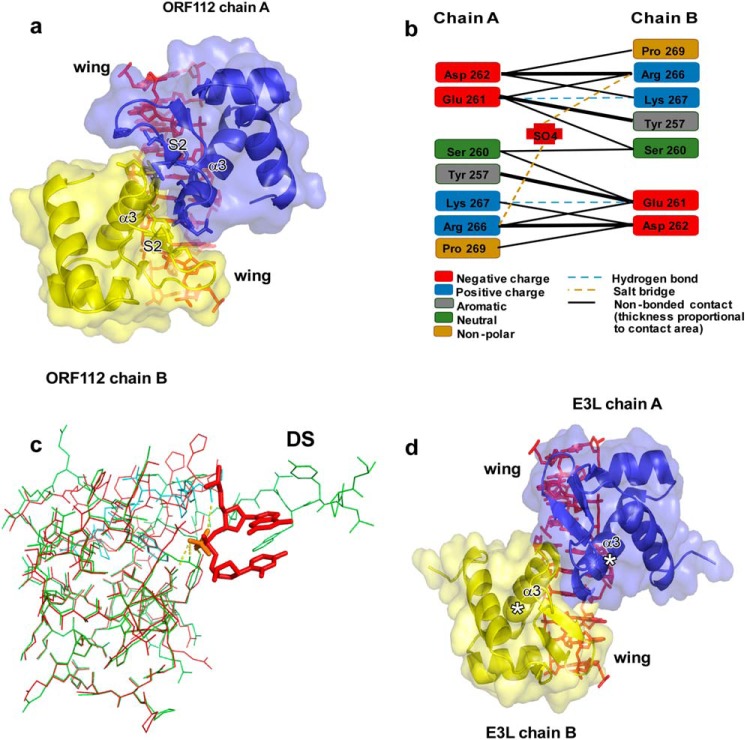
FIGURE 2.
DNA-mediated protein-protein interaction. a, representation of the protein-protein interface. CyHV-3 ORF112-Zα domain is shown as a blue and yellow cartoon with transparent surface. A 10-bp DNA was reconstructed from the asymmetric unit and is shown as a stick model. b, schematic representation of interactions between ORF112-Zα monomers. Hydrogen bonding involves the protein backbone. c, the structure of the free ORF112-Zα (green) (PDB code 4HOB) superimposed to the DNA bound form (red) with 0.33 Å RMSD. The C-terminal part of a second monomer (cyan) that is exchanged and completes the free protein structure is also shown. For clarity, only two DNA bases and their phosphate linkage from the complex structure are shown along with the sulfate ion S302 (orange) that occupies the same position as the phosphate in the free protein. In the free protein, the sulfate ion interacts with Tyr-257, Asn-253, and Arg-249, as well as with Lys-267 and the backbone of Gln-270 of the second monomer that are being exchanged. The C-terminal part of the domain that is involved in domain swapping is indicated (DS). d, putative complex formed by Yaba-like disease virus E3L-Zα monomers (modeled based on ORF112-Zα) with long Z-DNA. No clashes between backbone atoms of E3L-Zα monomers are observed.
All cellular proteins with Zα domains contain two or rarely three copies (Strongylocentrotus purpuratus ADAR1) of the domain. Studies of the PKZ-Zα suggest that efficient stabilization of the Z conformation requires binding of more than one domain (7). Only the viral proteins E3L encoded by poxviruses and the ORF112 encoded by CyHV-2 and CyHV-3 possess a single Zα domain and they may compensate for the lack of a covalently linked second domain through an on-DNA dimerization. We assessed whether Yaba-like disease virus E3L-Zα can potentially form the on-DNA dimer. After superimposing E3L-Zα monomers on the ORF112 structure, we do not observe backbone clashes between monomers (Fig. 2d); only few side chains overlap, but they could be accommodated in alternative conformations. Importantly, the C-terminal end of the Zα domain, which in the full-length E3L is physically linked to a dsRNA binding domain, does not interfere with the between-Zα interactions. This comparison suggests that E3L-Zα domains may also participate in protein-protein interaction when bound on DNA longer than the minimal (CG)3 binding site. This supports the idea that, although ORF112 and E3L contain one Zα domain, the functional unit interacting stably with DNA is a nucleic acid-dependent Zα dimer. This, on-DNA dimer formation may serve to provide additional stability to the complex and to block the Z-DNA/Z-RNA from reverting back to B or A conformation.
The Viral Zα Domains: Key Differences in the Protein-Nucleic Acid Interactions
Zα domains recognize the characteristic shape of the left-handed helix. This recognition involves sugar-phosphate backbone interactions and a unique CH-π contact formed by an absolutely conserved Tyr residue with a guanosine in syn conformation. The positioning of this critical Tyr residue is supported by an Asn and a Trp residue with all three residues forming a network of interactions that is critical for DNA binding. The residues engaged in the interactions with nucleic acids are located in helix 3 (α3) and the wing region (4). The ORF112-Zα domain follows the same rules, and the structure shows that the triad of conserved residues Tyr-257 (CH-π contact), Asn-253, and Trp-274 forms, also in this case, the core of the recognition mechanism.
Poxviruses encode E3L, a protein that contains a Zα domain and is crucial for the evasion of the host interferon responses. This protein inhibits PKR (18) and has been proposed to antagonize the DNA sensor DAI, another protein that contains Zα domains and drives the up-regulation of type I interferon genes upon activation (38). The crystal structure of the Zα domain of Yaba-like disease virus E3L and the NMR structure of vaccinia virus E3L demonstrated that these proteins interact with DNA in a very similar manner to that of ADAR1 and DAI Zα domains (39).
To understand better the relationship between E3L and ORF112, we compared the binding modes of these viral Zα domains in more detail (Fig. 3). The most striking differences between them are found in the wing region. In most Zα domains, the loop connecting the β-strands of the wing has one or two Pro residues that contribute to DNA binding through hydrophobic interactions. In addition, the wing contributes polar residues, such as Thr or Asn, that bind the DNA backbone through direct or water-mediated hydrogen bonds. The wing of ORF112-Zα is short, and the corresponding loop is oriented away from the DNA, as a result only Pro-272 (located in the γ-turn) contacts the DNA backbone (Fig. 3, a and c). In contrast, Yaba-like disease virus E3L-Zα uses four-wing residues to make contacts with DNA (Fig. 3, b and d). However, when we compare ORF112-Zα with the NMR structure of vaccinia virus E3L, determined in the absence of DNA, we find a much more similar wing region. This variation of the wing loop suggests a significant degree of flexibility and that its precise positioning may to some extent be dictated by the DNA backbone. This becomes important, because for Zα structures in complex with (CG)3, the wing contacts the edge of the DNA molecule, which often is distorted relative to the continuous helix. Moreover, in the ORF112 structure, the dimer formation contributes to the stabilization of the wing by introducing interactions with the C terminus of helix α3 of the second monomer. Thus, it is possible that other Zα domains will show changes in the wing orientation if bound to longer DNA sequences leading to a similar domain arrangement, such as the one we see for ORF112-Zα complex.
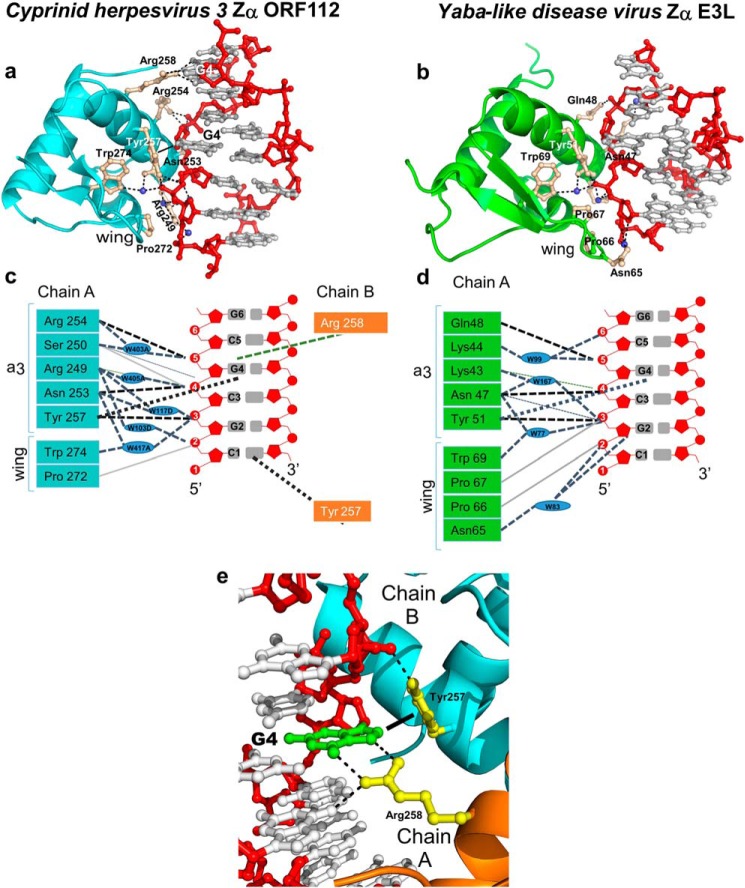
FIGURE 3.
Z-DNA binding modes of viral Zα domains from ORF112 (Cyprinid herpesvirus 3) and E3L (Yaba-like disease virus). a, binding interface between CyHV-3 ORF112-Zα and a reconstructed Z-DNA 7-bp duplex. The protein is represented as a light blue cartoon and with ball and stick key residues (wheat color) involved in the interaction with Z-DNA. Phosphate backbone and sugars are colored in red, and bases are in gray. Waters are depicted as the blue spheres. b, Z-DNA recognition by Yaba-like disease virus E3L-Zα (6 bp, PDB code 1SFU). The protein is shown as green cartoon; all other depictions are represented as in a. c and d, schematic representation of the interactions between ORF112-Zα (c) and Yaba-like disease virus E3L-Zα (d) with Z-DNA. The phosphate backbone and sugars are depicted in red, and bases are in gray. Direct hydrogen bonds are represented as black dashed lines, and water-mediated bonds shown as light blue dashed lines. Nonbonded contacts are drawn as solid light gray lines. The characteristic CH-π interaction between tyrosine and guanosine in syn conformation is shown as dotted black line. Blue ovals represent water molecules. e, close view of Arg-258A and Tyr-257B interactions with G4 (green) in syn conformation. The dotted lines indicate hydrogen bonds, whereas the filled line distinguishes the CH-π-bond of Tyr-257 to the G4 ring.
In the recognition helix α3 two absolutely conserved residues are identically positioned: Asn-253 and Tyr-257 for ORF112-Zα and Asn-47 and Tyr-51 for Yaba-like disease virus E3L-Zα. The rest of the interacting residues of α3 are distinct for these viral Zα domains. ORF112-Zα has three positively charged Arg residues (Arg-258, Arg-249, and Arg-254) interacting with the DNA. To our surprise, Arg-258 forms two direct bonds with the base of G4 on the strand opposite to the one where all other interactions occur (Fig. 3e). Such direct interaction with a DNA base has not been observed for Zα domains before and suggests a sequence specific recognition. Although this residue is not conserved among all Zα domains, Zα domains of DAI do have an arginine in equivalent position, whereas in Yaba-like disease virus E3L, the Lys-52 can play a similar role. This interaction extends beyond the minimal T(CG)3 binding site and could not be observed in previous structures. Arg-249 of chain A establishes a network of water-mediated hydrogen bonds with the phosphate backbone. Additionally, Arg-254 is among the most conserved backbone interactions and forms direct hydrogen bonds with the C5 phosphate in a similar manner as in the prototypic ADAR1-Zα. Yaba-like disease virus E3L has equivalent interactions, with Lys-43 and Gln-48 having similar roles to Arg-249 and Arg-254, whereas, as mentioned, Lys-52 could potentially form a similar base interaction as Arg-258. Finally, Ser-250, which adopts multiple conformations, concludes the DNA interacting residues of ORF112 interacting with G4 phosphate oxygen atoms. This interaction is conserved in PKZ-Zα but in equivalent position other Zα domains, including the prototypic ADAR1-Zα, present Lys residues interacting with the phosphate group one base pair away.
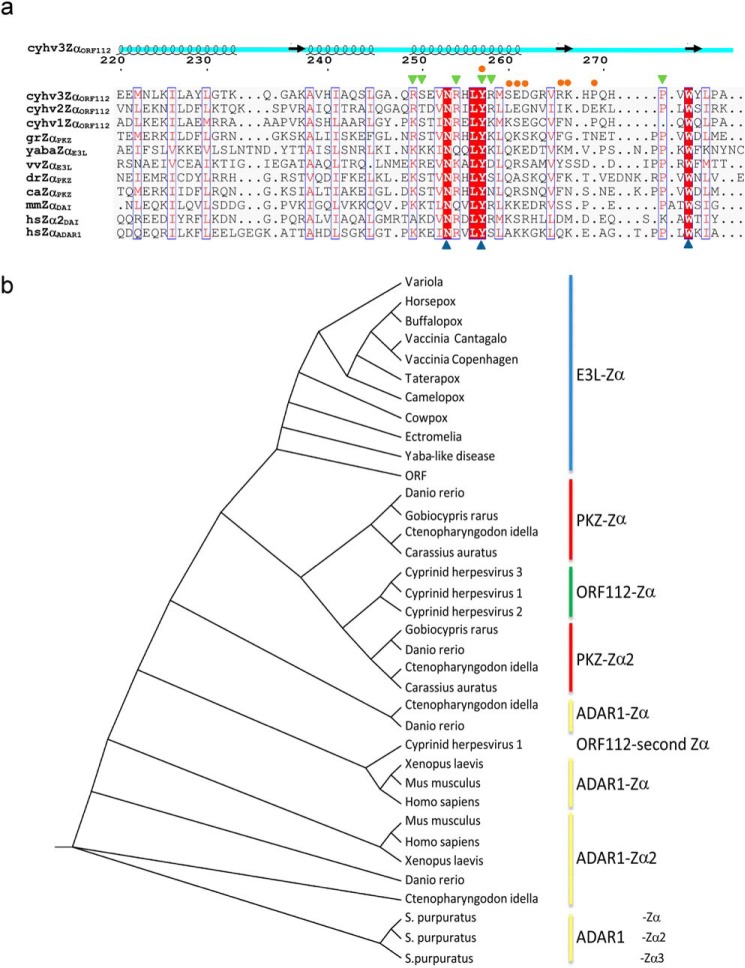
To complete our comparison, we performed structural alignments of ORF112-Zα with drPKZ-Zα, caPKZ-Zα, yabaE3L-Zα, and vvE3L-Zα, which yield 0.502, 0.581, 1.490, and 2.46 RMSD, respectively (Fig. 1b). We find that structurally, the most similar to ORF112-Zα is the zebrafish PKZ-Zα (drPKZ-Zα). Then we constructed a structure-guided sequence alignment (Fig. 4a), and we used a multiple sequence alignment of representative Zα domains as an input for phylogenetic analysis (Fig. 4b). In the resulting Zα cladogram, ORF112-Zα domains from all three cyprinid herpesviruses cluster together, and the analysis suggests that they share a common ancestor with the second Zα domain of PKZ. E3L-Zα domains form a clearly separate clade that appears to originate before the split of ADAR1 and PKZ and thus not directly linked to cyprinid herpesvirus domains. In agreement, the alignment shows that the highest sequence similarity of ORF112 is also with PKZ-Zα domains (Fig. 4a) (closest to Gobiocypris rarus PKZ Zα). Thus, the parsimonious explanation for the origin of CyHV-3 ORF112 is that this gene was independently co-opted from the host by the common ancestor of the three cyprinid herpesviruses rather than acquired by horizontal transfer from a poxvirus.
FIGURE 4.
ORF112-Zα phylogenetic analysis. a, structure-aided alignment of CyHV-3 ORF112-Zα with other Zα domains (for PDB codes see Fig. 1). Three sequences without structural information are included: Cyprinid herpesvirus 1 (cyhv1ZαORF112), Cyprinid herpesvirus 2 (cyhv2ZαORF112), and G. rarus PKZ-Zα (grZαPKZ). On the top of the alignment, the schematic representation of ORF112-Zα (cyhv3ZαORF112) secondary structure is drawn. Blue triangles below the alignment mark the triad of critical Z-DNA/Z-RNA binding residues: Asn, Tyr, and Trp. Blue boxes mark positions with conservation higher than 50%, and red shading highlights absolute conservation in this alignment. Residues involved in protein·DNA (green triangles) and protein-protein interactions (orange circles) are indicated above the alignment. b, cladogram generated based on curated (gaps removed) Muscle alignment with PhyML using Jones-Taylor-Thornton substitution model with the Shimodaira-Hasegawa approximate likelihood ratio test.
ORF112 and ADAR1 Zα Have Distinct Modes of DNA Binding
To substantiate the conclusions drawn from the structure of the complex, we designed mutants of ORF112 Zα and compared their binding affinities for dsDNA with the WT using isothermal titration calorimetry measurements. To understand these isothermal titration calorimetry experiments, one needs to consider that under the low salt conditions of our experiments, the B to Z equilibrium is highly shifted toward the B-form. Injection of Zα leads to Z-DNA binding and moves the equilibrium toward the Z-form. Thus, although the DNA concentrations are known, the concentration of the Z-form, which actually interacts with Zα, is lower. As a result, in the absence of a detailed model that includes the B to Z transition, the isothermal titration calorimetry experiments are only indicative of the relative affinities of the different proteins. The stoichiometries are systematically underestimated because of overestimation of the Z-DNA concentration.
WT ORF112-Zα binds (CG)6 with double the affinity shown for (CG)3 (Kd = 263 versus 538 nm), confirming the extension of the binding outside the (CG)3 minimal site. Interestingly, when we perform the analysis for the prototypic ADAR1 Zα under the same conditions, we find a slightly better binding than ORF112 to T(CG)3 but a completely different mode of binding against T(CG)6 interpreted as two binding sites one high (Kd 210 nm) and one low affinity (Kd 8.7 μm) (Fig. 5). These observations are consistent with the hypothesis that ORF112 forms a dimer covering the entire (CG)6 area, whereas ADAR1 may first bind the (CG)3 portion, creating a B-Z junction followed by an independent second binding event that leads to the Z conformation of the remaining half of the DNA.
FIGURE 5.
DNA binding affinities of ORF112 and ADAR1 Zα. a and b, isothermal titration calorimetry was used to determine the binding affinity of ORF112 Zα against a T(CG)3 (675 μm in the syringe versus 45 μm in the cell) (a) and a T(CG)6 (600 μm versus 20 μm) (b) double-stranded oligonucleotide. c and d, the same experiment but for ADAR1 Zα is shown in c (1200 μm versus 70 μm) and d (1200 μm versus 40 μm). A two-binding site model was used to fit the ADAR1 Zα against T(GC)6 data; for all other experiments, a one-site model was used.
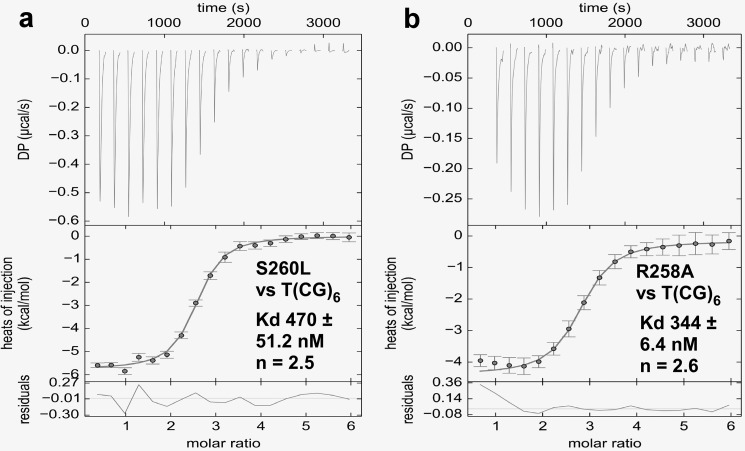
Two ORF112-Zα monomers are shown to have extensive contacts through their α3 helix and the wing region when bound to DNA, suggesting that such an interaction may contribute to the final stabilization of the complex. To test this hypothesis we mutated Ser-260, a residue located at the end of α3, which forms a symmetric contact, to either Leu or Glu. The first mutation was expected to interfere with dimer formation through steric hindrance because of the bulkiness of Leu, whereas the second mutation, in addition, should create repulsive forces between monomers. In agreement, we find that S260L mutant has a reduced affinity against T(CG)6 (Kd = 470 versus 263 nm (WT)) (Fig. 6a). As we expected, the drop in affinity was far more dramatic for S260E (below reliable determination levels); however, it is not clear whether this is only due to the inability to form the on-DNA dimer or because it also creates repulsion of the DNA backbone too. ORF112 also shows second strand interactions mediated by Arg-258. Indeed, a R258A mutant with a Kd of 344 nm for T(CG)6 shows a moderate affinity reduction in agreement with the notion that this represents a support contact (Fig. 6b).
FIGURE 6.
DNA binding affinities of ORF112 Zα mutants. a and b, isothermal titration calorimetry of S260L (1200 μm versus 40 μm) (a) and R258A (600 μm versus 20 μm) (b) mutants against T(CG)6. The results were interpreted using a one-site model.
ORF112 Localizes to Stress Granules during Cellular Stress in the Context of Viral Infection
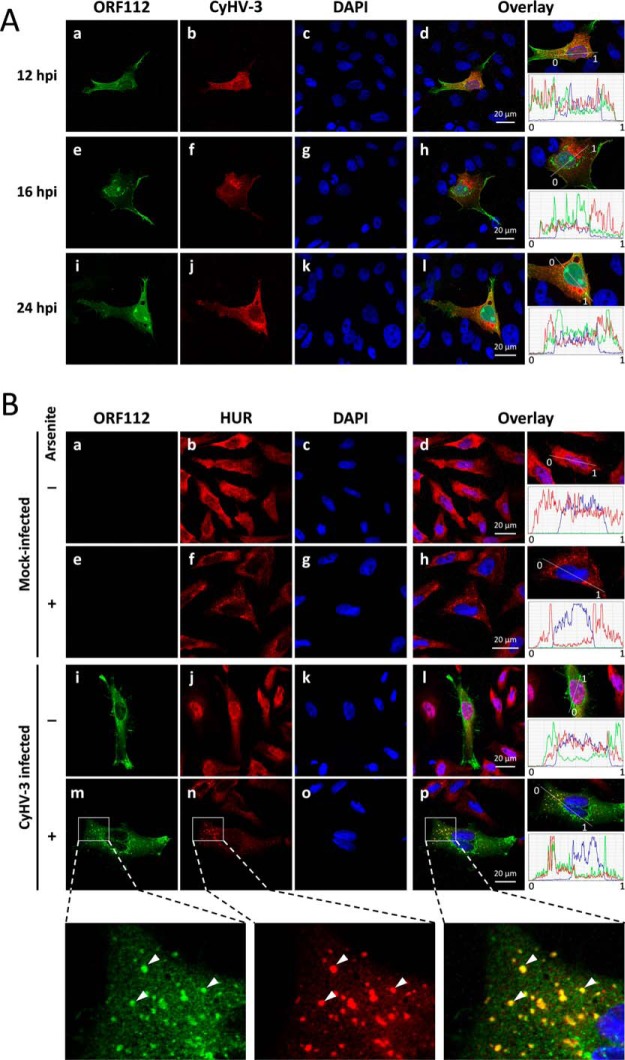
The results presented so far demonstrate that structurally and biochemically ORF112-Zα has many similarities but also distinct properties to Zα domains from mammalian ADAR1, DAI, and the poxvirus E3L. We then decided to study the behavior of the protein in cells. We investigated the expression of CyHV-3 ORF112 in the context of viral infection and characterized its subcellular localization and compared it with other Zα domains (9). First, we determined whether ORF112 is expressed as predicted by its gene structure encompassing an N-terminal repetitive sequence and a C-terminal Zα domain or as a shorter protein resulting from translation initiating at one of the internal methionine residues (Met-165, Met-189, and Met-224, American strain ABG42939.1) consisting mostly of the Zα domain. Analysis of purified CyHV-3 virions by two-dimensional LC MS/MS led to the identification of peptides, confirming that ORF112 is indeed expressed as a 280-residue-long protein (data not shown). The role of its N-terminal highly repetitive sequence, which is rich in Gln residues is unclear. Second, we investigated the kinetics of expression and the subcellular localization of ORF112 by confocal microscopy after infection of CCB cells with CyHV-3 (Fig. 7A). As early as 12 h postinfection, ORF112 was clearly detected using a specific polyclonal antibody. It localized mainly in the cytosol but also in the nucleus of infected cells. In the cytosol, most of the protein is concentrated in granular structures around the nucleus. However, the protein was also expressed throughout the cytoplasm and found associated with some regions of the plasma membrane. Within the nucleus, ORF112 was present in nucleoli and in the nucleoplasm (showing a granular structure). The granule structures detected in the cytoplasm and the nucleoplasm did not co-localize with CyHV-3 structural proteins (Fig. 7A, panels d, h, and l), indicating that they were not particle assembly sites or accumulations of virions. The ORF112 granules detected in the cytosol resembled stress granules in which Zα domains from ADAR1 and DAI have been shown to localize (9). However, immunofluorescent staining of CyHV-3-infected cells with antibodies raised against ORF112 (Fig. 7B, green signal) and the SG marker HuR (40) did not reveal co-localization or formation of SGs because of virus infection (Fig. 7B, panel l). Instead, when infected cells were treated with arsenite, SGs were indeed formed, and ORF112 was shown to localize in them (Fig. 7B, panel p). Thus, these results show that ORF112 is targeted to stress granules in relevant fish cells as has been shown in mammalian cells for the other Zα containing proteins (9).4 Thus, all available evidence supports the idea that ORF112-Zα is not only structurally but also functionally a Zα domain.
FIGURE 7.
ORF112 expression and subcellular localization during CyHV-3 infection and oxidative stress. A, CCB cells were infected with CyHV-3 at the multiplicity of infection of 0.01 plaque forming units per cell. At the indicated time postinfection, cells were treated for indirect immunofluorescent labeling of ORF112 (green signal; panels a, e, and i), CyHV-3 structural proteins (red signal; panels b, f, and j), and DNA staining (panels c, g, and k). Overlays of the three staining are shown (panels d, h, and l). The right column of panels illustrates the relative quantification of the intensities of the three fluorochromes assessed along the line indicated on the magnification of the overlay. B, CCB cells were mock infected or infected with CyHV-3 at the multiplicity of infection of 0.01 plaque forming units per cell. After an incubation of 16 h, cells were mock treated or treated with sodium arsenite (1 mm). Cells were then subjected to immunofluorescent staining of CyHV-3 ORF112 (green signal; panels a, e, i, and m), carp HuR/ELAVL1 (red signal; panels b, f, j, and n), and DNA staining (panels c, g, k, and o). Overlays of the three staining are shown (panels d, h, l, and p). The right column of panels illustrates the relative quantification of the intensities of the three fluorochromes assessed along the line indicated on the magnification of the overlay. The bottom row of panels represent magnification of a defined area of panels m, n, and p. Arrows indicate co-localizing signals. hpi, hours postinfection.
Discussion
In this study, we present the crystal structure of the Zα domain from CyHV-3 ORF112 in complex with T(CG)9 duplex DNA. The core recognition interactions between ORF112-Zα and Z-DNA are shared with Zα domains of ADAR1, DAI, PKZ, and E3L, with a triplet of conserved residues (Tyr-257, Asn-253, and Trp-274) maintaining almost identical conformations and interactions. The ORF112 of Cyprinid herpesvirus 1 and 2 conserve these residues too (Fig. 4) and likely our observations from the CyHV-3 ORF112-Zα structure also apply to these proteins. Importantly, the novel features of the ORF112 structure emerge from the fact that binding occurs on an 18-bp DNA fragment forming a continuous helix instead of the 6-bp CpG oligonucleotides present in the previous Zα structures with the exception of PKZ-Zα. Among such novel features is the fact that the wing region adopts an orientation pointing away from the DNA helix, and as a result, the only contact with the DNA is formed by Pro-272. This distinct orientation of the wing (in particular of the loop connecting β-strands S2 and S3) allows for protein-protein interactions with a second monomer bound 3 bp away on the opposite DNA strand, an interaction that can only form on a continuous helix. We propose that this on-DNA dimer formation is important for the stability of the complex for viral proteins, a proposal that is supported by our mutagenesis experiments. In the case of host proteins that all have two or more Zα domains, this stabilization can be provided by a covalently linked second domain of the same protein, as suggested by the structure of PKZ-Zα (7). An interesting exception to the presence of single Zα domains in viral proteins is CyHV-1, where ORF112 has a tandemly repeated Zα separated by a very short linker of just 2–3 amino acids. However, such short linkage makes unlikely the simultaneous association of both domains with the same DNA molecule. Structural and biochemical studies of this protein are required to clarify whether CyHV-1 has adopted a different binding mode and whether the two domains indeed form a functional unit.
Another unique feature of the ORF112 on-DNA dimer is the unique direct base contact of the Arg-258 of one monomer to the core guanine base bound by Tyr-257 of the second monomer. We found that such a base contact appears possible for other Zα domains if bound to DNA fragments longer than (CG)3. Minimal (CG)3 binding sites not only miss interactions that extend beyond the edges but also have small but perhaps significant distortions of the DNA helix toward both ends. Indeed, superposition of complexes of ORF112 with Yaba-like disease virus E3L shows for the last DNA phosphate a deviation of 0.9 Å compared with only 0.2 Å for the immediate previous base. Overall, although core recognition interactions are well represented in complexes with (CG)3, secondary interactions contributing to the stability of the complex may have been missed previously, and they may provide clues to explain differences in the affinity of different members of the family.
We have previously shown that free ORF112-Zα forms a dimer in solution in the presence of sulfate ions. The dimer of the free protein is formed through the exchange of the C-terminal 10 amino acids in a domain swapping arrangement. In solution, experiments showed an equilibrium between the monomeric protein and the domain swapped dimer, with the monomeric form being the dominant form, thus leaving unclear the significance of the observed dimer (17). The protein·DNA complex structure of ORF112-Zα confirms that the binding-competent form is the monomer, and no domain swapping is observed in the complex structure. Interestingly, the region that participates in protein-protein interactions in the complex structure marks the starting point of the domain swapping of the free protein (Fig. 2, a–c, DS). In the structure of the free ORF112-Zα, sulfate ions are located exactly at the position where DNA phosphates are found in the complex (Fig. 2c), and gel filtration experiments showed that the domain swapping is dependent on the presence of such sulfate ions (17). These observations led us to speculate that the key interaction of phosphate groups (sulfate in free ORF112-Zα) with Tyr-257 and neighboring amino acids causes destabilization/conformational flexibility of the wing region, which in the absence of actual DNA allows the observed domain swapping. Such conditional conformational flexibility suggests an induced fit mechanism for Zα domains.
Regarding the surprising presence of a characteristic poxvirus-like protein in cyprinid herpesviruses (41), the structure allows a comprehensive analysis of the similarities and differences of ORF112 and E3L-Zα, as well as that of the host PKZ. On one hand, our analysis clearly points to a close relationship between host PKZ- and viral ORF112-Zα domains. The significant similarities between ORF112- and PKZ-Zα suggest that either they have a common origin or that convergent evolution finely tunes the features of the two domains. On the other hand the relative divergence of E3L and ORF112 suggests that if the two proteins have a common origin, their split happened in a quite distant past.
The innate immune system relies on a large number of proteins that act as sensors of nucleic acids. Nucleic acids are detected based on their structure, their subcellular localization, or even their sequence (42, 43). According to these criteria, their detection can be interpreted by the innate immune system as the presence of a pathogen (pathogen-associated molecular pattern) or host cell damage (damage-associated molecular pattern). The description of Z-DNA/Z-RNA binding domains in proteins belonging to the host innate immune system and also in viral proteins involved in immune evasion mechanisms suggests that even a transient conformation of the nucleic acids could be detected and interpreted by the innate immune system as a pathogen-associated and/or damage-associated molecular pattern. ORF112 as E3L is expressed early during viral infection in agreement with a model that predicts the masking of corresponding pathogen-associated molecular patterns and the blocking of the activation of the innate immune responses as it is indicated by the absence of SG formation in infected cells. Nevertheless induction of SGs by artificial oxidative stress is enough to lead to accumulation of ORF112 to SGs, a behavior shared with other Zα domains (9). What is the identity of the nucleic acids targeted at SGs is currently unknown and clearly an important question to address.
The CyHV-3 (ORF112)-carp (PKZ) model provides a unique opportunity to study both in vitro and in vivo the roles in anti-viral innate immunity of proteins containing Zα domains. The crystal structure of CyHV-3 ORF112-Zα presented here defines critical amino acids for its interaction with Z-DNA/RNA. This knowledge is crucial for the construction of CyHV-3 mutants to be tested in vitro, but also in vivo by infection of the natural host. These experiments, which are in progress, will provide valuable information for fundamental immunology and virology of proteins containing Zα domain. These experiments could also generate important data for applied research if the constructed mutants express a safety-efficacy phenotype compatible with their use as live attenuated vaccines.
Author Contributions
A. A. conceived the project; A. A. and K. K. designed the experiments and analyzed the data; K. K. crystallized the ORF112 complex, collected data, determined the structure, and performed isothermal titration calorimetry experiments; A. V. guided the experiments using CyHV-3; K. R. and M. B. performed the CyHV-3 experiments; T. T. performed ORF112 purification; and L. G. identified the Zα localization to SGs in human cells.
Acknowledgments
We thank the personnel at Beamline ID29 of the European Synchrotron Radiation Facility (Grenoble, France) for support with data collection. We also thank Dr Lars Jansen for critical reading of the manuscript and Dr. Zbigniew Dauter for advice.
This work was supported by Fundaçâo para a Ciência e a Tecnologia Grants PTDC/BIA-PRO/112962/2009 and IF/00641/2013 (to A. A.) and SFRH/BD/51626/2011 (to K. K.). The research leading to these results received funding from European Community Seventh Framework Program Grant FP7/2007-2013 under BioStruct-X Grant Agreement 283570. The authors declare that they have no conflicts of interest with the contents of this article.
L. Gabriel and A. Athanasiadis, unpublished results.
- PKZ
- Z-DNA-dependent protein kinase
- CyHV-3
- cyprinid herpesvirus 3
- CCB
- common carp brain
- RMSD
- root mean square deviation
- dsRNA
- double-stranded RNA
- SG
- stress granule
- PDB
- Protein Data Bank.
References
- 1. Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., and Rich A. (1979) Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature 282, 680–686 [DOI] [PubMed] [Google Scholar]
- 2. Rich A., and Zhang S. (2003) Timeline: Z-DNA: the long road to biological function. Nat. Rev. Genet. 4, 566–572 [DOI] [PubMed] [Google Scholar]
- 3. Athanasiadis A. (2012) Zalpha-domains: at the intersection between RNA editing and innate immunity. Semin. Cell Dev. Biol. 23, 275–280 [DOI] [PubMed] [Google Scholar]
- 4. Schwartz T., Rould M. A., Lowenhaupt K., Herbert A., and Rich A. (1999) Crystal structure of the Zalpha domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science 284, 1841–1845 [DOI] [PubMed] [Google Scholar]
- 5. Schwartz T., Behlke J., Lowenhaupt K., Heinemann U., and Rich A. (2001) Structure of the DLM-1-Z-DNA complex reveals a conserved family of Z-DNA-binding proteins. Nat. Struct. Biol. 8, 761–765 [DOI] [PubMed] [Google Scholar]
- 6. Ha S. C., Lokanath N. K., Van Quyen D., Wu C. A., Lowenhaupt K., Rich A., Kim Y. G., and Kim K. K. (2004) A poxvirus protein forms a complex with left-handed Z-DNA: crystal structure of a Yatapoxvirus Zalpha bound to DNA. Proc. Natl. Acad. Sci. U.S.A. 101, 14367–14372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Rosa M., Zacarias S., and Athanasiadis A. (2013) Structural basis for Z-DNA binding and stabilization by the zebrafish Z-DNA dependent protein kinase PKZ. Nucleic Acids Res. 41, 9924–9933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Placido D., Brown B. A. 2nd, Lowenhaupt K., Rich A., and Athanasiadis A. (2007) A left-handed RNA double helix bound by the Z alpha domain of the RNA-editing enzyme ADAR1. Structure 15, 395–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ng S. K., Weissbach R., Ronson G. E., and Scadden A. D. (2013) Proteins that contain a functional Z-DNA-binding domain localize to cytoplasmic stress granules. Nucleic Acids Res. 41, 9786–9799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Deigendesch N., Koch-Nolte F., and Rothenburg S. (2006) ZBP1 subcellular localization and association with stress granules is controlled by its Z-DNA binding domains. Nucleic Acids Res. 34, 5007–5020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rothenburg S., Deigendesch N., Dittmar K., Koch-Nolte F., Haag F., Lowenhaupt K., and Rich A. (2005) A PKR-like eukaryotic initiation factor 2alpha kinase from zebrafish contains Z-DNA binding domains instead of dsRNA binding domains. Proc. Natl. Acad. Sci. U.S.A. 102, 1602–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cole J. L. (2007) Activation of PKR: an open and shut case? Trends Biochem. Sci. 32, 57–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bergan V., Jagus R., Lauksund S., Kileng O., and Robertsen B. (2008) The Atlantic salmon Z-DNA binding protein kinase phosphorylates translation initiation factor 2 alpha and constitutes a unique orthologue to the mammalian dsRNA-activated protein kinase R. FEBS J. 275, 184–197 [DOI] [PubMed] [Google Scholar]
- 14. Kim D., Hur J., Park K., Bae S., Shin D., Ha S. C., Hwang H. Y., Hohng S., Lee J. H., Lee S., Kim Y. G., and Kim K. K. (2014) Distinct Z-DNA binding mode of a PKR-like protein kinase containing a Z-DNA binding domain (PKZ). Nucleic Acids Res. 42, 5937–5948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rakus K., Ouyang P., Boutier M., Ronsmans M., Reschner A., Vancsok C., Jazowiecka-Rakus J., and Vanderplasschen A. (2013) Cyprinid herpesvirus 3: an interesting virus for applied and fundamental research. Vet. Res. 44, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Aoki T., Hirono I., Kurokawa K., Fukuda H., Nahary R., Eldar A., Davison A. J., Waltzek T. B., Bercovier H., and Hedrick R. P. (2007) Genome sequences of three koi herpesvirus isolates representing the expanding distribution of an emerging disease threatening koi and common carp worldwide. J. Virol. 81, 5058–5065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tomé A. R., Kuś K., Correia S., Paulo L. M., Zacarias S., de Rosa M., Figueiredo D., Parkhouse R. M., and Athanasiadis A. (2013) Crystal structure of a poxvirus-like zalpha domain from Cyprinid herpesvirus 3. J. Virol. 87, 3998–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Haig D. M., McInnes C. J., Thomson J., Wood A., Bunyan K., and Mercer A. (1998) The orf virus OV20.0L gene product is involved in interferon resistance and inhibits an interferon-inducible, double-stranded RNA-dependent kinase. Immunology 93, 335–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim Y. G., Muralinath M., Brandt T., Pearcy M., Hauns K., Lowenhaupt K., Jacobs B. L., and Rich A. (2003) A role for Z-DNA binding in vaccinia virus pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 100, 6974–6979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. White S. D., and Jacobs B. L. (2012) The amino terminus of the vaccinia virus E3 protein is necessary to inhibit the interferon response. J. Virol. 86, 5895–5904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kabsch W. (2010) XDS. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., and Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ban C., Ramakrishnan B., and Sundaralingam M. (1996) Crystalstructure of the self-complementary 5′-purine start decamer d(GCGCGCGCGC) in the Z-DNA conformation. I. Biophys. J. 71, 1215–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brennan R. G., Westhof E., and Sundaralingam M. (1986) Structure of a Z-DNA with two different backbone chain conformations. Stabilization of the decadeoxyoligonucleotide d(CGTACGTACG) by [Co(NH3)6]3+ binding to the guanine. J. Biomol. Struct. Dyn. 3, 649–665 [DOI] [PubMed] [Google Scholar]
- 26. Krissinel E., and Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 27. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 28. Guindon S., and Gascuel O. (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 [DOI] [PubMed] [Google Scholar]
- 29. Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J. F., Guindon S., Lefort V., Lescot M., Claverie J. M., and Gascuel O. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. DeLano W. L. (2002) PyMOL Molecular Graphics System, DeLano Scientific LLC, Palo Alto, CA [Google Scholar]
- 31. Neukirch M., Böttcher K., and Bunnajirakul S. (1999) Isolation of a virus from koi with altered gills. Bull. Eur. Assoc. Fish Pathol. 19, 221–224 [Google Scholar]
- 32. Costes B., Fournier G., Michel B., Delforge C., Raj V. S., Dewals B., Gillet L., Drion P., Body A., Schynts F., Lieffrig F., and Vanderplasschen A. (2008) Cloning of the koi herpesvirus genome as an infectious bacterial artificial chromosome demonstrates that disruption of the thymidine kinase locus induces partial attenuation in Cyprinus carpio koi. J. Virol. 82, 4955–4964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Michel B., Leroy B., Stalin Raj V., Lieffrig F., Mast J., Wattiez R., Vanderplasschen A. F., and Costes B. (2010) The genome of Cyprinid herpesvirus 3 encodes 40 proteins incorporated in mature virions. J. Gen. Virol. 91, 452–462 [DOI] [PubMed] [Google Scholar]
- 34. Stelter P., Huber F. M., Kunze R., Flemming D., Hoelz A., and Hurt E. (2015) Coordinated ribosomal L4 protein assembly into the pre-ribosome is regulated by its eukaryote-specific extension. Mol. Cell 58, 854–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Keller S., Vargas C., Zhao H., Piszczek G., Brautigam C. A., and Schuck P. (2012) High-precision isothermal titration calorimetry with automated peak-shape analysis. Anal. Chem. 84, 5066–5073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Houtman J. C., Brown P. H., Bowden B., Yamaguchi H., Appella E., Samelson L. E., and Schuck P. (2007) Studying multisite binary and ternary protein interactions by global analysis of isothermal titration calorimetry data in SEDPHAT: application to adaptor protein complexes in cell signaling. Protein Sci. 16, 30–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Le V. H., Buscaglia R., Chaires J. B., and Lewis E. A. (2013) Modeling complex equilibria in isothermal titration calorimetry experiments: thermodynamic parameters estimation for a three-binding-site model. Anal. Biochem. 434, 233–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Takaoka A., Wang Z., Choi M. K., Yanai H., Negishi H., Ban T., Lu Y., Miyagishi M., Kodama T., Honda K., Ohba Y., and Taniguchi T. (2007) DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448, 501–505 [DOI] [PubMed] [Google Scholar]
- 39. Kahmann J. D., Wecking D. A., Putter V., Lowenhaupt K., Kim Y. G., Schmieder P., Oschkinat H., Rich A., and Schade M. (2004) The solution structure of the N-terminal domain of E3L shows a tyrosine conformation that may explain its reduced affinity to Z-DNA in vitro. Proc. Natl. Acad. Sci. U.S.A. 101, 2712–2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kedersha N., and Anderson P. (2002) Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30, 963–969 [DOI] [PubMed] [Google Scholar]
- 41. Davison A. J., Kurobe T., Gatherer D., Cunningham C., Korf I., Fukuda H., Hedrick R. P., and Waltzek T. B. (2013) Comparative genomics of carp herpesviruses. J. Virol. 87, 2908–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gürtler C., and Bowie A. G. (2013) Innate immune detection of microbial nucleic acids. Trends Microbiol. 21, 413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Smith S., and Jefferies C. (2014) Role of DNA/RNA sensors and contribution to autoimmunity. Cytokine Growth Factor Rev. 25, 745–757 [DOI] [PubMed] [Google Scholar]