Abstract
The heterotetrameric (ϵ-β4-μ4-σ4) complex adaptor protein 4 (AP-4) is a component of a non-clathrin coat involved in protein sorting at the trans-Golgi network (TGN). Considerable interest in this complex has arisen from the recent discovery that mutations in each of its four subunits are the cause of a congenital intellectual disability and movement disorder in humans. Despite its physiological importance, the structure and function of this coat remain poorly understood. To investigate the assembly of the AP-4 coat, we dissected the determinants of interaction of AP-4 with its only known accessory protein, the ENTH/VHS-domain-containing protein tepsin. Using a variety of protein interaction assays, we found that tepsin comprises two phylogenetically conserved peptide motifs, [GS]LFXG[ML]X[LV] and S[AV]F[SA]FLN, within its C-terminal unstructured region, which interact with the C-terminal ear (or appendage) domains of the β4 and ϵ subunits of AP-4, respectively. Structure-based mutational analyses mapped the binding site for the [GS]LFXG[ML]X[LV] motif to a conserved, hydrophobic surface on the β4-ear platform fold. Both peptide-ear interactions are required for efficient association of tepsin with AP-4, and for recruitment of tepsin to the TGN. The bivalency of the interactions increases the avidity of tepsin for AP-4 and may enable cross-linking of multiple AP-4 heterotetramers, thus contributing to the assembly of the AP-4 coat. In addition to revealing critical aspects of this coat, our findings extend the paradigm of peptide-ear interactions, previously established for clathrin-AP-1/AP-2 coats, to a non-clathrin coat.
Keywords: adaptor protein, clathrin, intracellular trafficking, membrane trafficking, neurodevelopment, peptide interaction, protein complex, protein motif, protein sorting, protein-protein interaction
Introduction
Shuttling of cargo between membrane-bound compartments of the endomembrane system of eukaryotic cells is carried out by transport vesicles that bud from a donor compartment and fuse with an acceptor compartment (1). Cargo selection and vesicle budding are both mediated by protein coats that assemble on the cytosolic aspect of the donor compartment from components recruited from the cytosol. To date, about ten different protein coats have been described that participate in vesicular transport at various stages of the endomembrane system. Most of them consist of an outer “scaffold” layer and an inner “adaptor” layer, although in some cases both layers are tightly intertwined. Among the best-characterized protein coats are those containing clathrin as a scaffold and the AP-12 and AP-2 complexes as adaptors (2, 3, 4, 5). AP-1 and AP-2 are heterotetramers composed of homologous γ-β1-μ1-σ1 and α-β2-μ2-σ2 subunits, respectively (Fig. 1A). The large γ and β1 subunits of AP-1, and α and β2 subunits of AP-2, have a modular organization consisting of a folded N-terminal “trunk” domain, a largely unstructured “hinge” segment, and a folded C-terminal “ear” or “appendage” domain. The trunk domains of the large subunits, plus the whole μ and σ subunits, constitute a “core” that mediates membrane recruitment through interaction with specific phosphoinositides and/or Arf-family GTPases, as well as cargo selection through recognition of sorting signals in the cytosolic tails of transmembrane proteins. The hinge segments extend from the core and interact with the clathrin scaffold via LLDLD-type “clathrin-box” motifs (6). Finally, the ear domains interact with a variety of accessory proteins, some of which regulate coat assembly and interactions with the cytoskeleton, while others function as additional adaptors.
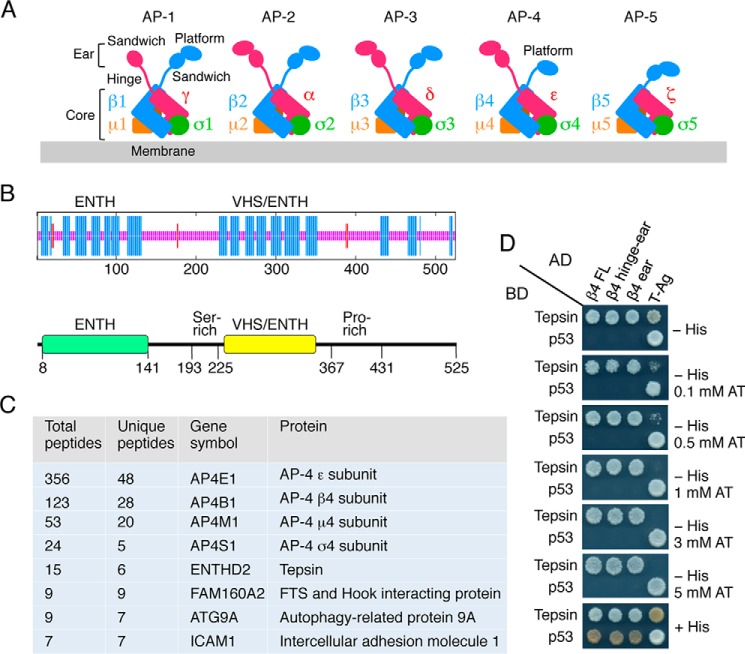
FIGURE 1.
Isolation of Tepsin as an AP-4 interactor. A, schematic representation of adaptor protein (AP) complexes, indicating their subunits and structural domains. Two other complexes not represented here, the COPI-F subcomplex and part of the TSET complex, have a similar structure. Notice that some ear domains comprise two subdomains referred to as sandwich and platform, while others consist of only a sandwich (e.g. AP-1γ) (16, 17) or platform (e.g. AP-4 β4) (41) subdomain. In addition, GGA proteins have a sandwich domain similar to the AP-1 γ-ear domain (18, 19) and the yeast exomer Chs5 subunit has a FBE domain that is structurally homologous to the sandwich-platform tandem of the AP-2 α-ear domain (50). B, top, consensus secondary structure prediction of tepsin by a combination of the MLRC, DSC, and PHD methods at the NPS@ server. Blue represents α-helix, red β-sheet, and magenta random coil structure. B, bottom, domain organization of human tepsin. Domain names and amino acid numbers are indicated. C, filtered results of the affinity purification and mass spectrometric analysis for proteins that co-purify with the human AP-4 complex. D, yeast two-hybrid (Y2H) analysis of the interaction of tepsin or p53 (control) fused to the GAL4 DNA binding domain (BD) with different AP-4 β4 constructs or the SV40 large T antigen (T-Ag) (control) fused to the GAL4 transcriptional activation domain (AD). Colonies were grown in the presence of varying concentrations of 3-amino-1,2,4-triazole (AT) to test the specificity and strength of the interactions. Colony growth in medium without histidine (−His) is indicative of interactions. Growth in complete medium (+His) is used as a control. All images are from plates grown for 4 days. The larger diameter of the growth areas on plates with higher concentrations of AT (particularly at 1–5 mm AT) stems from the fact that the yeast inoculum (5 μl drop) spreads over a larger area in these plates.
X-ray crystallographic analyses of the α- and β2-ear domains of AP-2 revealed that they have a similar bilobed structure comprising an N-terminal immunoglobulin-like β-sandwich subdomain and a C-terminal platform subdomain (7, 8, 9, 10) (Fig. 1A). Accessory proteins interact with the α and β2 ear domains via short peptide sequences or structural motifs that bind to sites on either the sandwich or platform subdomains. For instance, DPW (11) and WXX[FW]X[DE] motifs (12, 13, 14) (where X is any amino acid) (motifs denoted in PROSITE syntax) bind to the α-ear sandwich, while DP[FW] (7, 11) and FXDXF motifs (11, 14) bind to the α-ear platform, all in an extended conformation. On the other hand, a helical [DE]nX1–2FXX[FL]XXXR motif (15, 9, 10) and probably the extended DP[FW] motif as well (8) bind to the β2-ear platform. X-ray crystallography of the γ-ear and the related GGA-ear domains showed that they consist of only a sandwich subdomain (16, 17, 18, 19, 20) that binds accessory proteins via ψG[PDE][ψLM] motifs (where ψ is an aromatic amino acid) (21, 22, 23) or variants thereof (24, 25). The structure of the β1 ear has not been solved crystallographically, although, because of its homology to the β2 ear (∼75% amino acid sequence identity), it is expected to have a similar bilobed structure and to bind the same accessory proteins and motifs.
At least five other complexes (AP-3, AP-4, AP-5, COPI-F subcomplex, and part of TSET) (2, 3, 5, 26, 27) are known or predicted to have a structure similar to that of AP-1 and AP-2 (Fig. 1A). However, very little is known about their interactions with accessory proteins. We have been studying the AP-4 complex, a non-clathrin-associated heterotetramer of subunit composition ϵ-β4-μ4-σ4 (28, 29). This complex associates with the trans-Golgi network (TGN) (28, 29, 30) in an Arf-regulated manner (31), and participates in the sorting of several cargos, including glutamate receptors (32, 33) and members of the amyloid precursor protein family (34). Importantly, mutations in each of the subunits of AP-4 in humans have been found to cause an autosomal-recessive congenital disorder characterized by severe intellectual disability, progressive spastic paraplegia or tetraplegia, microcephaly, seizures, and growth retardation (35, 36, 37, 38, 39). Coated vesicle proteomics analyses by Borner et al. (40) identified the only currently known AP-4 accessory protein, tepsin (also known as the product of the ENTHD2 or AP4AT gene). Tepsin is encoded by several splice forms, the best characterized of which corresponds to a protein of 525 amino acids having an N-terminal ENTH domain, an intervening unstructured segment, a central VHS/ENTH domain, and a C-terminal unstructured segment (40) (Fig. 1B). Tepsin was shown to bind to the β4-ear domain of AP-4, and to be recruited to the TGN in manner dependent on this interaction (40). However, the structural determinants of this interaction and the possible existence of additional tepsin-AP-4 interactions remain to be investigated.
Herein we report the identification of a novel peptide motif, [GS]LFXG[ML]X[LV], in the C-terminal unstructured segment of tepsin that interacts with the β4-ear domain. A recent solution NMR structure of the β4 ear (PDB ID: 2MJ7) (41) showed that this domain consists of only a platform fold similar to that of the β2-ear platform subdomain. Mutational and binding analyses based on this structure allowed us to map the binding site for the [GS]LFXG[ML]X[LV] motif to the same surface that harbors the binding sites for the [DE]nX1–2FXX[FL]XXXR and DP[FW] motifs on the β2 ear (8, 9, 10, 15). Further analyses revealed a second motif at the C terminus of tepsin, S[AV]F[SA]FLN, which binds to the ϵ-ear domain of AP-4. Both motifs are phylogenetically conserved and contribute to the interaction of tepsin with AP-4 and its recruitment to the TGN. The bivalency of the interactions contributes to a higher avidity of tepsin for AP-4 and also has the potential to promote cross-linking of AP-4 heterotetramers in the process of AP-4-mediated protein sorting.
Experimental Procedures
DNA Recombinant Constructs
All mutations were generated by site-directed mutagenesis (QuickChange, Agilent) and confirmed by DNA sequencing.
Two Strep/One FLAG (TSF)-tagged Constructs
The pGADT7-human ϵ (NM_007347.4) vector (31) was subjected to mutagenesis to remove an internal XhoI site and used as a template for PCR amplification to generate a KpnI/XhoI fragment encoding full-length (FL) ϵ. Similarly, a KpnI/XhoI tepsin fragment encoding FL human tepsin (NM_144679.2) was PCR-amplified from pOTB7-tepsin (Thermo Scientific). The KpnI/XhoI ϵ and tepsin fragments were subcloned into the large fragment obtained after digestion of pcDNA 3.1-BLOS2 (42) with KpnI and XhoI. These ligations generated pcDNA 3.1-TSF-ϵ and pcDNA 3.1-TSF-tepsin, two constructs based on the pcDNA 3.1/myc-His A MCS plasmid (Invitrogen) that direct the expression of the AP-4 ϵ subunit and tepsin tagged at their N terminus with a TSF epitope.
Yeast Two-hybrid Vectors
The pGADT7-human β4 and pGADT7-based plasmids encoding FL human ϵ and the 1–138, 1–260, 1–545, 1–368, and 727–1137 ϵ fragments were previously described (31). An SfiI/PstI (filled in with T4 DNA polymerase) fragment encoding human β4 hinge-ear domains (residues 534–739), excised from pGBKT7-β4 hinge-ear, was subcloned into the SfiI/XhoI (filled in with T4 DNA polymerase) sites of pGADT7 (Clontech). The pGADT7-DNA β4 hinge-ear was subsequently spliced out by mutagenesis to generate pGADT7-β4 ear (residues 601–739). The above-described pcDNA 3.1-TSF-tepsin was mutagenized to remove an internal EcoRI site. An EcoRI/XhoI fragment encoding FL tepsin was subsequently generated by PCR amplification and subcloned into the EcoRI/SalI sites of pGBT9 (Clontech). The pGBT9-rat TGN38 tail construct (residues 324–353) was described previously (43).
pEGFP Constructs
Vectors encoding N-terminal or C-terminal fusions of tepsin with GFP were generated by subcloning an EcoRI/XhoI tepsin fragment into the EcoRI/SalI sites of pEGFP-C2 (Clontech) (GFP-tepsin construct) and by Gibson Assembly Master Mix (New England Biolabs)-directed cloning into the HindIII/EcoRI sites of pEGFP-N1 vector (Clontech) (tepsin-GFP construct).
GST Fusion Constructs
An NdeI/PstI fragment encoding the β4 hinge-ear domains was excised from pGBKT7-β4 hinge-ear, filled in-with T4 DNA polymerase and subcloned into the SmaI site of pGEX-4T-1 (GE Healthcare). The pGEX-5X-1-β4 ear construct was previously described (28). A BamH1-SalI fragment encoding the ϵ ear domain (residues 840–1137) was subcloned between the corresponding sites of pGEX-4T-1.
Antibodies
The mouse monoclonal anti-FLAG M2 antibodies used for immunofluorescence (affinity purified, F1804) and immunoblotting (F3165) were purchased from Sigma. The mouse monoclonal anti-ϵ antibody (BD612018) was from BD Biosciences, and the sheep polyclonal anti-TGN46 antibody (AHP500) was from AbD Serotec.
Cell Culture and Transfection
H4 and HeLa cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin and 100 μg/ml streptomycin (complete DMEM) at 37 °C and 5% CO2. Plasmid transfection was performed using Lipofectamine 2000 (Life Technologies) or X-tremeGENE 9 (Roche) as per the manufacturers' instructions.
Tandem Affinity Purification and Mass Spectrometry
H4 cells stably transfected with pcDNA 3.1-TSF-ϵ were selected by addition of 1.25 mg/ml G418 to the culture medium. Individual clones were isolated approximately 2 weeks after initial transfection, grown on 6-well plates and analyzed by immunofluorescence and immunoblotting using anti-FLAG and anti-ϵ antibodies. H4 cells stably expressing TSF-ϵ (grown on fifteen 150-mm culture dishes) were lysed in 15 ml of 50 mm Tris-HCl pH 7.4, 75 mm NaCl, 5 mm EDTA and 0.8% (v/v) Triton X-100 supplemented with proteinase inhibitor mixture (Roche). Cell lysates were cleared by centrifugation at 17,000 × g and 4 °C for 15 min and incubated with StrepTactin resin (IBA) for 2 h at 4 °C. The resin was washed twice with 50 mm Tris-HCl pH 7.4, 300 mm NaCl, 5 mm EDTA and 0.1% (v/v) Triton X-100 and once with phosphate-buffered saline (PBS). Bound proteins were eluted with 5 mm desthiobiotin-containing buffer (IBA) at room temperature. Eluates were supplemented with 0.8% (v/v) Triton X-100 and protease inhibitors and further purified by incubation for 2 h at 4 °C with FLAG antibody-coated beads (Sigma). Resin was washed as in the previous step and proteins eluted with 500 μg/ml FLAG peptide (Sigma) in 10 mm Tris-HCl pH 7.4, 150 mm NaCl. Eluted proteins were precipitated with 10% trichloroacetic acid, washed with acetone, air-dried, and analyzed by mass spectrometry at the Taplin Facility, Harvard University.
Yeast Two-Hybrid Assays
Assays were performed using the GAL4-based Matchmaker system (Clontech) and the AH109 reporter yeast strain as previously described (21).
GST Fusion Protein Expression and Purification
Recombinant protein expression in Escherichia coli strain BL21 (Novagen) transformed with pGEX-4T-1-β4 hinge-ear, pGEX-5X-1-β4 ear or pGEX-4T-1-ϵ ear plasmids was induced by addition of 0.5 mm IPTG followed by incubation for 3 h at 37 °C or overnight at 20 °C. Bacterial pellets were lysed by sonication in PBS supplemented with 10 μg/ml aprotinin, 4 μg/ml leupeptin, 1.7 μg/ml pepstatin, 1 mm 4-(2-aminoethyl)benzenesulfonyl, and 2 mm EDTA. After sonication, suspensions were supplemented with 1% (v/v) Triton X-100 followed by incubation for 30 min at 4 °C. Protein extracts were cleared by centrifugation at 14,000 × g and 4 °C for 30 min, and the supernatants subsequently bound to glutathione-Sepharose resin (GE Healthcare) and eluted with 20 mm glutathione.
Isothermal Titration Calorimetry (ITC)
GST-β4 hinge-ear, GST-ϵ ear and GST were dialyzed overnight against excess PBS pH 7.4 at 4 °C, followed by concentration on Centricon Ultracel YM-10 filters (EMD Millipore) to ∼100–180 μm. The peptides CAWSRDSLFAGMELVA and SEPSAFAFLNA (residues 463–478 and 515–525 in human tepsin) (New England Peptide) were dissolved to a final concentration of ∼1 and 1.4 mm, respectively. Immediately preceding experiments, the GST proteins and tepsin peptide solutions in PBS were supplemented with 0.5 mm DTT. Titration calorimetry was performed at 10 °C using an iTC200 system (Malvern Instruments). The tepsin peptides were injected over the GST-β4 hinge-ear and GST-ϵ ear (or a GST control) in 20 aliquots of 2 μl each at 150 s intervals. Data analysis was performed with Origin software. The affinity reported represents the mean ± S.D. of assays with three different preparations of recombinant protein.
Isolation of β4-ear Binding Peptides from a Phage-display Library
Previously described methods (44, 22) were used to construct a library of random dodecapeptides fused to the N terminus of the M13 bacteriophage major coat protein (2 × 1010 unique members). Phage pools displaying the library were cycled through rounds of binding selection with GST-β4 hinge-ear or GST-β4 ear fusion proteins coated on 96-well MaxiSorp immunoplates (Nalge Nunc) as capture targets. The library was sorted separately against each target, and phage were propagated in E. coli XL1-blue cells (Stratagene) supplemented with both M13-KO7 helper phage (New England Biolabs) to facilitate phage production and 10 μm IPTG to induce expression of the library. After three rounds of selection, phages from individual clones were analyzed in a phage enzyme-linked immunosorbent assay. Phages that bound to GST-β4-hinge-ear or GST-β4-ear fusion proteins but not to a control GST fusion were subjected to DNA sequence analysis.
StrepTactin Pulldowns
HeLa cells in 100-mm plates transiently transfected with different TSF-tagged tepsin constructs were washed twice with cold PBS and lysed in 0.8 ml of 50 mm Tris-HCl pH 7.4, 0.8% (v/v) Triton X-100, 75 mm NaCl (lysis buffer) with protease inhibitors (EDTA-free Complete, Roche). Extracts were centrifuged for 15 min at 21,000 × g and 4 °C, and supernatants incubated overnight at 4 °C with 60 μl of StrepTactin beads previously washed in PBS 0.01% (v/v) Triton X-100. Beads with immobilized complexes were washed three times with lysis buffer containing 0.1% (v/v) Triton X-100 and two times with PBS at 4 °C. Bound complexes were eluted from beads by incubation for 20 min at 25 °C with 50 μl of 2.5 mm desthiobiotin-containing buffer diluted in PBS. The suspensions were centrifuged at room temperature for 2 min at 16,000 × g and the supernatants analyzed by SDS-PAGE and immunoblotting.
Immunofluorescence Microscopy
HeLa cells were plated on 12-mm coverslips previously coated with fibronectin (Sigma) (0.1 mg/ml in PBS for 1 h at 37 °C) and grown to ∼30% confluency before transfection with the different pEGFP-tepsin constructs. At 24–36 h after transfection, coverslips were washed in PBS and permeabilized for 1 min with 25 mm HEPES-HCl, 150 mm potassium glutamate, 25 mm potassium chloride, 2.5 mm potassium acetate, 5 mm EDTA, and 0.00375% saponin at room temperature (21). Coverslips were washed in PBS and fixed for 12 min in 4% paraformaldehyde in PBS. Following fixation, coverslips were washed twice for 5 min in PBS, permeabilized for 15 min in 0.2% (v/v) Triton X-100 and blocked in 1% bovine serum albumin (BSA) in PBS for another 15 min. Staining was carried out for 30 min at 37 °C with sheep anti-TGN46 antibody diluted 1:1000 in 0.2% BSA in PBS (immunofluorescence buffer). Coverslips were then washed twice with PBS and incubated for 30 min at 37 °C with secondary Alexa Fluor 647 donkey anti-sheep (Life Technologies) antibody diluted 1:1000 in immunofluorescence buffer. Coverslips were next washed twice with PBS and mounted on slides using Fluoromount-G with DAPI (Electron Microscopy Sciences). Images were obtained on an inverted confocal laser scanning microscope (LSM780; Carl Zeiss) fitted with a 63X, 1.4 NA objective. Image analysis was performed using Image J (Rasband, W.S., ImageJ, NIH, Bethesda, Maryland).
Results
Isolation of Tepsin as an AP-4-interacting Protein
In experiments aimed at identifying proteins that interact with the AP-4 complex, detergent extracts of human H4 neuroglioma cells stably expressing the human ϵ subunit of AP-4 appended with an N-terminal Two-Strep/one FLAG (TSF) tag were subjected to tandem affinity purification (TAP) followed by mass spectrometry of the eluate. To limit our analysis to the most abundant, specific interactors, we set a threshold of four total peptides and filtered the results against the Contaminant Repository for Affinity Purification database (CRAPome). Only proteins that were not found as contaminants in this repository (0 spectral counts) were considered for further analysis. The top hits in order of total peptide number in the resulting short list were the four subunits of AP-4 (ϵ, β4, μ4, and σ4) (Fig. 1C), thus verifying the reliability of this approach. The fifth hit was the product of the ENTHD2 gene, tepsin (Fig. 1C), a protein that had been previously identified as an AP-4 interactor (40). The other proteins on this list were the products of the FAM160A2, ATG9A, and ICAM genes (Fig. 1C), the significance of which remains to be determined.
Identification of a Short Peptide Sequence in Tepsin That Is Required for Interaction with the AP-4 β4-Ear Domain
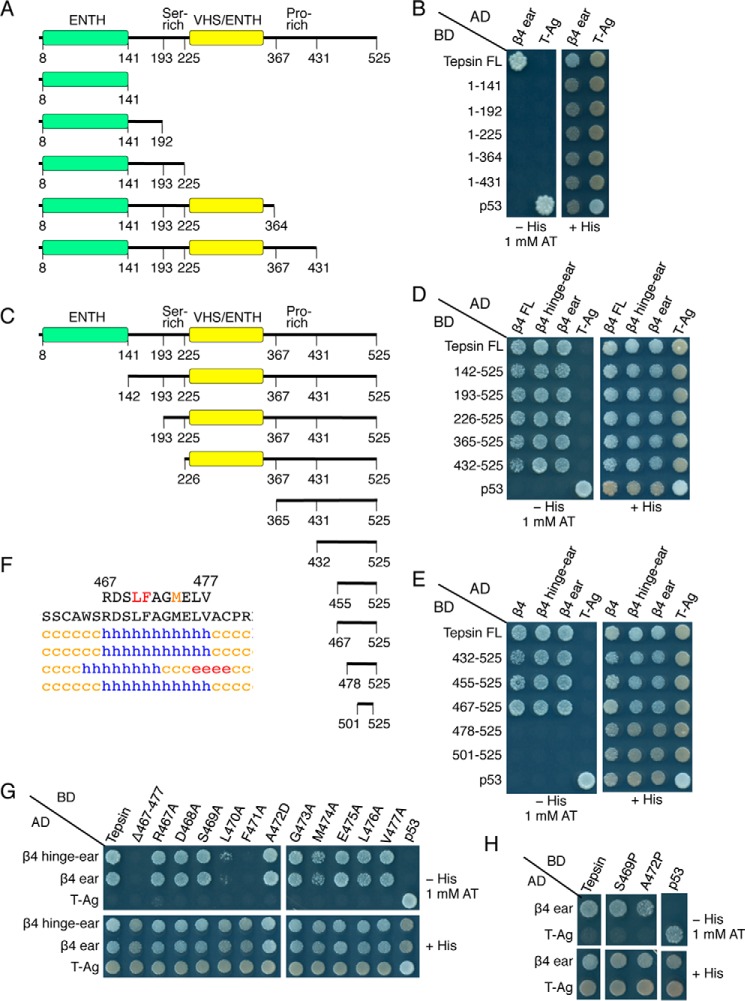
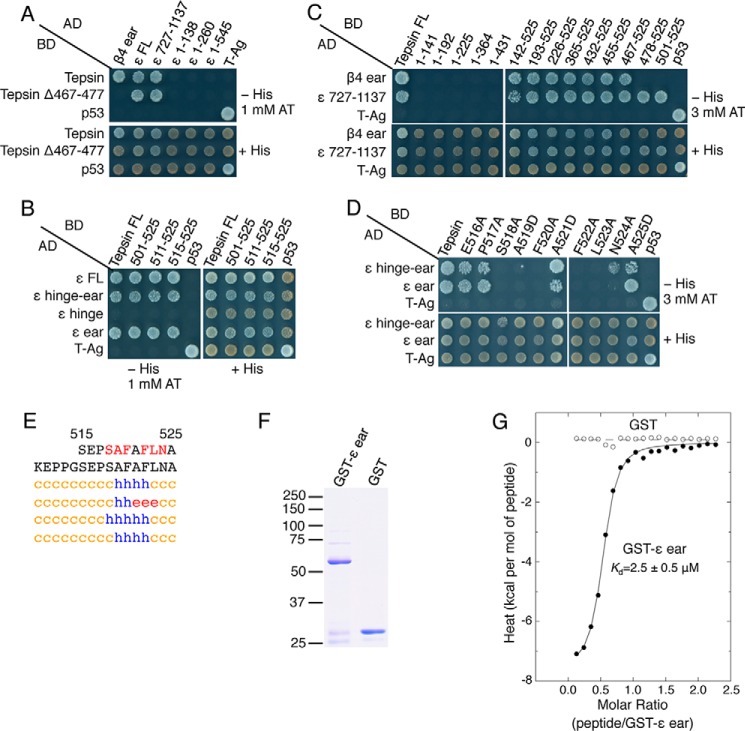
Because the structural determinants of the tepsin-AP-4 interaction were not known, we decided to undertake a systematic molecular dissection of AP-4-binding sequences in human tepsin using the yeast two-hybrid system (Y2H) as a reporter assay. In agreement with previous findings using other methodologies (40), we detected specific interaction of full-length tepsin with full-length β4, β4-hinge-ear, and β4-ear constructs (Fig. 1D). These interactions were relatively strong, as they withstood challenge with up to 5 mm 3-amino-1,2,4-triazole (AT), a competitive inhibitor of the His3 reporter in the Y2H system (Fig. 1D). We next tested a series of C-terminal (Fig. 2, A and B) and N-terminal truncation mutants (Fig. 2, C and D) of tepsin for their ability to interact with β4. We observed that deletion of 94 amino acids from the C terminus rendered tepsin unable to bind to the β4 ear (tepsin 1–431, Fig. 2B). Moreover, we found that these 94 amino acids were sufficient for interaction with the β4 ear (tepsin 432–525, Fig. 2D). Further deletion analyses delineated a determinant for interaction with the β4 ear to the segment spanning amino acids 467 to 477 of tepsin (Fig. 2E), having the sequence RDSLFAGMELV (Fig. 2F). An alanine-scan mutagenesis of this segment (except for Ala-472, which was substituted by aspartate) identified Leu-470, Phe-471, and Met-474 as critical elements of the interaction determinant (Fig. 2G). Although the 467–477 segment is contained within the largely unstructured C-terminal region of tepsin, theoretical analyses predicted that the segment itself could form a short α helix (Fig. 2F). However, substitution of a helix-breaking proline residue in place of Ser-469 or Ala-472 did not abrogate interaction of tepsin with the β4 ear (although the A472P mutant exhibited somewhat reduced binding) (Fig. 2H). Therefore, the 467–477 segment of tepsin is required for interaction with the β4 ear, possibly as part of an extended peptide sequence.
FIGURE 2.
Mutational and yeast two-hybrid analyses identify a short peptide sequence in tepsin that interacts with the AP-4 β4-ear domain. A, schematic representation of C-terminally truncated tepsin constructs, and B, testing for their interactions with the β4-ear domain using the Y2H system. C, schematic representation of N-terminally truncated tepsin constructs, and D and E, testing for their interactions with full-length (FL) β4, β4-hinge-ear and β4-ear constructs using the Y2H system. F, tepsin sequence required for interaction with the β4 ear, and secondary structure prediction by the methods described in the legend to Fig. 1B. Blue h represents α-helix, red e represents β-sheet, and orange c represents random coil structure. G and H, Y2H analysis of the interaction of tepsin mutants bearing single amino acid substitutions with the β4-hinge-ear and β4-ear domains. Y2H assays were performed as described in the legend to Fig. 1D.
The Core Residues LFAGM Define a Determinant That Is Sufficient for Interaction with the β4 Ear
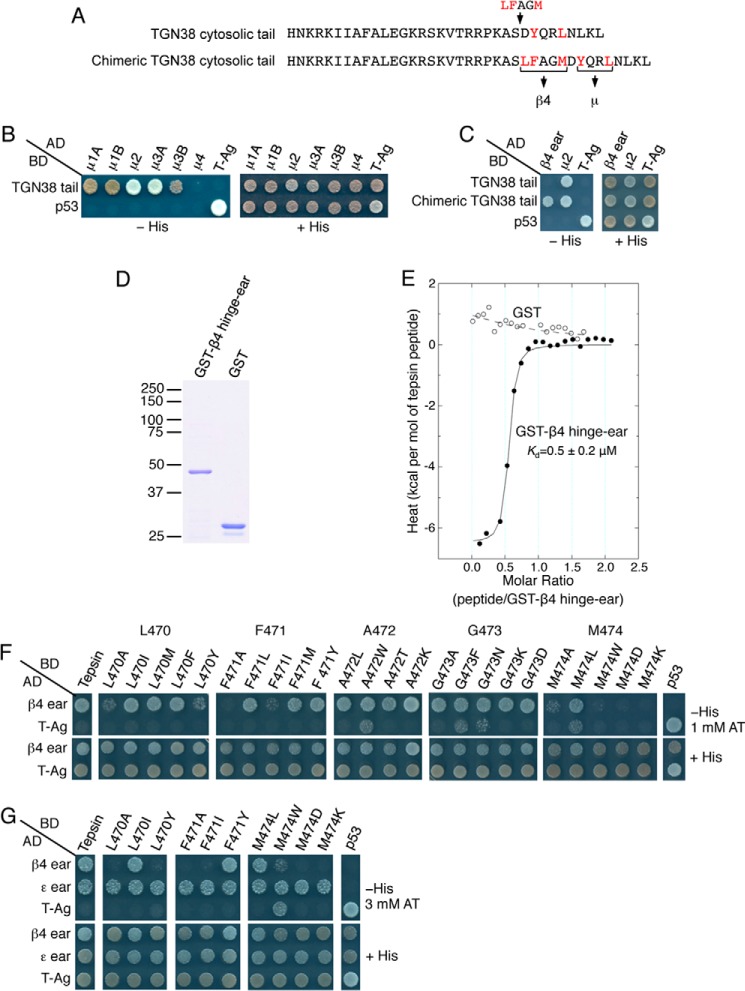
To determine if the 467–477 segment contained sufficient information for interaction with the β4 ear, we inserted the LFAGM sequence from this segment into the cytosolic tail of another protein, TGN38 (Fig. 3A), and again used the Y2H assay to test for interactions. The TGN38 tail has an YXXØ-type sorting signal (YQRL, residues 350–353) (Fig. 3A) that interacts with the μ subunits of several AP complexes (45, 46, 47) (Fig. 3B), but not with the β4 ear (Fig. 3C). The sequence LFAGM was placed between residues 348 and 349 of the TGN38 cytosolic tail, in an unstructured region immediately upstream of the YQRL sequence recognized by AP μ subunits. Importantly, insertion of the LFAGM sequence was sufficient to confer on the TGN38 tail the ability to interact with the β4 ear (Fig. 3C). These results thus demonstrated that the LFAGM sequence constitutes the core of an autonomous β4-ear-binding determinant. Because Leu-470 is the first essential hydrophobic residue within this sequence, we designated it as position zero of an interaction motif.
FIGURE 3.
The hydrophobic core of the short tepsin sequence is sufficient for interaction with the AP-4 β4 ear and tolerates substitutions by other hydrophobic amino acids. A, sequences of the rat TGN38 cytosolic tail and a chimeric TGN38 cytosolic tail having an insertion of the LFAGM sequence from tepsin. Key amino acids for interaction with AP μ subunits and the AP-4 β4 ear are highlighted in red. B, Y2H analysis of the interaction of the TGN38 cytosolic tail with μ subunits of AP complexes. C, Y2H analysis of the interaction of the TGN38 cytosolic tail and the chimeric TGN38 cytosolic tail with the AP-4 β4 ear and the AP-2 μ2 subunit. D, SDS/PAGE and Coomassie Blue staining of GST-β4 hinge-ear and GST proteins used in ITC assays. The positions of molecular mass markers (in kDa) are indicated at left. E, ITC analysis of the interaction of β4-ear-binding peptide CAWSRDSLFAGMELVA with GST-β4 hinge-ear. The binding isotherm depicts the normalized integrated heat change (kcal/mol of injected peptide) as a function of the molar ratio of peptide to GST-β4 hinge-ear. Data were fit to a one-site binding model. The indicated Kd value for the tepsin peptide-GST-β4 hinge-ear interaction represents the mean ± S.D. of three determinations performed with different preparations of recombinant protein. F, Y2H analysis of the interaction of tepsin mutants having single amino acid substitutions in the LFAGM sequence with the β4 ear. Y2H assays were performed as described in the legend to Fig. 1D. G, diminished avidity for β4 ear seen with tepsin mutants L470A, L470Y, F471A, F471I, M474W, M474D, and M474K in panel F is not the due to differences in their levels of expression, stability or folding, as demonstrated by the ability of these mutant constructs to interact with the AP-4 ϵ ear, a second tepsin partner identified in this study (see Fig. 6). The assay shown was performed at higher stringency conditions (3 mm AT).
In Vitro Binding of an LFAGM-containing Peptide to the β4-ear
To independently confirm that a segment containing the LFAGM sequence is sufficient for binding to β4, and to quantitatively assess this interaction, we analyzed by isothermal titration calorimetry (ITC) the binding of the tepsin peptide CAWSRDSLFAGMELVA to purified GST-β4-hinge-ear or GST (control) proteins (Fig. 3D). We observed that this peptide bound to GST-β4-hinge-ear with a Kd of 0.5 ± 0.2 μm (mean ± S.D., n = 3) but not to GST (Fig. 3E).
Sequence Requirements of the β4-ear-binding Determinant
To characterize further the β4-ear-binding sequence, we substituted each residue within the LFAGM core by different amino acids, and analyzed the interactions using the Y2H system. We found that Leu-470 (position 0) could be substituted by bulky hydrophobic residues such as Ile, Met or Phe, but not Ala or Tyr, without loss in interaction with the β4 ear (Fig. 3F). Phe-471 (position 1) could be substituted by Leu, Met, or Tyr, but not Ala or Ile (Fig. 3F). In contrast, Ala-472 (position 2) and Gly-473 (position 3) tolerated various substitutions (Fig. 3F). Finally, Met-474 (position 4) could only be substituted by Leu among all the amino acids tested (Fig. 3F). These observations confirmed that the bulky hydrophobic Leu-470, Phe-471, and Met-474 are the most critical elements of the interaction motif, although substitutions by chemically similar amino acids are tolerated in some cases.
Definition of a Consensus Motif for Interaction with the β4 Ear by Screening of a Phage Display Peptide Library
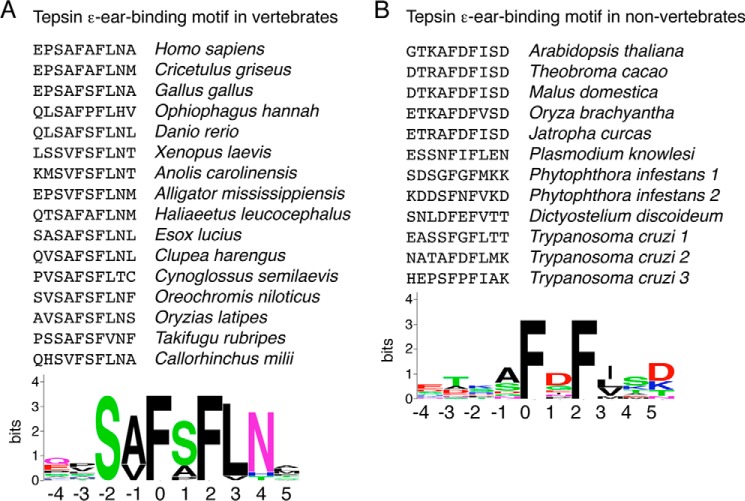
To search more broadly for peptide sequences that bind to the β4 ear, we screened a phage display library encoding random dodecameric peptides using GST-β4-hinge-ear and GST-β4-ear fusion proteins as baits. The peptide sequences isolated by both β4 constructs all shared an 8-amino acid core fitting a [GS]LFXG[ML]X[LV] consensus motif (Fig. 4, A and B). Reassuringly, this motif was in accordance with the LFAGM sequence identified in human tepsin as a β4-ear ligand in our Y2H assays (Figs. 2 and 3), as well as with the homologous sequences in tepsins from all eukaryotic super-groups (Fig. 4C). Less frequent variants of this consensus motif displayed Ile, Met, or Phe in place of Leu(0), Leu, or Tyr in place of Phe(+1), Val in place of [ML](+4), and Ile, Trp, or Phe in place of [LV](+6), consistent with our amino acid substitution analyses (Fig. 3F) and with the presence of some of these rarer variants in orthologous tepsins (Fig. 4C).
FIGURE 4.
Combinatorial peptide screens and sequence comparisons define a consensus motif for binding to the AP-4 β4 ear. A and B, peptides selected by GST-β4 hinge-ear (A) and GST-β4 ear (B) from a phage display library of random dodecapeptides (12-mers) fused to the N terminus of the M13 bacteriophage major coat protein. An 8-amino acid conserved core is indicated by the shaded box. C, conservation of the β4-ear-binding sequence in species from different eukaryotic super-groups. Consensus motifs for all three sequence lists are represented by sequence logos created with WebLogo.
Mapping of a Conserved Binding Site for the Tepsin Motif on the β4 Ear
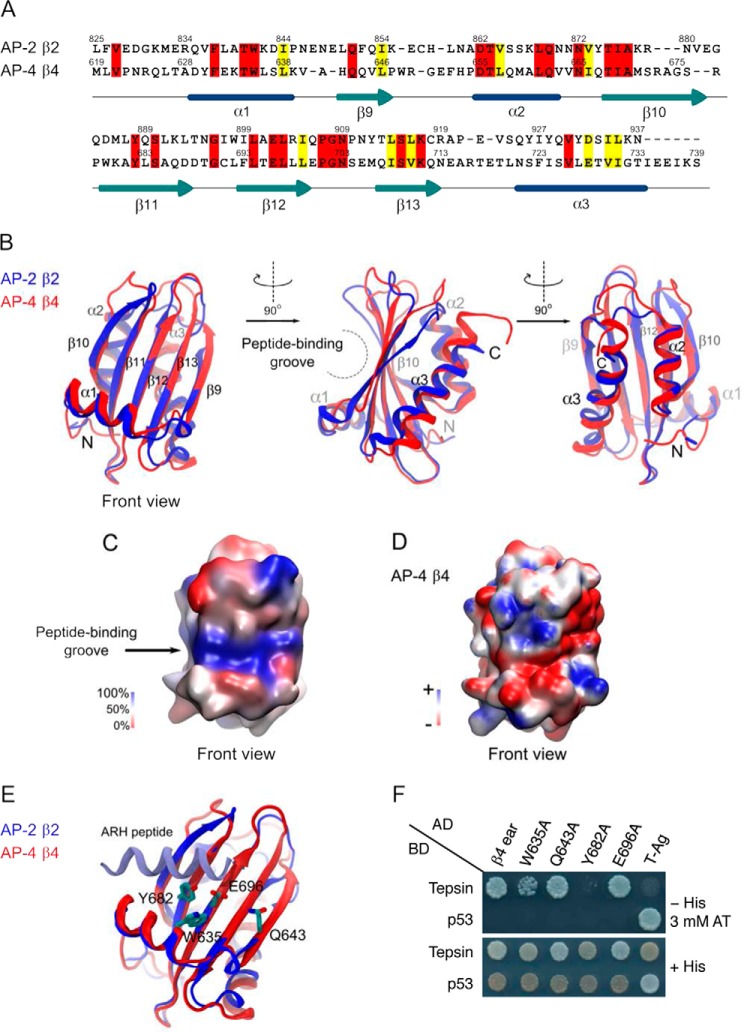
The AP-4 β4 ear exhibits only a low degree (19%) of amino acid sequence identity to the platform subdomain of the AP-2 β2 ear (Fig. 5A). However, a recently reported NMR structure of the β4 ear (PDB ID: 2MJ7) (41) shows that its three-dimensional structure is very similar to that of the β2 platform subdomain (PDB ID: 1E42) (8) (Fig. 5B). Indeed, the root-mean-square deviation of atoms between both structures is 2.15 Å, indicative of a high degree of structural homology. Both structures consist of a five-stranded β-sheet flanked on either side by one and two α-helices (Fig. 5B). Analysis of sequence conservation between both structures using the BLOSUM 60 matrix revealed a highly conserved groove (highlighted in blue in Fig. 5C) that corresponds to the binding site for the α-helical [DE]nX1–2FXX[FL]XXXR motif on the β2 platform subdomain (9, 10, 15). On the β4 ear, this groove features a combination of hydrophobic and acidic surface potential (white and red, respectively, in Fig. 5D), and contains conserved residues shown to be important for [DE]nX1–2FXX[FL]XXXR binding to the β2 platform (Fig. 5E) (9, 10, 15). Among these conserved β4 residues are Trp-635, Gln-643, Tyr-682, and Glu-696, homologous to β2 residues Trp-841, Gln-851, Tyr-888 and Glu-902, respectively (Fig. 5E). Single substitution of these residues by alanine in the β4 ear followed by Y2H analyses showed that Tyr-682 was essential and Trp-635 partially required for interaction with tepsin (Fig. 5F), as was also the case for the corresponding substitutions in the interaction of the β2 ear with the [DE]nX1–2FXX[FL]XXXR motif (9, 15). The β4 residues Gln-643 and Glu-696, on the other hand, were unimportant for binding to tepsin (Fig. 5F), likely because the homologous residues in β2 engage the arginine residue in the [DE]nX1–2FXX[FL]XXXR motif (9, 10), which is not present in the tepsin [GS]LFXG[ML]X[LV] motif. From these experiments we concluded that the tepsin [GS]LFXG[ML]X[LV] motif likely binds to a site on the β4 ear that is similar to that for the [DE]nX1–2FXX[FL]XXXR motif on the β2 ear, albeit with differences that impart specificity to the interactions.
FIGURE 5.
Structure-based mutagenesis of the AP-4 β4 ear identifies residues required for interaction with the tepsin motif. A, sequence alignment of the AP-2 β2 ear platform subdomain and AP-4 β4 ear domain. Identical residues are highlighted in red and chemically conserved residues are highlighted in yellow. Secondary structure elements are indicated. B, comparison of the crystal structure of the AP-2 β2 ear platform subdomain (PDB ID: 1E42) (8) and the solution NMR structure of the AP-4 β4 ear domain (PDB ID: 2MJ7) (41). The positions of the N and C termini are indicated. The first of the three structures is considered the front view. C, sequence conservation between the AP-2 β2 ear platform subdomain and the AP-4 β4 ear domain (front view). The similarity index was calculated using the BLOSUM 60 distance matrix and plotted on the surface of the AP-4 β4 ear domain. Blue indicates 100% similarity, white 50%, and red no similarity in amino acids at the same position. Notice the high degree of similarity, particularly in the peptide-binding groove. D, surface representation of the AP-4 β4 ear domain colored for electrostatic potential (positive (+5 kT e−1), in blue, to negative (−5 kT e−1) in red). This surface was created using the APBS algorithm plug-in implemented in VMD (51, 52, 53). E, comparison of the structure of the AP-2 β2 ear platform subdomain in complex with a bound [DE]nX1–2FXX[FL]XXXR-type helical peptide from ARH (PDB ID: 2G30) (9) with the structure of the AP-4 β4 ear domain (PDB ID: 2MJ7) (41), indicating residues on β4 that are homologous to those involved in peptide binding in β2. F, Y2H analysis of the interaction of tepsin with β4-ear constructs having mutations in the residues indicated in E. Y2H assays were performed as described in the legend to Fig. 1D.
Identification of a Second Motif at the C Terminus of Tepsin that Mediates Interaction with the AP-4 ϵ Ear
Because some accessory proteins bind to both ears of an AP complex, as is the case for the binding of Epsin, Eps15, and AP180 to the α and β2 ear domains of AP-2 (8), we next sought to determine if tepsin also binds to the ϵ ear of AP-4. Y2H analyses showed that tepsin indeed bound to full-length ϵ as well as to a fragment containing part of the hinge and all of the ϵ ear (727–1137), but not to constructs spanning different portions of the trunk domain (residues 1 to 545) (Fig. 6A). Further analyses showed that tepsin was recognized by the ϵ-hinge-ear or ϵ-ear, but not ϵ-hinge, domains (Fig. 6B). Analysis of a series of tepsin N- and C-terminal truncation mutants revealed that the ϵ-ear-binding determinant was different from the β4-ear-binding fragment (residues 467–477) and was contained within the last 11 amino acids (515–525) (Fig. 6, A–C), comprising the sequence SEPSAFAFLNA. Single alanine substitutions in this sequence (except for alanines 519, 521, and 525, which were replaced by aspartate) revealed a requirement for Ser-518, Ala-519, Phe-520, Phe-522, Leu-523, and Asn-524 for interaction with the ϵ ear (Fig. 6D). Theoretical analyses of the SEPSAFAFLNA sequence predict that it occurs within a largely unstructured context, although, similarly to the β4-ear-binding sequence, the critical residues themselves have the potential to form a short α-helix (Fig. 6E). ITC analysis showed that a SEPSAFAFLNA peptide bound to the GST-ϵ ear (Fig. 6F) with a Kd of 2.5 ± 0.5 μm (mean ± S.D., n = 3) (Fig. 6G), 5-fold weaker than the binding of the CAWSRDSLFAGMELVA peptide to the GST-β4-hinge-ear (Fig. 3E).
FIGURE 6.
Identification of a second tepsin motif that mediates interactions with the AP-4 ϵ-ear domain. A–C, Y2H analysis of the interaction of full-length and N- and C-terminally truncated forms of AP-4 ϵ with full-length tepsin and a series of N- and C-terminally truncated forms of tepsin (schemes shown in Fig. 2, A and C). D, Y2H analysis of the interaction of ϵ-hinge-ear and ϵ-ear constructs with tepsin mutants having single amino acid substitutions in the segment spanning amino acids 516 to 525. E, secondary structure prediction of the tepsin C-terminal region including segment 515–525 by the methods described in the legend to Fig. 1B. F, SDS/PAGE analysis and Coomassie Blue staining of GST-ϵ ear and GST proteins used in ITC assays. The positions of molecular mass markers (in kDa) are indicated at left. The GST-ϵ ear migrates with an apparent molecular mass of 60 kDa. G, ITC analysis of the interaction of ϵ-ear-binding peptide SEPSAFAFLNA with GST-ϵ ear. The binding isotherm depicts the normalized integrated heat change (kcal/mol of injected peptide) as a function of the molar ratio of peptide to GST-ϵ ear. Data were fit to a one-site binding model. The indicated Kd value for the tepsin peptide-GST-ϵ ear interaction represents the mean ± S.D. of three determinations performed with different preparations of recombinant protein.
The ϵ-ear-binding sequence is quite conserved at the C terminus of all vertebrate tepsins, fitting the consensus motif S[AV]F[SA]FLN (Fig. 7A). This structural element was more difficult to recognize in non-vertebrate tepsins, but closer inspection revealed that they all contained a more degenerate, shorter version of the motif, FXF[LIMV] (Fig. 7B). This motif was also found within the C-terminal unstructured segment of the non-vertebrate tepsins, albeit not at the very C terminus. From these experiments, we concluded that tepsin has the ability to bind to both the β4 ear via a [GS]LFXG[ML]X[LV] motif and to the ϵ ear via a distinct S[AV]F[SA]FLN motif (in vertebrates).
FIGURE 7.
Consensus motif for binding to the AP-4 ϵ ear. A and B, conservation of the ϵ-ear-binding sequence in vertebrate (A) and non-vertebrate species (B). Consensus motifs are represented by sequence logos created with WebLogo.
Both Tepsin Motifs Contribute to Interaction with AP-4 and Recruitment of Tepsin to the TGN in Cells
To assess the importance of both interactions for association of tepsin with AP-4 in human cells, we transfected HeLa cells with plasmids encoding TSF-tagged WT or mutant tepsin constructs, and examined their interaction with endogenous AP-4 by co-precipitation analysis (Fig. 8). We observed that both full-length tepsin and a truncation mutant lacking the N-terminal ENTH domain (tepsin 142–525) co-precipitated AP-4, as detected by immunoblotting with an antibody to endogenous AP-4 ϵ. Deletion of both the N-terminal ENTH and the central VHS/ENTH domains (tepsin 357–525), however, resulted in a great decrease in AP-4 co-precipitation, revealing that the central VHS/ENTH domain is required for tepsin-AP-4 interactions. Deletion of either the β4-binding [GS]LFXG[ML]X[LV] motif (tepsinΔ467–477) or ϵ-binding S[AV]F[SA]FLN motif (tepsin 1–515) also greatly reduced AP-4 co-precipitation, and combined mutation of both motifs completely eliminated it (Fig. 8). These experiments thus demonstrated that both ear-binding motifs are required for efficient tepsin-AP-4 association and, additionally, uncovered a requirement of the central VHS/ENTH domain for this association in human cells.
FIGURE 8.
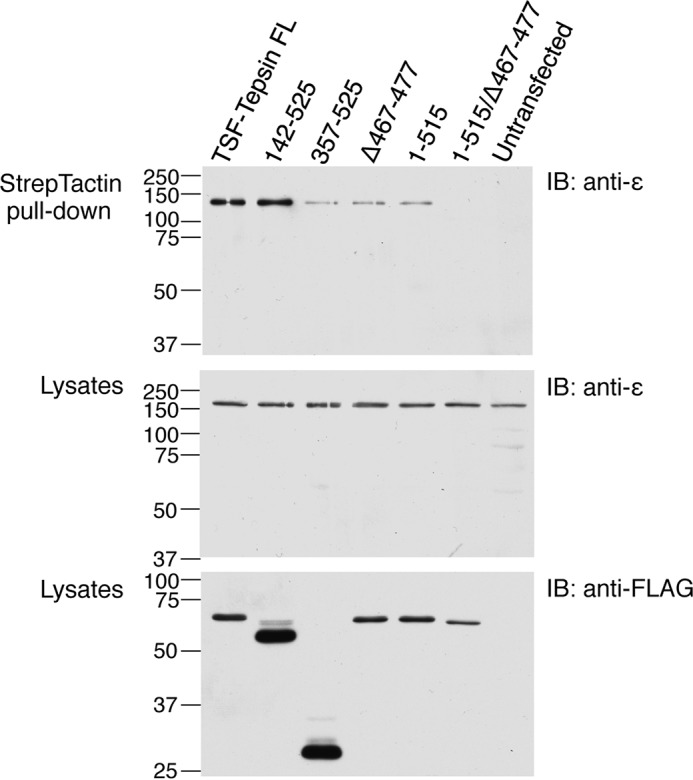
Both the β4-ear- and ϵ-ear-binding motifs are required for interaction of tepsin with AP-4 in HeLa cells. A, HeLa cells were transfected with plasmids encoding various TSF-tagged tepsin constructs, including full length, 145–525 (missing the N-terminal ENTH domain), 367–525 (missing both the N-terminal ENTH domain and central VHS/ENTH domain), Δ467–477 (missing the β4-ear-binding [GS]LFXG[ML]X[LV] motif), 1–515 (missing the ϵ-ear-binding S[AV]F[SA]FLN motif), and 1–515/Δ467–477 (missing both motifs) (see Fig. 1B for schematic representation of tepsin), or were left untransfected. Cell lysates were subjected to StrepTactin pulldown followed by SDS-PAGE and immunoblotting (IB) with antibody to endogenous AP-4ϵ. Cell lysates were also analyzed by SDS-PAGE and immunoblotting for AP-4ϵ and the FLAG epitope contained within the TSF tag. The positions of molecular mass markers (in kDa) are indicated on the left of each immunoblot.
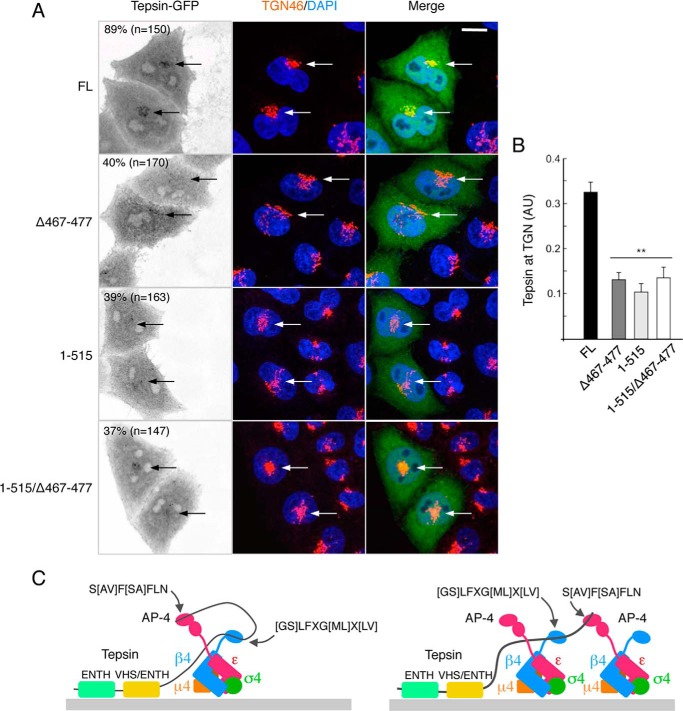
A previous study showed that tepsin localizes to the TGN in an AP-4-dependent manner (40). To test for the importance of both ear-binding motifs for this localization, we first examined the distribution of different tagged forms of tepsin in transfected HeLa cells. We observed that tepsin tagged at the C terminus with GFP (tepsin-GFP) showed localization to the TGN in 89% of the cells, as determined by co-staining for TGN46, although the protein was also found in the cytosol of all cells (Fig. 9A). Tagging of tepsin at the N terminus with either GFP or TSF resulted in lower percentages of cells exhibiting TGN staining (data not shown), indicating that a free N-terminal ENTH domain is required for optimal TGN association. Mutational analyses were consequently conducted using tepsin-GFP as a template. Deletion of the [GS]LFXG[ML]X[LV] or S[AV]F[SA]FLN motifs, individually or in combination, reduced the TGN localization of tepsin-GFP to ∼40% of the cells (Fig. 9A) and ∼40% overall intensity (Fig. 9B). We concluded that interactions of both tepsin motifs with the corresponding AP-4 β4 and ϵ ears are required for efficient association of tepsin with the TGN. The residual TGN association of the mutant constructs, however, suggests that other parts of tepsin also contribute to its recruitment to the TGN.
FIGURE 9.
Role of β4-ear- and ϵ-ear-binding motifs in the recruitment of tepsin to the TGN. A, HeLa cells expressing FL, Δ467–477, 1–515 or 1–515/Δ467–477 tepsin-GFP constructs were immunostained for endogenous TGN46 and imaged by confocal fluorescence microscopy. The percentage of cells exhibiting TGN fluorescence for each tepsin-GFP construct is shown (n indicates the number of cells analyzed in each group). Bars: 10 μm. B, quantitation of the fraction of tepsin-GFP fluorescence associated with TGN (envelope defined by TGN46 staining) in cells transfected with the different constructs. TGN association was calculated as the difference between the GFP fluorescence in the TGN envelope minus the GFP fluorescence in an area of the cytosol identical to the TGN (and contiguous to the TGN) divided by the GFP fluorescence in the cytosol ((GFP in TGN minus GFP in cytosol)/GFP in cytosol) (tepsin at TGN, expressed in arbitrary units, AU). Statistical significance was calculated by ANOVA followed by Tukey's test (**, p < 0.01 when compared with FL construct; 40 cells were analyzed in each group). C, scheme depicting the bivalent interaction of one tepsin molecule with one or two AP-4 complexes.
Discussion
Protein coats involved in cargo sorting and vesicle budding are formed by cooperative assembly of multiple components that interact with each other via low-to-moderate affinity interactions (in the order of 1–100 μm) (3, 4). Many of these interactions involve short peptide motifs within unstructured parts of proteins binding to folded domains of other proteins. The interaction networks that contribute to coat assembly and dynamics have been most extensively characterized for clathrin coats containing the AP-2 or AP-1 adaptors, but remain poorly understood for most other coats. Here we describe two novel peptide motifs within the C-terminal unstructured part of the accessory protein tepsin that mediate interactions with the β4-ear and ϵ-ear domains of the AP-4 adaptor. Moreover, we show that both interactions are required for high-avidity association of tepsin with AP-4 and for efficient recruitment of tepsin to the TGN in human cells. These findings demonstrate that the paradigm of peptide motifs in accessory proteins binding to ear domains of AP complexes, previously established for clathrin-AP-1/AP-2 coats, also applies to a non-clathrin coat containing AP-4.
The β4-ear-binding motif is contained within the segment spanning residues 467 to 477 of the 525-amino acid isoform of human tepsin. The sequence of this segment is RDSLFAGMELV, and its most critical residues for binding to the β4 ear are Leu, Phe, and Met within the LFAGM core (Figs. 2 and 3). Insertion of this core sequence into an unrelated protein confers the ability to bind to the β4 ear (Fig. 3), indicating that it can function as an autonomous interaction determinant. Mutational analyses of this sequence (Figs. 2 and 3), screening of a phage display peptide library (Fig. 4), and comparison of orthologous tepsin sequences from all eukaryotic super-groups (Fig. 4) contribute to defining the 8-amino acid consensus motif [GS]LFXG[ML]X[LV], in which the first Leu corresponds to position 0.
The ϵ-ear-binding motif is found right at the C terminus of human tepsin, within the segment comprising amino acids 515 to 525. The sequence of this segment is SEPSAFAFLNA and its most critical elements are the SAF and FLN tripeptide sequences (Fig. 6). To generalize this finding, we attempted the identification of peptides that bind to the ϵ-ear by screening of a phage display peptide library, but we only isolated a few nonspecific clones.3 Sequence comparisons to tepsin orthologs, however, were instructive. We found that all vertebrate tepsins share a conserved sequence at their C terminus that fits the 7-amino acid consensus motif S[AV]F[SA]FLN, in which the first Phe is considered position 0 (Fig. 7A). In non-vertebrate species we found a conserved but more degenerate motif that fits the consensus FXF[LIMV] (Fig. 7B). Although this motif is shorter than the vertebrate motif, it is worth noting that the bulky hydrophobic residues at positions 0, 2, and 3 are conserved. Whether this motif mediates interactions of the non-vertebrate protein orthologs remains to be experimentally tested.
Both tepsin motifs are different from previously-described motifs that mediate interactions with the ear domains of other adaptors. Although they all share the property of having bulky hydrophobic or aromatic amino acids as their key residues, the exact amino acids, their position within the sequence, the nature of the intervening amino acids, and the secondary structure of the sequence likely confer specificity to the interactions. Solution NMR studies (41) have shown that the β4-ear domain has a fold similar to that of the platform subdomain of the AP-2 β2 ear (8, 9, 10). Mutational analysis based on these structures allowed us to map the binding site for the [GS]LFXG[ML]X[LV] motif to a surface of the β4 ear (Fig. 5) that is at a location similar to the binding site for the α-helical [DE]nX1–2FXX[FL]XXXR motif (9, 10) and the linear DP[FW] motif (8) on the β2 ear. The bulky hydrophobic residues of the [GS]LFXG[ML]X[LV] motif are likely to anchor this sequence by fitting into hydrophobic pockets similar to thosethat accommodate the F and [FL] residues of the [DE]nX1–2FXX[FL]XXXR motif on the β2 ear (9, 10). The lack of a requirement of β4 residues Gln-643 and Glu-696 for tepsin binding (Fig. 5), however, differs from the involvement of the homologous β2 residues Gln-851 and Glu-902 in binding the Arg residue of the helical [DE]nX1–2FXX[FL]XXXR motif (9, 10). These differences likely account, at least in part, for the specificity of different motif-ear interactions. Although the RDSLFAGMELV sequence from tepsin has some probability of adopting an α-helical structure (Fig. 2F), the fact that the short sequence LFAGM is sufficient to confer binding to the β4 ear (Fig. 3C) suggests that it could bind in an extended conformation. Elucidation of the active conformation of the [GS]LFXG[ML]X[LV] motif will require the resolution of its atomic structure in complex with the β4 ear. To date, the atomic structure of the ϵ ear has not been reported, precluding structure-based mutational analyses of the S[AV]F[SA]FLN-binding site. The ϵ-ear is predicted to have both sandwich and platform domains homologous to those of the AP-2 α and β2 ears, and it is possible that the S[AV]F[SA]FLN motif will bind to sites similar to those that bind DPW (7, 11), WXX[FW]X[DE] (12, 13, 14) or FXDXF (11, 14) motifs on the AP-2 subunits.
The presence of two motifs, [GS]LFXG[ML]X[LV] and S[AV]F[SA]FLN, could enhance the avidity and specificity of tepsin for binding to a single AP-4 molecule (Fig. 9C). Indeed, co-precipitation analysis showed that both motifs are required for optimal interaction of tepsin with AP-4 in human cells (Fig. 8). In addition, these assays revealed that the central VHS/ENTH domain in tepsin also contributes to interaction with AP-4 in cells (Fig. 8), although such interaction was not evident in the Y2H assays using single β4 or ϵ subunits (Figs. 2 and 6). Perhaps both subunits, or other parts of the AP-4 complex, are required for interaction with the central VHS/ENTH domain. Previous affinity purifications using GST fused to the AP-4 β4- or ϵ-ear domains failed to identify any specific AP-4-binding partners, even when similar methods isolated a plethora of AP-2 interactors (10). In light of our findings on tepsin-AP-4 interactions, we think that both AP-4 ear domains are likely needed for the affinity purification approach to work. In addition to increasing avidity, the linkage of two motifs that bind to different ear domains could promote cross-linking of different AP-4 heterotetramers, thus contributing to the assembly of AP-4 coats in cells (Fig. 9C).
Both motifs were also found to contribute to recruitment of tepsin to the TGN, consistent with the fact that this recruitment requires binding to AP-4 (40). The cellular function of tepsin remains to be determined, although by analogy with ENTH/VHS-domain-containing proteins that are components of other coats (48, 49), it could contribute to stabilization of the AP-4 coat, vesicle budding, or recognition of additional cargos. The definition of consensus binding motifs reported here could allow the identification of additional AP-4 accessory proteins using bioinformatics approaches, as we previously did for the AP-1γ- and GGA-ear domains (22). A pattern search of eukaryotic genomes using the MyHits web server yielded 339 hits for the [GS]LFXG[ML]X[LV] motif. The same analysis for the S[AV]F[SA]FLN and FXF[LIMV] motifs produced 24 and “too many matches,” respectively. Without independent information on AP-4 interactors from other physical or genetic interaction studies, at present it is difficult to ascertain if any of these hits are relevant. Nevertheless, the identification of the determinants of tepsin interaction with AP-4 reported here should aid in the elucidation of the molecular and cellular mechanisms by which these proteins participate in cargo trafficking and development of the central nervous system.
Author Contributions
R. M. and J. S. B. conceived the project. R. M. performed most of the experiments. C. M. G. conducted the structural and immunofluorescence microscopy analyses. S. S. S. performed the phage display peptide screen. All authors contributed to the writing of the manuscript.
Acknowledgments
We thank Xiaolin Zhu for expert technical assistance, Tal Keren-Kaplan for gift of reagents, and David Gershlick for helpful discussions.
This work was funded by the Intramural Program of NICHD, National Institutes of Health (ZIA HD001607). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
S. S. Sidhu, unpublished results.
- AP
- adaptor protein
- AD
- GAL4 transcriptional activation domain
- AT
- 3-amino-1,2,4-triazole
- BD
- GAL4 DNA binding domain
- ITC
- isothermal titration calorimetry
- T-Ag
- SV40 large T antigen
- TAP
- tandem affinity purification
- TGN
- trans-Golgi network
- TSF
- Two-Strep/FLAG
- Y2H
- yeast two-hybrid.
References
- 1. Bonifacino J. S., and Glick B. S. (2004) The mechanisms of vesicle budding and fusion. Cell 116, 153–166 [DOI] [PubMed] [Google Scholar]
- 2. Robinson M. S. (2004) Adaptable adaptors for coated vesicles. Trends Cell Biol. 14, 167–174 [DOI] [PubMed] [Google Scholar]
- 3. Traub L. M., and Bonifacino J. S. (2013) Cargo recognition in clathrin-mediated endocytosis. Cold Spring Harb. Perspect. Biol. 5, a016790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kirchhausen T., Owen D., and Harrison S. C. (2014) Molecular structure, function, and dynamics of clathrin-mediated membrane traffic. Cold Spring Harb. Perspect. Biol. 6, a016725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Park S. Y., and Guo X. (2014) Adaptor protein complexes and intracellular transport. Biosci. Rep. 34, pii: e00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dell'Angelica E. C., Klumperman J., Stoorvogel W., and Bonifacino J. S. (1998) Association of the AP-3 adaptor complex with clathrin. Science 280, 431–434 [DOI] [PubMed] [Google Scholar]
- 7. Owen D. J., Vallis Y., Noble M. E., Hunter J. B., Dafforn T. R., Evans P. R., and McMahon H. T. (1999) A structural explanation for the binding of multiple ligands by the alpha-adaptin appendage domain. Cell 97, 805–815 [DOI] [PubMed] [Google Scholar]
- 8. Owen D. J., Vallis Y., Pearse B. M., McMahon H. T., and Evans P. R. (2000) The structure and function of the β2-adaptin appendage domain. EMBO J. 19, 4216–4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Edeling M. A., Mishra S. K., Keyel P. A., Steinhauser A. L., Collins B. M., Roth R., Heuser J. E., Owen D. J., and Traub L. M. (2006) Molecular switches involving the AP-2 beta2 appendage regulate endocytic cargo selection and clathrin coat assembly. Dev Cell 10, 329–342 [DOI] [PubMed] [Google Scholar]
- 10. Schmid E. M., Ford M. G., Burtey A., Praefcke G. J., Peak Chew S. Y., Mills I. G., Benmerah A., and McMahon H. T. (2006) Role of the AP2 beta-appendage hub in recruiting partners for clathrin coated vesicle assembly. PLos Biol. 4, e262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brett T. J., Traub L. M., and Fremont D. H. (2002) Accessory protein recruitment motifs in clathrin-mediated endocytosis. Structure 10, 797–809 [DOI] [PubMed] [Google Scholar]
- 12. Jha A., Agostinelli N. R., Mishra S. K., Keyel P. A., Hawryluk M. J., and Traub L. M. (2004) A novel AP-2 adaptor interaction motif initially identified in the long-splice isoform of synaptojanin 1, SJ170. J. Biol. Chem. 279, 2281–2290 [DOI] [PubMed] [Google Scholar]
- 13. Mishra S. K., Hawryluk M. J., Brett T. J., Keyel P. A., Dupin A. L., Jha A., Heuser J. E., Fremont D. H., and Traub L. M. (2004) Dual engagement regulation of protein interactions with the AP-2 adaptor alpha appendage. J. Biol. Chem. 279, 46191–46203 [DOI] [PubMed] [Google Scholar]
- 14. Praefcke G. J., Ford M. G., Schmid E. M., Olesen L. E., Gallop J. L., Peak-Chew S. Y., Vallis Y., Babu M. M., Mills I. G., and McMahon H. T. (2004) Evolving nature of the AP2 alpha-appendage hub during clathrin-coated vesicle endocytosis. EMBO J. 23, 4371–4383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mishra S. K., Keyel P. A., Edeling M. A., Dupin A. L., Owen D. J., and Traub L. M. (2005) Functional dissection of an AP-2 β2 appendage-binding sequence within the autosomal recessive hypercholesterolemia (ARH) protein. J. Biol. Chem. 280, 19270–19280 [DOI] [PubMed] [Google Scholar]
- 16. Kent H. M., McMahon H. T., Evans P. R., Benmerah A., and Owen D. J. (2002) Gamma-adaptin appendage domain: structure and binding site for Eps15 and γ-synergin. Structure 10, 1139–1148 [DOI] [PubMed] [Google Scholar]
- 17. Nogi T., Shiba Y., Kawasaki M., Shiba T., Matsugaki N., Igarashi N., Suzuki M., Kato R., Takatsu H., Nakayama K., and Wakatsuki S. (2002) Structural basis for the accessory protein recruitment by the γ-adaptin ear domain. Nat. Struct. Biol. 9, 527–531 [DOI] [PubMed] [Google Scholar]
- 18. Collins B. M., Praefcke G. J., Robinson M. S., and Owen D. J. (2003) Structural basis for binding of accessory proteins by the appendage domain of GGAs. Nat. Struct. Biol. 10, 607–613 [DOI] [PubMed] [Google Scholar]
- 19. Miller G. J., Mattera R., Bonifacino J. S., and Hurley J. H. (2003) Recognition of accessory protein motifs by the γ-adaptin ear domain of GGA3. Nat. Struct. Biol. 10, 599–606 [DOI] [PubMed] [Google Scholar]
- 20. Jürgens M. C., Vörös J., Rautureau G. J., Shepherd D. A., Pye V. E., Muldoon J., Johnson C. M., Ashcroft A. E., Freund S. M., and Ferguson N. (2013) The hepatitis B virus preS1 domain hijacks host trafficking proteins by motif mimicry. Nat. Chem. Biol. 9, 540–547 [DOI] [PubMed] [Google Scholar]
- 21. Mattera R., Arighi C. N., Lodge R., Zerial M., and Bonifacino J. S. (2003) Divalent interaction of the GGAs with the Rabaptin-5-Rabex-5 complex. EMBO J. 22, 78–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mattera R., Ritter B., Sidhu S. S., McPherson P. S., and Bonifacino J. S. (2004) Definition of the consensus motif recognized by γ-adaptin ear domains. J. Biol. Chem. 279, 8018–8028 [DOI] [PubMed] [Google Scholar]
- 23. Kametaka S., Moriyama K., Burgos P. V., Eisenberg E., Greene L. E., Mattera R., and Bonifacino J. S. (2007) Canonical interaction of cyclin G associated kinase with adaptor protein 1 regulates lysosomal enzyme sorting. Mol. Biol. Cell 18, 2991–3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Duncan M. C., Costaguta G., and Payne G. S. (2003) Yeast epsin-related proteins required for Golgi-endosome traffic define a γ-adaptin ear-binding motif. Nat Cell Biol. 5, 77–81 [DOI] [PubMed] [Google Scholar]
- 25. Mills I. G., Praefcke G. J., Vallis Y., Peter B. J., Olesen L. E., Gallop J. L., Butler P. J., Evans P. R., and McMahon H. T. (2003) EpsinR: an AP1/clathrin interacting protein involved in vesicle trafficking. J. Cell Biol. 160, 213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hirst J., Irving C., and Borner G. H. (2013) Adaptor protein complexes AP-4 and AP-5: new players in endosomal trafficking and progressive spastic paraplegia. Traffic 14, 153–164 [DOI] [PubMed] [Google Scholar]
- 27. Hirst J., Schlacht A., Norcott J. P., Traynor D., Bloomfield G., Antrobus R., Kay R. R., Dacks J. B., and Robinson M. S. (2014) Characterization of TSET, an ancient and widespread membrane trafficking complex. Elife 3, e02866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dell'Angelica E. C., Mullins C., and Bonifacino J. S. (1999) AP-4, a novel protein complex related to clathrin adaptors. J. Biol. Chem. 274, 7278–7285 [DOI] [PubMed] [Google Scholar]
- 29. Hirst J., Bright N. A., Rous B., and Robinson M. S. (1999) Characterization of a fourth adaptor-related protein complex. Mol. Biol. Cell 10, 2787–2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Simmen T., Höning S., Icking A., Tikkanen R., and Hunziker W. (2002) AP-4 binds basolateral signals and participates in basolateral sorting in epithelial MDCK cells. Nat Cell Biol. 4, 154–159 [DOI] [PubMed] [Google Scholar]
- 31. Boehm M., Aguilar R. C., and Bonifacino J. S. (2001) Functional and physical interactions of the adaptor protein complex AP-4 with ADP-ribosylation factors (ARFs). EMBO J. 20, 6265–6276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yap C. C., Murate M., Kishigami S., Muto Y., Kishida H., Hashikawa T., and Yano R. (2003) Adaptor protein complex-4 (AP-4) is expressed in the central nervous system neurons and interacts with glutamate receptor δ2. Mol. Cell Neurosci. 24, 283–295 [DOI] [PubMed] [Google Scholar]
- 33. Matsuda S., Miura E., Matsuda K., Kakegawa W., Kohda K., Watanabe M., and Yuzaki M. (2008) Accumulation of AMPA receptors in autophagosomes in neuronal axons lacking adaptor protein AP-4. Neuron 57, 730–745 [DOI] [PubMed] [Google Scholar]
- 34. Burgos P. V., Mardones G. A., Rojas A. L., daSilva L. L., Prabhu Y., Hurley J. H., and Bonifacino J. S. (2010) Sorting of the Alzheimer's disease amyloid precursor protein mediated by the AP-4 complex. Dev. Cell 18, 425–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Verkerk A. J., Schot R., Dumee B., Schellekens K., Swagemakers S., Bertoli-Avella A. M., Lequin M. H., Dudink J., Govaert P., van Zwol A. L., Hirst J., Wessels M. W., Catsman-Berrevoets C., Verheijen F. W., de Graaff E., de Coo I. F., Kros J. M., Willemsen R., Willems P. J., van der Spek P. J., and Mancini G. M. (2009) Mutation in the AP4M1 gene provides a model for neuroaxonal injury in cerebral palsy. Am. J. Hum. Genet. 85, 40–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Abou Jamra R., Philippe O., Raas-Rothschild A., Eck S. H., Graf E., Buchert R., Borck G., Ekici A., Brockschmidt F. F., Nöthen M. M., Munnich A., Strom T. M., Reis A., and Colleaux L. (2011) Adaptor protein complex 4 deficiency causes severe autosomal-recessive intellectual disability, progressive spastic paraplegia, shy character, and short stature. Am. J. Hum. Genet. 88, 788–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moreno-De-Luca A., Helmers S. L., Mao H., Burns T. G., Melton A. M., Schmidt K. R., Fernhoff P. M., Ledbetter D. H., and Martin C. L. (2011) Adaptor protein complex-4 (AP-4) deficiency causes a novel autosomal recessive cerebral palsy syndrome with microcephaly and intellectual disability. J. Med .Genet. 48, 141–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bauer P., Leshinsky-Silver E., Blumkin L., Schlipf N., Schröder C., Schicks J., Lev D., Riess O., Lerman-Sagie T., and Schöls L. (2012) Mutation in the AP4B1 gene cause hereditary spastic paraplegia type 47 (SPG47). Neurogenetics 13, 73–76 [DOI] [PubMed] [Google Scholar]
- 39. Hardies K., May P., Djémié T., Tarta-Arsene O., Deconinck T., Craiu D., AR working group of the EuroEPINOMICS RES Consortium, Helbig I., Suls A., Balling R., Weckhuysen S., De Jonghe P., and Hirst J. (2015) Recessive loss-of-function mutations in AP4S1 cause mild fever-sensitive seizures, developmental delay and spastic paraplegia through loss of AP-4 complex assembly. Hum. Mol. Genet. 24, 2218–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Borner G. H., Antrobus R., Hirst J., Bhumbra G. S., Kozik P., Jackson L. P., Sahlender D. A., and Robinson M. S. (2012) Multivariate proteomic profiling identifies novel accessory proteins of coated vesicles. J. Cell Biol. 197, 141–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Eletsky A., Rotshteyn D. J., Pederson K., Shastry R., Maglaqui M., Janjua H., Xiao R., Everett J. K., Montelione G. T., Prestegard J. H., Szyperski T., and Northeast Structural Genomics Consortium, N. S. G. C.) (2013) Solution NMR structure of β-adaptin appendage domain of human adaptor protein complex 4 subunit β. DB ID: 2MJ7 [Google Scholar]
- 42. Pu J., Schindler C., Jia R., Jarnik M., Backlund P., and Bonifacino J. S. (2015) BORC, a multiprotein complex that regulates lysosome positioning. Dev. Cell 33, 176–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ohno H., Fournier M. C., Poy G., and Bonifacino J. S. (1996) Structural determinants of interaction of tyrosine-based sorting signals with the adaptor medium chains. J. Biol. Chem. 271, 29009–29015 [DOI] [PubMed] [Google Scholar]
- 44. Sidhu S. S., Lowman H. B., Cunningham B. C., and Wells J. A. (2000) Phage display for selection of novel binding peptides. Methods Enzymol. 328, 333–363 [DOI] [PubMed] [Google Scholar]
- 45. Ohno H., Stewart J., Fournier M. C., Bosshart H., Rhee I., Miyatake S., Saito T., Gallusser A., Kirchhausen T., and Bonifacino J. S. (1995) Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science 269, 1872–1875 [DOI] [PubMed] [Google Scholar]
- 46. Owen D. J., and Evans P. R. (1998) A structural explanation for the recognition of tyrosine-based endocytotic signals. Science 282, 1327–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mardones G. A., Burgos P. V., Lin Y., Kloer D. P., Magadán J. G., Hurley J. H., and Bonifacino J. S. (2013) Structural Basis for the Recognition of Tyrosine-based Sorting Signals by the μ3A Subunit of the AP-3 Adaptor Complex. J. Biol. Chem. 288, 9563–9571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Legendre-Guillemin V., Wasiak S., Hussain N. K., Angers A., and McPherson P. S. (2004) ENTH/ANTH proteins and clathrin-mediated membrane budding. J. Cell Sci. 117, 9–18 [DOI] [PubMed] [Google Scholar]
- 49. Sen A., Madhivanan K., Mukherjee D., and Aguilar R. C. (2012) The epsin protein family: coordinators of endocytosis and signaling. Biomol. Concepts 3, 117–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Paczkowski J. E., Richardson B. C., Strassner A. M., and Fromme J. C. (2012) The exomer cargo adaptor structure reveals a novel GTPase-binding domain. EMBO J. 31, 4191–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Humphrey W., Dalke A., and Schulten K. (1996) VMD: visual molecular dynamics. J. Mol. Graph 14, 33- 8, 27–28 [DOI] [PubMed] [Google Scholar]
- 52. Dolinsky T. J., Nielsen J. E., McCammon J. A., and Baker N. A. (2004) PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 32, W665–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dolinsky T. J., Czodrowski P., Li H., Nielsen J. E., Jensen J. H., Klebe G., and Baker N. A. (2007) PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 35, W522–5 [DOI] [PMC free article] [PubMed] [Google Scholar]