Background: The genome of Streptococcus pneumoniae encodes a second uncharacterized family 20 glycoside hydrolase.
Results: GH20C displays activity on both terminal β-linked N-acetylglucosamine and N-acetylgalactosamine.
Conclusion: GH20C is an enzyme able to cleave a wide variety of N-acetylhexosamine-terminating sugars.
Significance: S. pneumoniae has the biochemical ability to act on a wide variety of sugars that it would encounter in the human body.
Keywords: enzyme inhibitor, glycobiology, glycoside hydrolase, Streptococcus, x-ray crystallography
Abstract
An important facet of the interaction between the pathogen Streptococcus pneumoniae (pneumococcus) and its human host is the ability of this bacterium to process host glycans. To achieve cleavage of the glycosidic bonds in host glycans, S. pneumoniae deploys a wide array of glycoside hydrolases. Here, we identify and characterize a new family 20 glycoside hydrolase, GH20C, from S. pneumoniae. Recombinant GH20C possessed the ability to hydrolyze the β-linkages joining either N-acetylglucosamine or N-acetylgalactosamine to a wide variety of aglycon residues, thus revealing this enzyme to be a generalist N-acetylhexosaminidase in vitro. X-ray crystal structures were determined for GH20C in a ligand-free form, in complex with the N-acetylglucosamine and N-acetylgalactosamine products of catalysis and in complex with both gluco- and galacto-configured inhibitors O-(2-acetamido-2-deoxy-d-glucopyranosylidene)amino N-phenyl carbamate (PUGNAc), O-(2-acetamido-2-deoxy-d-galactopyranosylidene)amino N-phenyl carbamate (GalPUGNAc), N-acetyl-d-glucosamine-thiazoline (NGT), and N-acetyl-d-galactosamine-thiazoline (GalNGT) at resolutions from 1.84 to 2.7 Å. These structures showed N-acetylglucosamine and N-acetylgalactosamine to be recognized via identical sets of molecular interactions. Although the same sets of interaction were maintained with the gluco- and galacto-configured inhibitors, the inhibition constants suggested preferred recognition of the axial O4 when an aglycon moiety was present (Ki for PUGNAc > GalPUGNAc) but preferred recognition of an equatorial O4 when the aglycon was absent (Ki for GalNGT > NGT). Overall, this study reveals GH20C to be another tool that is unique in the arsenal of S. pneumoniae and that it may implement the effort of the bacterium to utilize and/or destroy the wide array of host glycans that it may encounter.
Introduction
Glycoside hydrolase family 20 (GH20)3 is a large family whose members are all N-acetylhexosaminidases that use a substrate-assisted catalytic mechanism. Detailed studies of GH20 enzymes, and members of other families using the same general mechanism, show this neighboring group catalytic mechanism to proceed in two steps with the acetamido group of the hexosamine participating as a nucleophile, resulting in the formation of a transient oxazoline intermediate, with the product ultimately retaining the stereochemistry of the anomeric carbon (1–6). The catalytic residues in GH20 enzymes are found as a conserved Asp/Glu pair. The Glu residue acts as the general catalytic acid/base, whereas the Asp is thought to stabilize the oxazoline intermediate (5, 7). Although the catalytic mechanism of this family is well understood and there are currently over 100 characterized family members, including x-ray crystal structures of 14 members, the molecular details imparting glycon (terminal hexosamine) and aglycon (leaving group on reducing side of the terminal sugar) specificities among the family are less well known. Indeed, some family members are N-acetylgalactosaminidases, whereas others are N-acetylglucosaminidases; most are exo-acting but some are endo-acting (8, 9). Aglycon specificity is impossible to predict in this family, but a relevant example where aglycon recognition is an important factor in biological processes includes the human hexosaminidases A and B (HexA and HexB). HexA and HexB are responsible for the cleavage of terminal N-acetylgalactosamine (GalNAc) from the GM2 ganglioside (GalNAcβ(1–4)-[N-acetylneuraminic acid α(2–3)-]-Galβ(1–4)-Glc-ceramide)within the lysosome (10, 11). Mutations in the genes encoding either of these enzymes can lead to lysosomal storage disorders with sometimes severe pathologies (12, 13).
GH20 enzymes are common in microbes where their known or predicted specificities suggest a wide variety of biological roles. A notable example whose discovery dates back over 50 years is StrH, which is an exo-β-d-N-acetylglucosaminidase able to release terminal N-acetylglucosamine (GlcNAc) residues from complex carbohydrates. It was first purified from Streptococcus pneumoniae culture supernatants by Hughes and Jeanloz (14), later characterized in detail by Yamashita et al. (15), and the gene encoding StrH was ultimately identified by Clarke et al. (16). This enzyme contains two tandem catalytic GH20 modules; together, these modules and their structural features give StrH specificity for the mannose arms of N-linked glycans (i.e. the aglycon) making the enzyme very specific for terminal GlcNAc residues in complex N-linked glycans (9). In addition to its defining specificity, this cell wall-bound surface protein is implicated in virulence through a potential role in host complement modulation and nutrient acquisition (9).
The role of StrH in the host-S. pneumoniae interaction is an example of an expanding paradigm of how this bacterium attacks and utilizes host glycans to inhabit and invade human tissues (9, 17). S. pneumoniae is a commensal bacterium inhabiting the human upper respiratory tract of healthy individuals, but in some circumstances it can progress beyond the nasopharynx to other normally sterile sites and cause diseases ranging from comparatively minor infections, like otitis media, to lethal infections such as pneumonia, bacteremia, and meningitis (18). The occurrence of S. pneumoniae infections remains high in both developed and developing countries with high-risk groups, including children, the elderly, and immuno-compromised individuals. Approximately 15 million cases of invasive S. pneumoniae infections are reported annually with ∼1.6 million of these resulting in death (19). Given that the primary route of entry and/or colonization niche is the human nasopharynx, S. pneumoniae encounters an environment that is rich in the complex carbohydrates displayed by the host tissues. In keeping with this, large scale screening has suggested that a significant number of genes encoding putative and known glycoside hydrolases, including StrH, may participate in virulence (20–22). These are increasingly being validated as important components of the host-bacterium interaction, such as nutritional support, enhanced adherence, and immune system modulation (9, 17, 23–28).
It was from the perspective of the importance of carbohydrate-processing enzymes to S. pneumoniae virulence and the role of StrH in this that we noted the presence in the S. pneumoniae genome of a second gene encoding a family 20 glycoside hydrolase. This gene appears to be present in all sequenced strains of the bacterium. The putative product of this gene, referred to as GH20C, displays ∼70% amino acid sequence identity with the structurally characterized Streptococcus gordonii GcnA, which displays both N-acetylgalactosaminidase and N-acetylglucosaminidase activity on synthetic aryl glycosides (29, 30). Although the relatively high amino acid sequence identity between GcnA and GH20C lends itself to inferring similar abilities to cleave N-acetylhexosaminides, GH20C remains uncharacterized. Likewise, the aglycon specificities of both GcnA and GH20C are unknown. Given the presence of an N-glycan-specific N-acetylglucosaminidase, StrH, in S. pneumoniae, we hypothesized that GH20C is present to expand the ability of the bacterium to process N-acetylhexosamine-containing host glycans, rather than having an overlapping specificity. Through structural and biochemical analyses of recombinant GH20C, we show that this enzyme is able to cleave terminal GalNAc and GlcNAc residues and that it has only limited selectivity for aglycon sugars. These results suggest that rather than being redundant with StrH, GH20C is likely responsible for the cleavage of terminal N-acetylhexosamine residues that StrH cannot hydrolyze and, in particular, provides N-acetyl-β-d-galactosaminidase activity that does not appear to be present in any other known or predicted S. pneumoniae enzymes.
Experimental Procedures
Materials
Oligosaccharides were obtained from V-labs (Covington, LA). PUGNAc and NGT synthesized in-house were kind gifts from Dr. David Vocadlo. GalPUGNAc and GalNGT synthesized in-house were kind gifts from Dr. Keith Stubbs. All other reagents, including GlcNAc and GalNAc, were purchased from Sigma unless otherwise specified.
Cloning
The gene fragment encoding GH20C (amino acids 1–626) was PCR-amplified from S. pneumoniae TIGR4 genomic DNA using the oligonucleotide primers GH20C-Fw (5′-CATATCGCTAGCATGGTAAGATTTACAG) and GH20C-Rv (5′-GGTGGTCTCGAGTTAAGTCGTATAAATC). The amplified DNA fragment was cloned using standard procedures into pET 28a (Novagen) using 5′ NheI and 3′ XhoI restriction sites, respectively. The resulting plasmid, pGH20C, encoded the desired polypeptide fused to an N-terminal hexa-histidine tag by a thrombin protease cleavage site. Standard PCR site-directed mutagenesis procedures were used to introduce the E223Q substitution into pGH20C using the oligonucleotide primers GH20CE223Q-Fw (5′-ATCGGGATGGACCAGGCCCACTTGGTT) and GH20CE223Q-Rv (5′-AACCAAGTGGGCCTGGTCCATCCCGAT) (52). The DNA sequence fidelity of all constructs was verified by bidirectional sequencing.
Protein Production and Purification
All constructs were transformed into the expression strain, Escherichia coli BL21 Star (DE3) strain. Six liters of yeast/tryptone broth, containing 50 μg/ml kanamycin, was inoculated with the transformed cells and incubated at 37 °C. Once an optical density of ∼1 at 595 nm was reached, protein production was induced by the addition of isopropyl 1-thio-β-d-galactopyranoside to a final concentration of 0.5 mm. Incubation of the cultures was continued overnight with shaking at 16 °C. Cells were harvested by centrifugation at 5000 × g for 10 min and ruptured by chemical lysis. The cleared supernatant of the cell lysate was loaded onto a Ni2+-nitrilotriacetic acid-immobilized metal affinity chromatography column. Polypeptide was eluted with binding buffer (20 mm Tris, pH 8.0) containing increasing concentrations of imidazole (0–500 mm). GH20C was concentrated and buffer-exchanged into 20 mm Tris, pH 8.0, in a stirred ultrafiltration unit (Amicon). Purified protein was further purified by size exclusion chromatography using a Sephacryl S-200 column (GE Healthcare) and concentrated using a stirred cell ultrafiltration device (Amicon) with a 10,000-Da molecular mass cutoff membrane (Millipore).
Determination of Protein Concentration
Protein concentration was determined by UV absorbance at 280 nm using the calculated extinction coefficient (31) of 94,200 m−1 cm−1.
Enzyme Kinetics and Inhibition
All steady state kinetic studies were performed at 37 °C in a Cary/Varian 300 Bio UV-visible spectrophotometer. The pH optimum was determined using an end-point assay (stopped with 100 mm NaOH) in McIIvaine buffer at pH values ranging from 2.25 to 9.2; the optimum activity was found at 6.5 (data not shown). Determination of kinetic constants was done in triplicate 1-ml assays containing 50 mm citrate buffer, pH 6.5, 0.1% BSA, 2 nm GH20C, and 0–0.9 mm 4-nitrophenyl-N-acetylglucosaminide (pNP-GlcNAc)or 0–0.45 mm 4-nitrophenyl-N-acetylgalactosaminide (pNP-GalNAc). Nitrophenolate production was monitored at 400 nm; rates were calculated using an extinction coefficient, ϵ400 nm, of 6090 m−1 cm−1. Michaelis-Menten parameters were determined using GraphPad by nonlinear curve fitting. The Ki values for the inhibition by PUGNAc, GalPUGNAc, NGT, and GalNGT were determined by Dixon plots analysis using pNP-GlcNAc as a substrate.
Activity Screen
Qualitative analysis of GH20C activity on di- and trisaccharides was done by thin layer chromatography using a variety of oligosaccharides containing non-reducing terminal GlcNAc or GalNAc. Reactions were done in a 20-μl final volume containing 50 mm Tris, pH 8, 30 nm GH20C, and 3 mm sugar. Reactions were incubated at 37 °C for 1 h and then spotted on a silica plate. TLC was run for 2 h in n-propyl alcohol/H2O/ethanol (7:2:1) solvent, and oligosaccharides were revealed by using 5% sulfuric acid and a 30-min incubation at 110 °C.
General Crystallography Procedures
Crystals were obtained at 18 °C using sitting-drop vapor diffusion for screening and hanging drop vapor diffusion for optimization. For data collection, single crystals were flash-cooled with liquid nitrogen in crystallization solution previously dehydrated to increase cryoprotectant content for each crystal form as given below. Diffraction data were collected either on a “home-beam” comprising a Rigaku R-AXIS 4++ area detector coupled to an MM-002 x-ray generator with Osmic “blue” optics and an Oxford Cryostream 700, Beamline 9-2 of the Stanford Linear Accelerator Center (SLAC, Stanford Synchrotron Radiation Lightsource (SSRL), Stanford, CA), or 08ID-1 at the Canadian Light Source (Saskatoon, Saskatchewan) as indicated in Table 1. Most diffraction data were processed using MOSFLM and SCALA (32). However, the NGT and PUGNAc complexes were processed with HKL2000 and XDS, respectively. All data collection and processing statistics are shown in Table 1. For all structures, manual model building was performed with COOT (33), and refinement of atomic coordinates was performed with REFMAC (34). The addition of water molecules was performed in COOT with FINDWATERS and manually checked after refinement. In all data sets, refinement procedures were monitored by flagging 5% of all observations as “free” (35). Model validation was performed with MOLPROBITY (36).
TABLE 1.
Data collection and structure refinement statistics
Values in parentheses are for the highest resolution shell. r.m.s.d., root-mean-square deviation.
| Native | GlcNAc | GalNAc | NGT | GalNGT | PUGNAc | GalPUGNAc | |
|---|---|---|---|---|---|---|---|
| Data collection statistics | |||||||
| Wavelength | 1.5418 | 1.03322 | 0.97946 | 0.97926 | 0.97965 | 1.12709 | 1.12709 |
| Beamline | MM-002 | CLS ID-1 | SSRL BL9-2 | CLS ID-1 | CLS ID-1 | SSRL BL9-2 | SSRL BL9-2 |
| Processing software | MOSFLM | MOSFLM | MOSFLM | HKL2000 | MOSFLM | XDS | MOSFLM |
| Space group | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 | C2221 |
| Resolution (Å) | 50.70–1.94 (2.05–1.94) | 46.60–1.84 (1.94–1.84) | 79.54–2.08 (2.19–2.08) | 40.00–2.70 (2.75 − 2.70) | 95.69–2.10 (2.21–2.10) | 38.66–1.78 (1.81–1.78) | 19.93–2.20 (2.28–2.20) |
| Cell dimension a, b, c (Å) | 92.8, 136.9, 202.3 | 87.7, 98.1, 132.9 | 95.6, 99.1, 133.5 | 83.5, 96.7, 132.6 | 95.7, 99.4, 133.2 | 93.7, 136.9, 201.4 | 85.8, 95.6, 131.6 |
| Rmerge | 0.106 (0.337) | 0.079 (0.323) | 0.101 (0.389) | 0.076 (0.423) | 0.115 (0.243) | 0.042 (0.329) | 0.079 (0.480) |
| Completeness (%) | 97.4 (96.9) | 100.0 (100.0) | 98.2 (96.0) | 95.4 (94.2) | 92.9 (84.5) | 99.6 (93.2) | 99.8 (98.6) |
| I/σI | 9.2 (3.9) | 18.0 (6.2) | 20.1 (6.2) | 24.5 (5.2) | 11.1 (6.0) | 28.2 (3.9) | 11.0 (2.8) |
| Redundancy | 3.8 (3.7) | 8.1 (8.0) | 11.1 (9.8) | 7.9 (8.3) | 6.4 (5.9) | 7.8 (4.8) | 6.4 (6.4) |
| Total reflections | 704,154 ( 98,506) | 816,738 (115,612) | 833,007 (102,853) | 227,615 (11,512) | 444,859 (52,848) | 1,943,027 (54,993) | 175,575 (17,093) |
| Unique reflections | 184,285 (26,532) | 100,651 (14,523) | 74,999 (10,549) | 28,812 (1,387) | 69,114 (9,010) | 248,507 (11,400) | 27,418 (2,499) |
| Refinement statistics | |||||||
| Rwork/Rfree (%) | 0.21/0.26 | 0.17/0.20 | 0.16/0.20 | 0.21/0.28 | 0.17/0.22 | 0.16/0.20 | 0.23/0.29 |
| r.m.s.d. | |||||||
| Bond lengths (Å) | 0.009 | 0.008 | 0.010 | 0.008 | 0.010 | 0.019 | 0.013 |
| Bond angles (degrees) | 1.278 | 1.149 | 1.334 | 1.218 | 1.329 | 1.828 | 1.526 |
| Average B-factors (Å2) | |||||||
| Protein molecules | 20.1 | 18.3 | 17.8 | 40.8 | 14 | 24.8 | 46.3 |
| Solvent molecules | 23.2 (H2O) | 21.4 (H2O) | 18.8 (H2O) | 27.6 (H2O) | 21.0 (H2O) | 32.3 (H2O) | 37.6 (H2O) |
| 25.1 (EDO) | 35.7 (GOL) 33.2 (EDO) | 60.6 (EDO) | 23.8 (MG) | ||||
| Ligand | 25.7 | 13.1 | 26.4 | 8.6 | 25.4 | 43.4 | |
| No. of atoms | |||||||
| Protein atoms | 19,641 | 9941 | 10,071 | 9108 | 10,011 | 19,394 | 4380 |
| Solvent atoms | 1618 (H2O) | 592 (H2O) | 621 (H2O) | 79 (H2O) | 1095 | 1958 (H2O) | 38 (H2O) |
| 4 (EDO) | 4 (EDO) 12 (GOL) | 4 EDO | 3 (MG) | ||||
| Ligand atoms | 30 | 30 | 28 | 28 | 100 | 25 | |
| Ramachandran statistics | |||||||
| Most favored (%) | 96.7 | 97.4 | 97.3 | 94.9 | 97.4 | 97.1 | 93.3 |
| Additional allowed (%) | 3.1 | 2.4 | 2.6 | 5.2 | 2.5 | 2.7 | 6.3 |
| Disallowed (%) | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 | 0.4 |
GH20C Structural Determination
All crystals of GH20C (20 mg/ml) were grown in 12% (w/v) PEG 3350, 0.2 m magnesium chloride, 0.05 m CAPS, pH 10.2, and 1% glycerol. Crystals were soaked for 5 min to several hours in the crystallization solution containing an excess of reaction products and inhibitors. Data sets were collected as above on a single crystal that was cryoprotected by exposure of the crystallization drop to air to dehydrate it. The structure of GcnA (Protein Data Bank code 2EPN) was used to solve the native structure of GH20C by molecular replacement using PHASER. For all the complex structures, GH20C native structure was used as a search model for molecular replacement using PHASER.
Generation of Δgh20C Strain
A PCR ligation technique was used to replace gh20C with a chloramphenicol-resistant cassette (37). Regions flanking the gh20C gene and the chloramphenicol-resistant cassette were amplified by PCR with primers containing specific restriction sites. The flanking region 1 was amplified using the forward primer 5′-AATACTAAGCCTACACAGACTGATTTCAA-3′ and the reverse primer 5′-CATATGCATATGCAGCGTCGACGATTTATACG-3′ containing the NdeI restriction site. The second flanking region was amplified using the forward primer 5′-CATATGCTCGAGCCTCAATAGCTTGCGTTTGTT-3′ containing the XhoI restriction site and the reverse primer 5′-TGGCTAGATCAACGCTTGTCAGAACAGGA-3′. The chloramphenicol-resistance cassette was amplified with the CAM-Fw primer (5′-TTTGGACATATGGATGAAAATTTGTTTGAT-3′) containing an NdeI restriction site and the CAM-Rv (5′-CTTTGTCTCGAGGTACAGTCCGGCATTATC-3′) containing XhoI restriction site. Each amplicon was digested with the appropriate restriction enzyme and ligated together. This ligation reaction was subsequently used to transform S. pneumoniae TIGR4 creating the Δgh20C strain where the gh20C gene was replaced by the chloramphenicol-resistance gene. The S. pneumoniae transformation protocol was adapted from Bricker and Camilli (38). Bacteria were grown for about 9 h in Todd Hewitt broth to an approximate A600 of 0.8. A 1:50 dilution was then made into the pre-induction growth medium of Todd Hewitt broth, 0.5% (w/v) glycine, 11 mm HCl and grown at 37 °C in a candle jar until an A600 of 0.03 was reached. The media were then supplemented with 10 mm NaOH, 0.2% (w/v) BSA, 1 mm CaCl2, and 100 ng/ml competence-stimulating peptide 2 (CSP-2; synthesized by GenScript) in their respective order and incubated in a candle jar at 37 °C for 14 min. The entire ligation reaction was then added to the media and subsequently incubated for 1 h at 37 °C in a candle jar. The transformation reaction was then diluted 1:4 with Todd Hewitt broth and incubated further for 2 h. The transformations were plated on sheep blood agar supplemented with chloramphenicol and incubated overnight in a candle jar at 37 °C. Colonies were re-streaked onto new blood agar plates, and the genomic position of the chloramphenicol cassette was confirmed by several PCR analyses.
RT-PCR and Western Blotting
S. pneumoniae GH20C mRNA levels when exposed to different carbohydrates were determined by quantitative reverse transcription PCR. S. pneumoniae TIGR4 was grown in a casein hydrolysate-based medium with 0.3% sucrose and 0.25% yeast extract (AGCHY) (39, 40) supplemented with 0.5% glucose at 37 °C with 5% CO2 to an A600 of 0.4. Bacteria were pelleted and washed with AGCHY, then resuspended in AGCHY or AGCHY with 0.5% sugar, and incubated at 37 °C with 5% CO2 for 30 min. Bacteria were lysed using lysozyme and mutanolysin, and RNA was extracted using the Qiagen RNeasy Plus mini kit. Removal of genomic DNA contamination was carried out by DNase I treatment (Roche Applied Science) with subsequent quantitative RT-PCR performed using SuperScript III one-step RT-PCR kit (Invitrogen) on an LC480 real time cycler (Roche Applied Science). GH20C transcription levels were analyzed using primers CTGGTTGGCTGTAGTTGCTG and CGAGGTTTATCTGGCTGGTC and data normalized to expression levels for 16S rRNA using primers CATGCAAGTAGAACGCTGAA and TGTCATGCAACATCCACTCT. Each condition was performed in biological triplicates, and the fold change relative to expression levels when the bacterium was not exposed to sugar was calculated.
S. pneumoniae TIGR4 was streaked out on trypticase soy agar plates supplemented with defibrinated sheep blood and incubated overnight at 37 °C in a candle jar. Culture was started by inoculating 5 ml of AGCHY media supplemented with 1% d-galactose and incubated for 6 h in a candle jar at 37 °C. The S. pneumoniae TIGR4 bacterial pellet was resuspended in phosphate-buffered saline, pH 7.4 (PBS), with protease inhibitor (Roche Applied Science) and lysed by sonication to yield the lysate fraction. Supernatant from the S. pneumoniae TIGR4 culture was filter-sterilized to remove any residual pneumococcal cells. Trichloroacetic acid precipitation was performed to concentrate secreted proteins. For that, 1 part 100% trichloroacetic acid (w/v) was added to 4 parts supernatant, left on ice for ∼30 min, and centrifuged for 5 min at 14,000 rpm. The precipitated pellet was washed twice with cold acetone, dried at room temperature, and resuspended in SDS-PAGE loading buffer yielding the extracellular fraction. Laemmli buffer was added to both extracellular and lysate samples, and samples were incubated at 100 °C for 10 min. Samples were run on a 10% SDS-PAGE, transferred to a nitrocellulose membrane (Whatman), and analyzed by Western blot using a 1:50 dilution of rabbit anti-serum raised against purified recombinant GH20C from TIGR4 strain (antibodies were raised by ImmunoPrecise Antibodies Ltd., Victoria, British Columbia, Canada). Horseradish peroxidase-conjugated anti-rabbit (GE NA934) secondary antibody was used at 1:5000. Proteins were detected by chemiluminescence (Millipore WBLUF0500) and exposed to film.
Results
gh20C Gene Is Transcribed and a Protein Product Produced
Using quantitative PCR, we detected the presence of gh20C transcripts in response to exposure to different monosaccharides as the only source of carbon and determined the change in expression of the transcript relative to expression in the absence of a carbohydrate source (Fig. 1A). We observed a statistically significant reduction in the detection of transcript in the presence of glucose and mannose, and the largest reduction was in the presence of GlcNAc. No change was observed in the presence of galactose, fucose, or GalNAc. This result indicates under the conditions used that, in general, the gh20C gene is expressed and that it is expressed in the absence of added carbohydrate but is repressed in the presence of glucose, mannose, and GlcNAc. To confirm that the production of gh20C transcript also results in the production of a protein product, we performed Western blotting for GH20C using a polyclonal antibody raised against recombinant GH20C. The detection of a protein with similar electrophoretic migration to recombinant GH20C protein was detected by Western blotting in the lysate and supernatant fractions of wild-type S. pneumoniae TIGR4 cultures (Fig. 1B). Although antibody-reactive bands were also detected in the lysate and supernatant fractions of a Δgh20C mutant strain created from the S. pneumoniae TIGR4 parent, the band with the same migration properties as the recombinant GH20C was absent in both the lysate and supernatant fractions of this mutant (Fig. 1B). Thus, the gh20C gene is transcribed and translated in this bacterium with the regulation being, at least in part, consistent with catabolite repression. Our crude fractionation indicates that GH20C is found associated with the cells and in the culture supernatant, albeit to a lesser degree in the latter fraction. The detection of GH20C in the supernatant fraction is not consistent with the absence of known motifs that target the protein for secretion; however, the ability of S. pneumoniae to non-classically secrete proteins that do not possess a signal peptide or a known anchoring motif is well documented (41).
FIGURE 1.
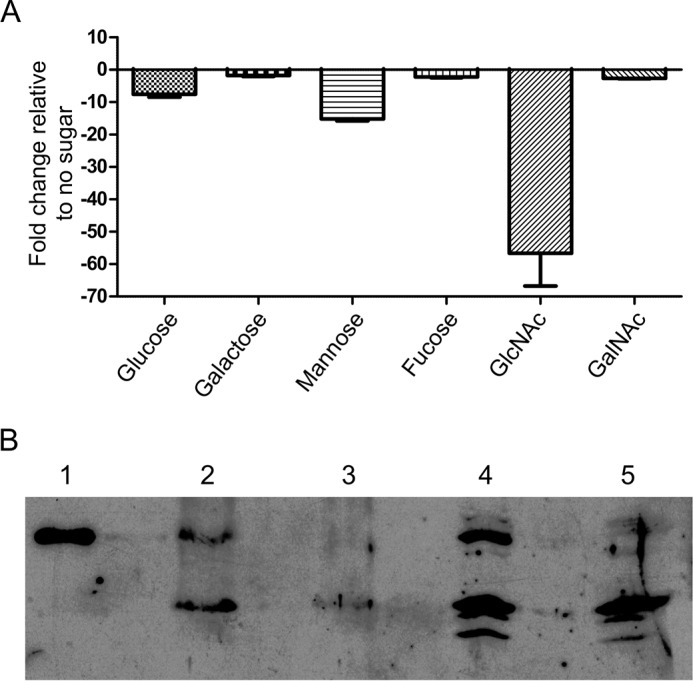
GH20C expression. A, fold changes of GH20C transcript were measured by quantitative RT-PCR after exposure of TIGR4 to different carbohydrates. The fold change relative to expression levels when the bacterium was not exposed to sugar was calculated, and standard deviations of three replicates are indicated on each bar. B, production of GH20C protein by S. pneumoniae was detected by Western blotting using a polyclonal antibody raised against recombinant GH20C. Wild-type and Δgh20C mutant of S. pneumoniae TIGR4 were grown for 6 h at 37 °C in AGCH media and harvested by centrifugation. The cells were then lysed by sonication, and the culture supernatant and lysed cellular fractions were analyzed by Western blotting. Lane 1, recombinant GH20C; lane 2, TIGR4 wild-type supernatant; lane 3, Δgh20C mutant supernatant; lane 4, TIGR4 wild-type lysate; lane 5, Δgh20C mutant lysate.
Activity of GH20C
The complete open reading frame of the gh20C gene (1878 bp) was cloned in-frame with an N-terminal hexa-histidine tag. The product of this gene fusion, referred to as GH20C, was produced in E. coli and purified to apparent homogeneity, as judged by SDS-PAGE, by immobilized metal affinity chromatography and size exclusion chromatography. In qualitative analyses, recombinant GH20C displayed activity on both 4-nitrophenyl N-acetyl-β-d-glucosaminide (pNP-GlcNAc) and 4-nitrophenyl N-acetyl-β-d-galactosaminide (pNP-GalNAc). Using pNP-GlcNAc, the pH optimum of the enzyme was determined to be 6.5, and subsequent kinetic characterization at this pH revealed a Km of 560 (±60) μm, a kcat of 3650 (±330) min−1, and a kcat/Km of 6.5 (±0.6) min−1·μm−1. Kinetic analysis using pNP-GalNAc gave a Km of 262 (±14) μm, a kcat of 1100 (±59) min−1, and a kcat/Km of 4.5 (±0.2) min−1·μm−1. Thus, these two synthetic substrates displayed compensatory changes in Km and kcat to result in similar overall efficiency of cleavage, as represented by kcat/Km, for the two substrates.
Given the apparent lack of preference of GH20C for GlcNAc or GalNAc as the glycon, we probed potential aglycon selectivity using thin layer chromatography to qualitatively examine the activity of GH20C on a variety of biologically relevant GlcNAc- and GalNAc-terminating sugars. Consistent with its activity on pNP-GlcNAc and pNP-GalNAc, GH20C displayed the ability to cleave a variety of sugars with terminal β-linked GlcNAc and GalNAc (Table 2). The tested substrates encompassed galactose (Gal), mannose (Man), and GalNAc as aglycon residues linked β1–2, β1–3, β1–4, or β1–6 to the glycon, indicating broad substrate cleavage capabilities. Notably, GH20C was able to cleave the trisaccharide β-d-GlcNAc-(1→6)-[β-d-Gal-(1→3)]-α-d-GalNAc, suggesting the ability to accommodate more complex aglycons. The only substrate that was not observed to be cleaved was β-d-GalNAc-(1→4)-β-d-Gal. Because of the β-1,4-linkage and axial configuration of C4 in the Gal residue of this disaccharide, the sugar would adopt a conformation with a significant angle between the planes of the pyranose rings (i.e. a kinked or pleated conformation). This feature separates it from the other tested sugars where the composite pyranose rings would be roughly coplanar resulting in an extended substrate conformation.
TABLE 2.
Activity screen of GH20C
| Compound | Oligosaccharide | Activity |
|---|---|---|
| 1 | β-d-GlcNAc- (1→6)-[β-d-Gal-(1→3)]-α-d-GalNAc | + |
| 2 | β-d-GalNAc-(1→3)-β-d-Gal | + |
| 3 | β-d-GalNAc-(1→4)-β-d-Gal | − |
| 4 | β-d-GlcNAc-(1→3)-β-d-GalNAc | + |
| 5 | β-d-GlcNAc-(1→6)-β-d-GalNAc | + |
| 6 | β-d-GlcNAc-(1→3)-β-d-Man | + |
| 7 | β-d-GlcNAc-(1→2)-β-d-Man | + |
| 8 | β-d-GlcNAc-(1→3)-β-d-Gal | + |
| 9 | β-d-GlcNAc-(1→4)-β-d-GlcNAc | + |
Structure of GH20C
In an effort to provide molecular level insight into substrate recognition and turnover by GH20C, we pursued structural studies by x-ray crystallography. An unliganded x-ray crystal structure of the enzyme crystallized in the space group P212121 was determined to 1.9 Å resolution by molecular replacement using the S. gordonii GcnA structure (Protein Data Bank code 2EPN) as a search model. The final refined model of GH20C contained four molecules of the protein in the asymmetric unit. The structure of the GH20C monomer comprises three domains, referred to as domains I–III ordered from the N terminus (Fig. 2A). Domain I is a β-sheet of parallel β-strands, packed against which is a pair of almost parallel α-helices. In turn, the α-helices sit against the outer rim of domain II. Domain II is a (β/α)8 barrel, or TIM barrel, which is not only the core fold of GH family 20 but is one of the most common folds among glycoside hydrolases in general (42). Domain III is all α-helical with four anti-parallel helices organized in a near planar arrangement. Domain III is unusual among structurally characterized GH20 enzymes and has only been observed in GcnA (30, 43). Indeed, GH20C and GcnA share the same domain organization and an overall root mean square deviation of 0.79 Å over 619 matched Cαs. Being the core catalytic domain of the GH20s, domain II is well conserved among all of the structurally characterized members of the family, whereas domain I is present in all but Aggregatibacter actinomycetemcomitans DspB and the two GH20s found in StrH (43, 44). In these latter enzymes domain I is replaced by a FIVAR-like (found in various architectural regions) domain (9).
FIGURE 2.

X-ray crystal structure of GH20C. A, GH20C monomer with the loops represented in gray and the domains represented in green schematic for the N-terminal domain, yellow for the central (β/α)8 domain, and purple for the C-terminal domain. B, GH20C dimer with one monomer colored as in A and the other monomer represented as a slate blue surface. C, overlay of the GH20C active site (yellow stick representation for side chains and transparent gray schematic representation of the backbone) with GcnA (cyan stick representation for side chains) in complex with NGT (salmon stick representation). The numbering refers to that of the GH20C polypeptide.
The four molecules of GH20C present in the asymmetric unit were modeled as two identical homodimers. The primary interface of the dimer is at domain III where the C-terminal α-helices of each domain in the dimer pack against each other, although the distal end of the domain reaches over to interact with the edge of the (β/α)8 barrel in the adjacent monomer (Fig. 2B). The dimer interface, which exceeds 3000 Å2 of buried surface area, is completed by loops extending from the (β/α)8 barrel to interact with the same loops on the adjacent monomer, although this interaction contributes the minority of the dimer interface. In this dimer, there are predicted to be 45 hydrogen bonds and 19 ion pairs. Notably, this same dimer is present in the four different crystal forms obtained for GH20C in this study. Furthermore, with a root mean square deviation of 0.85 Å over 1228 matched Cαs, it is extremely similar to the dimer formed by GcnA. The active site of GcnA, identified by the acquisition of a structure in complex with NGT, resides in the center of the (β/α)8 barrel on the side exposed to the solvent channel bisecting the dimer. This active site is conserved in GH20C with residues Asp-222 and Glu-223 as the putative catalytic residues (Fig. 2C). These results support the idea that dimerization may be a conserved feature of GH20 enzymes possessing domain III; however, there is presently no evidence supporting dimerization as being necessary for catalysis.
Structure of GH20C in Complex With Reaction Products
Toward understanding the interaction of GH20C with its substrates, we sought to determine its structure in complex with relevant carbohydrates and carbohydrate analogs of both gluco- and galacto-configured chemistries. Initially, we soaked crystals of GH20C with GlcNAc to obtain a product complex. This crystal form contained only a single dimer of GH20C in the asymmetric unit, and we found electron density in Fo − Fc maps consistent with GlcNAc in each of the predicted active sites, although the electron density indicated that the complexes were not fully occupied or were partially disordered (Fig. 3A). We could not model GlcNAc in the expected 4C1 conformation, but rather we were able to model the electron density with a GlcNAc conformation resembling the “4-sofa” conformation expected for the Michaelis complex, as observed in other GH20 enzymes (3, 9, 45). In this conformation the acetamido group is bent underneath the pyranose ring with acetamido oxygen positioned on its potential trajectory for nucleophilic attack on C1 (Fig. 3B). In the active site of one GH20C monomer, Asp-222 and Glu-223 engage the substrate with Glu-223 appropriately positioned 2.7 Å from O1 of the GlcNAc to act as the acid base and Asp-222 interacting with the acetamido nitrogen of the monosaccharide (Fig. 3B). In the other active site, however, the loop comprising residues 219–225, and thus containing the catalytic residues, is retracted from the GlcNAc residue preventing engagement of the catalytic residues by the sugar and creating what is presumably a catalytically incompetent conformation of the enzyme (Fig. 3C). Notably, the retracted conformation of this loop is present in all four of the monomers in the unliganded structure. In GcnA, the retracted conformation was observed in the ligand-free form of the enzyme, whereas the engaged conformation was found for an NGT-bound complex (30).
FIGURE 3.

Structure of GH20C in complex with reaction products. A, maximum likelihood σA-weighted Fo − Fc electron density maps contoured at 2.5σ for GlcNAc (left; represented in orange sticks) and GalNAc (right; represented in green sticks). B, specific interactions between the GH20C active site (side chains represented as yellow sticks with the catalytic residues as orange sticks) in its engaged form and GlcNAc (in magenta stick representation). Waters involved in the hydrogen bonding network are shown as red spheres. The loop carrying the catalytic residues is represented as orange schematic. C, overlay of two GH20C monomers from the GlcNAc complex structure. The engaged form is shown in yellow for the protein and with GlcNAc as magenta sticks. The form of the protein with the catalytic loop retracted is shown in green with the GlcNAc shown as cyan sticks. Relative active site residues are shown as sticks. D, overlay of GH20C in complex with GlcNAc (orange sticks with black dashed lines for hydrogen bonds) and GalNAc (green sticks with green dashed lines for hydrogen bonds).
In the active site where the catalytic residues are fully engaged, the GlcNAc sits in a cradle of aromatic residues formed primarily by Trp-266, Trp-306, Tyr-308, and Trp-373, although its polar groups are coordinated by a series of direct and water-mediated hydrogen bonds between the sugar and enzyme (Fig. 3B). All of these interactions, except those mediated by Asp-222 and Glu-223, are maintained in the active site even when the catalytic residues are not engaged (Fig. 3C).
In an effort to better understand both the recognition of a GalNAc glycon and other potential aglycon components of substrates, we approached obtaining non-hydrolyzed substrate complexes by inactivating GH20C through mutation of Glu-223, the acid/base catalyst, to a glutamine residue. The E223Q mutant retained only ∼6% of the wild-type enzyme activity on pNP-GlcNAc; however, soaking crystals of GH20CE223Q with β-d-GalNAc-(1→3)-β-d-Gal resulted in electron density in both active sites of the dimer that could only be modeled as monomeric GalNAc. The electron density in Fo − Fc maps terminated at O1 of the GalNAc, suggesting that either the aglycon was entirely disordered or the substrate was hydrolyzed (Fig. 3A). Given that the electron density terminated clearly and cleanly, we favor the possibility that the substrate was cleaved over the 30-s time scale of the crystal soaking experiment, despite the reduced activity of the mutant. Unlike the GlcNAc product complex, this GalNAc complex showed the active sites of both monomers to be in the conformation that fully engages the catalytic residues. Both GalNAc residues were in the relaxed 4C1 chair conformation and made the same suite of interactions with the enzyme as the distorted GlcNAc (Fig. 3D).
Inhibition of GH20C
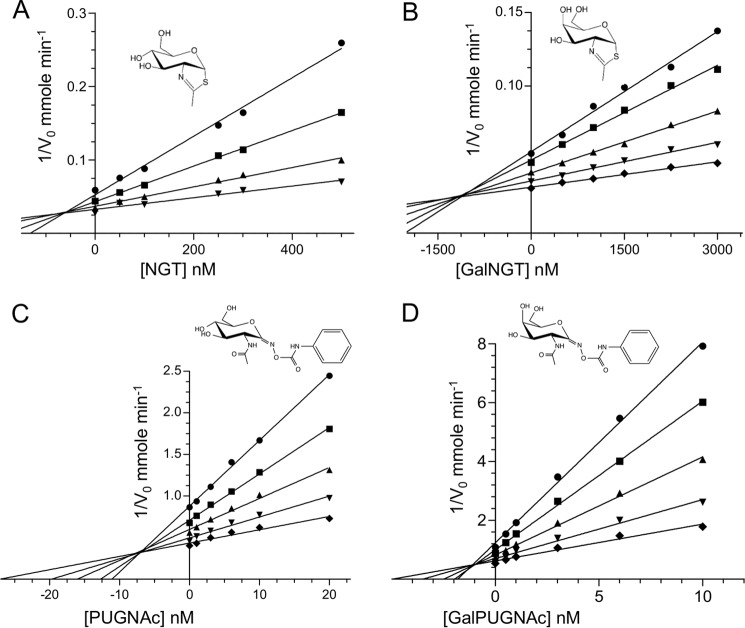
The Km and kcat values for pNP-GlcNAc and pNP-GalNAc appeared to display compensating changes resulting in a similar overall efficiency for the hydrolysis of both these substrates by GH20C. To more effectively probe the role of the stereochemistry at C4, independent of substrate turnover, we examined the ability of two pairs of compounds to inhibit GH20C by a kinetic approach using pNP-GlcNAc as a substrate. One pair of related compounds, O-(2-acetamido-2-deoxy-d-glucopyranosylidene)amino N-phenyl carbamate (PUGNAc) and O-(2-acetamido-2-deoxy-d-galactopyranosylidene)amino N-phenyl carbamate (GalPUGNAc), which differ only by being C4 epimers, have an sp2-hybridized center at C1 that resembles the planar oxocarbenium ion of the predicted transition state. This feature, along with fortuitous interactions between the enzyme and the phenylcarbamate group of these compounds, often make them very effective N-acetylhexosaminidase inhibitors (46). N-Acetyl-d-glucosamine-thiazoline (NGT) and GalNGT, which also only differ by being C4 epimers, were chosen for their ability to mimic the oxazoline intermediate of the substrate-assisted catalytic mechanism utilized by GH20 enzymes (47). A Dixon-plot analysis showed that all four compounds were competitive inhibitors of GH20C (Fig. 4). The PUGNAc-based compounds were the most potent with Ki values of 7.9 (±0.7) nm for PUGNAc and an ∼8-fold better Ki value for Gal-PUGNAc of 1.1 (±0.1) nm (Fig. 4, A and B). NGT and Gal-NGT yielded Ki values of 60.6 (±3.0) and 1130 (±110) nm, respectively (Fig. 4, C and D). The obtained pattern for general inhibitor efficacy (i.e. GalPUGNAc > PUGNAc > NGT > GalNGT) is consistent with previous inhibition data obtained for the same set of inhibitors against human and bacterial GH20 enzymes (4). The observation that the Ki of GalPUGNAc was eight times better than that for PUGNAc, but the Ki for GalNGT was 20 times weaker than for NGT, indicates the enzyme does not consistently favor an axial or equatorial O4 in these pairs of inhibitors. This does not resolve the ambiguity in the role of O4 in substrate recognition by GH20C and suggests that the O4 is not the only feature that discriminates recognition of these inhibitor pairs.
FIGURE 4.
Inhibition of GH20C. Dixon plot analyses used to determine the inhibition constant of NGT (A), GalNGT (B), PUGNAc (C), and GalPUGNAc (D). Insets show the chemical structures of the inhibitors. The concentration of enzyme used were 100 nm (circle), 150 nm (square), 200 nm (triangle), 350 nm (inverted triangle), and 500 nm (diamond).
We determined the structures of GH20C in complex with all four compounds to examine the molecular basis underlying the relative selectivity of these inhibitors. For both GalNGT and NGT, single dimers were modeled in the asymmetric unit with each monomer containing clear electron density for the inhibitor, promoting easy modeling of the compounds (Fig. 5, A and B). Similar to the GlcNAc product complex, which was obtained from an orthorhombic crystal with the same unit cell dimensions as the NGT complex, one of the GH20C monomers in the NGT complex had an active site conformation with the catalytic residues retracted; the other monomer was found in the expected “active” conformation. As anticipated based on other studies of GH20 enzymes, both GalNGT and NGT adopted the 4C1 conformation with the catalytic intermediate-mimicking thiazoline ring accommodated in a pocket formed primarily by Trp-266, Trp-306, Tyr-308, and Trp-373. Overall, when comparing the engaged conformations of the GalNGT and NGT complexes, the active site residues were in the same conformations, within positional error of the structures (Fig. 5C). The only outlier to this observation is the side chain of Arg-94, which is positioned differently in the NGT complex to accommodate hydrogen bonding with the equatorial O4 of this ligand. Similarly, the GalNGT and NGT compounds were in very similar positions with the primary difference being the positions of the O4 hydroxyls in these C4 epimers.
FIGURE 5.

Structures of GH20C in complex with inhibitors. Maximum likelihood σA-weighted Fo − Fc electron density maps for NGT (A) and GalNGT (B) contoured at 2.5σ. C, overlay of the active site of GH20C bound to NGT (yellow sticks) and GalNGT (blue sticks). Hydrogen bonds are represented by green dashed lines for the NGT complex and dashed black lines for the GalNGT complex. D, maximum likelihood σA-weighted Fo − Fc electron density maps for GalPUGNAc contoured at 2.5σ. E, overlay of the active site of GH20C bound to Gal-PUGNAc (salmon sticks) and PUGNAc (cyan sticks) found in the engaged conformations of the enzyme. Hydrogen bonds are represented by dashed black lines for the PUGNAc complex structure and dashed green lines for the Gal-PUGNAc complex structure. F and G, maximum likelihood σA-weighted Fo − Fc electron density maps for PUGNAc contoured at 2.5σ showing PUGNAc when bound to the three retracted conformation GH20C molecules in the asymmetric unit (F) and when bound the single “engaged” conformation GH20C molecules in the asymmetric unit (G).
The single molecule of GH20C in the GalPUGNAc complex was found with the active site in the fully engaged conformation and with clear electron density for the inhibitor (Fig. 5D). The GalNAc portion of GalPUGNAc made the same set of interactions with the protein as observed for GalNAc and Gal-NGT (Fig. 5E). Along with mimicry of the putative transition state conformation, the additional hydrogen bonding interactions between the phenyl carbamate moiety and Tyr-309 and Glu-223 likely contribute to the high affinity of this inhibitor. Of the four molecules of GH20C in the PUGNAc-bound complex, three were found with the active site occupied but in the retracted conformation. These three inhibitors were found with the phenyl carbamate group in the same orientation as that of Gal-PUGNAc, but due to the retraction of the loop carrying the catalytic residues, only Tyr-309 hydrogen bonds with this chemical group (Fig. 5F). The fourth molecule in the asymmetric unit was found in the bound and fully engaged state. This was modeled with the phenyl carbamate group in a reversed conformation relative to the other bound inhibitors (Fig. 5G), yet with a similar complement of hydrogen bonds as for the recognition of GalPUGNAc (Fig. 5E). However, the electron density for the phenyl moiety of this molecule was not clear suggesting disorder in this region and the potential for the inhibitor to adopt more than a single conformation (Fig. 5G).
Discussion
Many β-N-acetyl-hexosaminidases have some activity on both terminal GlcNAc and GalNAc residues (e.g. human HexA and HexB); however, usually such enzymes display specificity for one configuration of sugar or the other (9, 44, 48). GH20C from S. pneumoniae is among only a small number of characterized enzymes that have similar activities on both types of terminal sugar. In our hands, recombinant GH20C showed different Km and kcat values for the cleavage of synthetic GlcNAc and GalNAc β-aryl glycosides. However, the differences in these two kinetic parameters for the two substrates, which differ only by being C4 epimers, were compensating, resulting in quite similar kcat/Km values and thus representing a similar overall specificity for the two substrates. GH20C shows selectivity for GlcNAc and GalNAc through a set of interactions in its glycon-binding site that is common to the GH20 enzymes, most conspicuously through a generally conserved aromatic nest in the active site. Notably, interactions between Asp-222 and the acetamido nitrogen of the sugar and Tyr-308 and the acetamido oxygen of the sugar provide specificity for the 2-acetamido group. Despite the different stereochemistries of GlcNAc and GalNAc at C4, the Asp-375 side chain is positioned such that it is able to maintain potential hydrogen bonding if the O4 of the sugar is axial or equatorial. Thus, the ability of the enzyme to accommodate these two sugars with very similar sets of molecular interactions is consistent with the roughly equal ability of GlcNAc or GalNAc substrates to be hydrolyzed by GH20C.
In keeping with its ability to hydrolyze both tested synthetic substrates, GH20C was able to cleave di- and trisaccharides that terminated in either GlcNAc or GalNAc and did so with little apparent selectivity for the aglycon moiety, at least from a qualitative perspective. This suggests that unlike StrH, which is highly specific for terminal β(1–2)-linked GlcNAc residues in N-glycans, GH20C is able to cleave a relatively wide range of N-acetylhexosamine terminating sugars. Indeed, GH20C appears able to cleave motifs found in O-glycans (compounds 1, 2, 4, 5, and 8 in Table 2; compound 8 is not normally found in glycans but is an approximation of β-d-GlcNAc-(1→3)-β-d-GalNAc found in O-glycans) and N-glycans (compound 7 in Table 2). GH20C was also able to cleave chitobiose (β-d-GlcNAc-(1→4)-β-d-GlcNAc; compound 9), which is found in the core structure of N-glycans and in chitin. It was unable to cleave compound 3, a motif found in glycosphingolipids (Table 2). This is consistent with the architecture of its active site, which is very open at the potential aglycon-binding site and would likely afford only limited, if any, additional interactions with a specific aglycon residue(s) (Fig. 6A). In contrast, the GH20A domain of StrH, which has a very constrained specificity, has a narrow active site in the aglycon binding region, providing specificity for the terminal β-d-GlcNAc-(1→2)-β-d-Man motif in complex N-linked glycans (Fig. 6B) (9). In the case of β-d-GalNAc-(1→4)-β-d-Gal, the only sugar tested that GH20C did not have activity on, the axial O4 of the Gal residue results in the sugar having a bent conformation, which we predict would result in steric clash with Glu-223, the amino acid acting as the catalytic acid/base.
FIGURE 6.
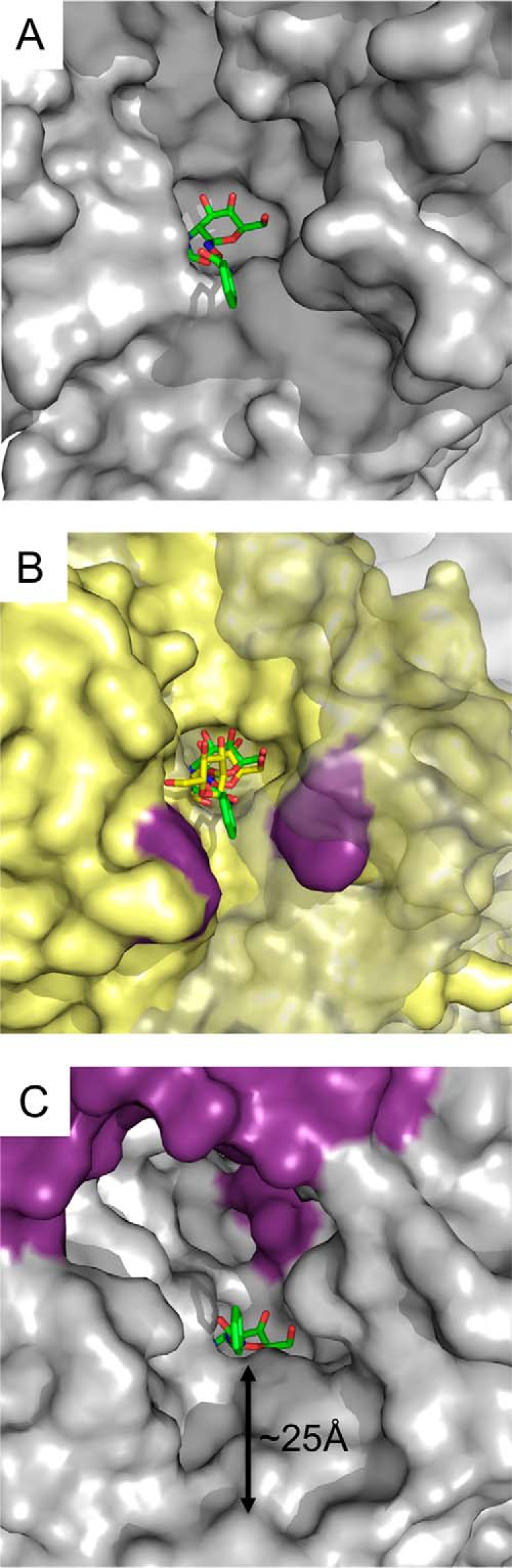
Surface contours of the GH20C active site. A, solvent-accessible surface of the active site of the GH20C monomer, with GalPUGNAc shown as green sticks, shows the open nature of the active site. B, overlay of GH20C (gray transparent surface with GalPUGNAc show in green sticks) with the GH20A module of StrH. The surface of GH20A is shown in yellow with critical aromatic amino acids flanking the entrance to the active site shown in purple. The β-d-GlcNAc-(1→2)-β-d-Man disaccharide bound to GH20A is shown as yellow sticks. C, solvent-accessible surface of the active site of the GH20C dimer, with GalPUGNAc shown as green sticks, shows the tunnel of the active site, which is ∼25 Å from the entrance to the subsite accommodating the glycon (−1 subsite).
In vitro, GH20C displayed little selectivity for the cleavage of di- and trisaccharides, suggesting the potential for broad specificity in a biological setting. This leads one to rationalize how such small soluble sugars may be provided to the bacterium in an environment where such sugars are only found attached to host tissues. The release of soluble GH20C substrates would require endo-acting glycoside hydrolases to release them from those glycans. At present, however, the repertoire of such known or predicted enzymes in S. pneumoniae is extremely limited as the bacterium appears to favor an approach whereby host glycans are trimmed from the non-reducing ends by exo-acting glycoside hydrolases (27). Related to this, should GH20C be an intracellular enzyme, as predictions suggest, its substrate(s) would have to be imported by specific carbohydrate transporters. This in turn implies a need for a diversity of transporters to import the potentially wide variety of carbohydrate substrates for GH20C. Indeed, contradicting this possibility, the carbohydrate transporters of S. pneumoniae have primarily been associated with transport of monosaccharides, consistent with the predominance of exo-acting glycoside hydrolases produced by this bacterium (49). Based on this, we presently favor the proposal that the most logical location for GH20C is outside of the bacterial cell, where due to its extracellular location the enzyme would have easy access to the non-reducing terminal GlcNAc or GalNAc residues of a wide variety of substrates, independent of the presence of particular endo-acting enzymes or specific carbohydrate transporters. Indeed, our detection of GH20C in culture supernatants provides some support for our proposed extracellular location of the enzyme, despite its lack of known motifs directing the secretion of this protein. Furthermore, there is precedent for such non-classically secreted glycoside hydrolases in S. pneumoniae of which the most relevant to this scenario is the exo-β-1,3-galactosidase BgaC that was found localized to the cell surface of this bacterium (50).
The capability of GH20C to tolerate a broad range of aglycon residues is likely related to its very open active site, which therefore results in a potential lack of specific interactions with an aglycon. This interpretation, however, does not take into account the quaternary structure of the enzyme. By virtue of dimer formation, the entrance to the active site becomes a tunnel that extends ∼25 Å along the dimer interface until the glycon-binding site is reached (Fig. 6C). If GH20C does operate in some capacity on tissue/cell-tethered glycans as an extracellular exo-glycosidase, rather than on soluble glycans released by another enzyme, the architecture of the active site entrance may influence the biological specificity of the enzyme. First, the depth of the entrance suggests that the glycan must reach ∼25 Å into the active site, which suggests a glycan of at least four or five sugar residues adopting an extended conformation would be required to access the catalytic site. Second, the shape of the active site entrance may place additional constraints on the substrate by selecting for substrates with an overall tertiary conformation that can be accommodated by the opening to the active site. Precedent for this is seen in the two catalytic modules of StrH (9). These catalytic modules have N-acetylglucosaminidase activity but limited N-acetylgalactosaminidase activity, and they are able to release GlcNAc from a variety of disaccharides. However, when terminal GlcNAc or GalNAc residues are considered in the context of their display on biological glycans, not small soluble sugars, the catalytic modules of StrH become extremely specific for the terminal GlcNAc residues on complex N-glycans as the distal aglycon-binding sites, and overall active site architecture select for these large glycans and legislate against accommodation of GlcNAc or GalNAc residues displayed on different glycans. Therefore, although GH20C appears to have limited substrate selectivity in vitro, we acknowledge the possibility that its substrate in vivo may indeed be more specific, but it is very likely different from the substrate of StrH.
The catalytic mechanism utilized by members of glycoside hydrolase family 20 is well established, and our structural studies of GH20C provided results that are entirely consistent with the features expected for the substrate-assisted catalytic mechanism. Our structures, however, also provided unexpected insight into some of the nuances of substrate recognition and cleavage by this enzyme. Structural changes that can range from small and subtle to quite large in glycoside hydrolase active sites upon substrate binding is not the rule nor is it uncommon. Indeed, the S. gordonii homolog of GH20C, GcnA, undergoes a change in the active site upon inhibitor binding whereby the catalytic residues go from a retracted state to engaging the inhibitor in a manner consistent with being positioned for catalysis (30). A nearly identical structural change was observed for GH20C upon binding both products of catalysis and inhibitors, indicating a trajectory where the loop containing the catalytic residues (the catalytic loop) goes from retracted and disordered to engaged and ordered. Unexpectedly, however, the retracted form of the catalytic loop was also observed in product- and inhibitor-bound forms of the enzyme, indicating that engagement of the catalytic loop is not necessarily a prerequisite to the enzyme binding its ligands.
In the catalytic landscape, the substrate-stabilized and engaged form of the enzyme would typically lead to formation of the Michaelis complex, which in the case of β-glycoside hydrolases involves distortion of the glycon residue to generate a sugar conformation poised to promote hydrolysis of the glycosidic bond. In the case of GH20C, the GlcNAc in the product complex with this sugar was observed to be in a distorted 4E half-envelope conformation, despite the absence of an aglycon, whereas the GalNAc product complex was observed to be in the lower energy chair conformation. Surprisingly, the GlcNAc residue present in the retracted conformation of the active site was also in the distorted conformation, suggesting in this enzyme that engagement of the catalytic residues is not requisite for substrate distortion in preparation for formation of the Michaelis complex. Thus, although we acknowledge that the somewhat locked forms of the protein in the crystal lattice(s) may bias the states of the trapped complexes, the results nevertheless suggest a complex trajectory of substrate recognition whereby the enzyme may be able to form multiple pre-Michaelis complexes. At present it is unclear whether these may represent intermediates along a pathway to formation of the fully engaged and catalytically competent conformation or whether they are non-productive complexes.
GH20C was able to catalyze the cleavage of pNP-GlcNAc and pNP-GalNAc with similar efficiency, as judged by kcat/Km, but with somewhat different yet compensating values for the kinetic parameters kcat and Km. To provide insight into this, we examined the inhibition of the enzyme by pairs of galacto-configured and gluco-configured inhibitors. In doing this, we revealed a pattern of Ki values that was previously observed for GH20 enzymes as follows: Gal-PUGNAc < PUGNAc < NGT < GalNGT (4). The significance of this is there is no consistent preference for the axial or equatorial C4 hydroxyl. However, what the relationship does suggest is that when an aglycon (e.g. the phenylcarbamate of GalPUGNAc and PUGNAc) is present (i.e. as in the Michaelis complex), the axial O4 is preferred for binding. Conversely, when the aglycon is absent (i.e. after departure of the aglycon leaving group), as represented by the thiazoline inhibitors, the equatorial O4 is favored. Thus, on this basis, we tentatively suggest that although GH20C converts GlcNAc and GalNAc substrates to products with generally similar overall efficiency, this may be achieved through slightly different energy landscapes leading to the catalyzed release of the two different terminal sugar epimers.
The potential impact and role that GH20C may have on pneumococcal virulence is presently unknown. Although other carbohydrate-degrading enzymes from S. pneumoniae have demonstrated roles in a variety of aspects of the host-bacterium interaction, such as adherence and innate immune response evasion (17, 24, 51), a common observation is their participation in nutrient acquisition (23, 25), which is thought to help support microbial persistence in the host. Based on our biochemical results, we may speculate that this enzyme complements the highly specific StrH potentially providing an overall greater ability to process the hexosaminide sugars from host glycans thus affording an expanded nutrient reservoir in the host. This, however, and other possible contributions to the host-bacterium interaction remain to be determined.
Author Contributions
M. R. designed, performed, and analyzed the x-ray crystallographic and biochemical experiments. C. S. R. assisted with the determination and completion of three x-ray crystal structures. M. A. H. performed and analyzed the quantitative PCR experiments. J. K. H. performed enzyme activity studies. J. C. P. contributed to experimental design and manuscript editing. A. B. B. conceived and coordinated the study and performed data analysis. M. R. and A. B. B. wrote the paper.
Acknowledgments
We thank the staff at the Canadian Light Source and the Stanford Synchrotron Radiation Lightsource (SSRL) beamlines where diffraction data were collected. The Canadian Light Source is supported by the Natural Sciences and Engineering Research Council of Canada, the National Research Council Canada, the Canadian Institutes of Health Research, the Province of Saskatchewan, Western Economic Diversification Canada, and the University of Saskatchewan. SSRL is a Directorate of SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the United States Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the Department of Energy Office of Biological and Environmental Research and by the National Institutes of Health, NCRR, Biomedical Technology Program Grant P41RR001209, and the NIGMS. We thank Dr. David Vocadlo for helpful discussion.
This work was supported by Canadian Institute of Health Research Operating Grant MOP 130305. The authors declare that they have no conflicts of interest with the contents of this article.
- GH20
- glycoside hydrolase family 20
- GalNAc
- N-acetylgalactosamine
- GalNGT
- N-acetyl-d-galactosamine-thiazoline
- NGT
- N-acetyl-d-glucosamine-thiazoline
- PUGNAc
- O-(2-acetamido-2-deoxy-d-glucopyranosylidene)amino N-phenyl carbamate
- GalPUGNAc
- O-(2-acetamido-2-deoxy-d-galactopyranosylidene)amino N-phenyl carbamate
- SSRL
- Stanford Synchrotron Radiation Lightsource
- CAPS
- 3-(cyclohexylamino)propanesulfonic acid
- Fw
- forward
- Rv
- reverse
- pNP
- 4-nitrophenyl.
References
- 1. Bøhle L. A., Mathiesen G., Vaaje-Kolstad G., and Eijsink V. G. (2011) An endo-β-N-acetylglucosaminidase from Enterococcus faecalis V583 responsible for the hydrolysis of high-mannose and hybrid-type N-linked glycans. FEMS Microbiol. Lett. 325, 123–129 [DOI] [PubMed] [Google Scholar]
- 2. Jiang Y.-L., Yu W.-L., Zhang J.-W., Frolet C., Di Guilmi A.-M., Zhou C.-Z., Vernet T., and Chen Y. (2011) Structural basis for the substrate specificity of a novel β-N-acetylhexosaminidase StrH protein from Streptococcus pneumoniae R6. J. Biol. Chem. 286, 43004–43012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Prag G., Papanikolau Y., Tavlas G., Vorgias C. E., Petratos K., and Oppenheim A. B. (2000) Structures of chitobiase mutants complexed with the substrate di-N-acetyl-d-glucosamine: the catalytic role of the conserved acidic pair, aspartate 539 and glutamate 540. J. Mol. Biol. 300, 611–617 [DOI] [PubMed] [Google Scholar]
- 4. Sumida T., Stubbs K. A., Ito M., and Yokoyama S. (2012) Gaining insight into the inhibition of glycoside hydrolase family 20 exo-β-N-acetylhexosaminidases using a structural approach. Org. Biomol. Chem. 10, 2607–2612 [DOI] [PubMed] [Google Scholar]
- 5. Williams S. J., Mark B. L., Vocadlo D. J., James M. N., and Withers S. G. (2002) Aspartate 313 in the Streptomyces plicatus hexosaminidase plays a critical role in substrate-assisted catalysis by orienting the 2-acetamido group and stabilizing the transition state. J. Biol. Chem. 277, 40055–40065 [DOI] [PubMed] [Google Scholar]
- 6. Drouillard S., Armand S., Davies G. J., Vorgias C. E., and Henrissat B. (1997) Serratia marcescens chitobiase is a retaining glycosidase utilizing substrate acetamido group participation. Biochem. J. 328, 945–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fernandes M. J., Yew S., Leclerc D., Henrissat B., Vorgias C. E., Gravel R. A., Hechtman P., and Kaplan F. (1997) Identification of candidate active site residues in lysosomal β-hexosaminidase A. J. Biol. Chem. 272, 814–820 [DOI] [PubMed] [Google Scholar]
- 8. Sakurama H., Kiyohara M., Wada J., Honda Y., Yamaguchi M., Fukiya S., Yokota A., Ashida H., Kumagai H., Kitaoka M., Yamamoto K., and Katayama T. (2013) Lacto-N-biosidase encoded by a novel gene of Bifidobacterium longum subspecies longum shows unique substrate specificity and requires a designated chaperone for its active expression. J. Biol. Chem. 288, 25194–25206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pluvinage B., Higgins M. A., Abbott D. W., Robb C., Dalia A. B., Deng L., Weiser J. N., Parsons T. B., Fairbanks A. J., Vocadlo D. J., and Boraston A. B. (2011) Inhibition of the pneumococcal virulence factor StrH and molecular insights into N-glycan recognition and hydrolysis. Structure 19, 1603–1614 [DOI] [PubMed] [Google Scholar]
- 10. Hepbildikler S. T., Sandhoff R., Kolzer M., Proia R. L., and Sandhoff K. (2002) Physiological substrates for human lysosomal β-hexosaminidase S. J. Biol. Chem. 277, 2562–2572 [DOI] [PubMed] [Google Scholar]
- 11. Kresse H., Fuchs W., Glössl J., Holtfrerich D., and Gilberg W. (1981) Liberation of N-acetylglucosamine-6-sulfate by human β-N-acetylhexosaminidase A. J. Biol. Chem. 256, 12926–12932 [PubMed] [Google Scholar]
- 12. Mahuran D. J. (1999) Biochemical consequences of mutations causing the GM2 gangliosidoses. Biochim. Biophys. Acta 1455, 105–138 [DOI] [PubMed] [Google Scholar]
- 13. Keilani S., Lun Y., Stevens A. C., Williams H. N., Sjoberg E. R., Khanna R., Valenzano K. J., Checler F., Buxbaum J. D., Yanagisawa K., Lockhart D. J., Wustman B. A., and Gandy S. (2012) Lysosomal dysfunction in a mouse model of Sandhoff disease leads to accumulation of ganglioside-bound amyloid-peptide. J. Neurosci. 32, 5223–5236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hughes R. C., and Jeanloz R. W. (1964) The extracellular glycosidases of Diplococcus pneumoniae. II. Purification and properties of a β-N-acetylglucosaminidase. Action on a derivative on the α-1-acid glycoprotein of human plasma. Biochemistry 3, 1535–1543 [DOI] [PubMed] [Google Scholar]
- 15. Yamashita K., Ohkura T., Yoshima H., and Kobata A. (1981) Substrate specificity of Diplococcal β-N-acetylhexosaminidase, a useful enzyme for the structural studies of complex type asparagine-linked sugar chains. Biochem Biophys Res Commun. 100, 226–232 [DOI] [PubMed] [Google Scholar]
- 16. Clarke V. A., Platt N., and Butters T. D. (1995) Cloning and expression of the β-N-acetylglucosaminidase gene from Streptococcus pneumoniae. Generation of truncated enzymes with modified aglycon specificity. J. Biol. Chem. 270, 8805–8814 [DOI] [PubMed] [Google Scholar]
- 17. Dalia A. B., Standish A. J., and Weiser J. N. (2010) Three surface exoglycosidases from Streptococcus pneumoniae, NanA, BgaA, and StrH, promote resistance to opsonophagocytic killing by human neutrophils. Infect. Immun. 78, 2108–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McCullers J. A., and Tuomanen E. I. (2001) Molecular pathogenesis of pneumococcal pneumonia. Front. Biosci. 6, 877–889 [DOI] [PubMed] [Google Scholar]
- 19. Bogaert D., De Groot R., and Hermans P. W. (2004) Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect. Dis. 4, 144–154 [DOI] [PubMed] [Google Scholar]
- 20. Hava D. L., and Camilli A. (2002) Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45, 1389–1406 [PMC free article] [PubMed] [Google Scholar]
- 21. Polissi A., Pontiggia A., Feger G., Altieri M., Mottl H., Ferrari L., and Simon D. (1998) Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect. Immun. 66, 5620–5629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Orihuela C. J., Radin J. N., Sublett J. E., Gao G., Kaushal D., and Tuomanen E. I. (2004) Microarray analysis of pneumococcal gene expression during invasive disease. Infect. Immun. 72, 5582–5596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Burnaugh A. M., Frantz L. J., and King S. J. (2008) Growth of Streptococcus pneumoniae on human glycoconjugates is dependent upon the sequential activity of bacterial exoglycosidases. J. Bacteriol. 190, 221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Limoli D. H., Sladek J. A., Fuller L. A., Singh A. K., and King S. J. (2011) BgaA acts as an adhesin to mediate attachment of some pneumococcal strains to human epithelial cells. Microbiology 157, 2369–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. King S. J., Hippe K. R., and Weiser J. N. (2006) Deglycosylation of human glycoconjugates by the sequential activities of exoglycosidases expressed by Streptococcus pneumoniae. Mol. Microbiol. 59, 961–974 [DOI] [PubMed] [Google Scholar]
- 26. Uchiyama S., Carlin A. F., Khosravi A., Weiman S., Banerjee A., Quach D., Hightower G., Mitchell T. J., Doran K. S., and Nizet V. (2009) The surface-anchored NanA protein promotes pneumococcal brain endothelial cell invasion. J. Exp. Med. 206, 1845–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. King S. J. (2010) Pneumococcal modification of host sugars: a major contributor to colonization of the human airway? Mol. Oral Microbiol. 25, 15–24 [DOI] [PubMed] [Google Scholar]
- 28. Manco S., Hernon F., Yesilkaya H., Paton J. C., Andrew P. W., and Kadioglu A. (2006) Pneumococcal neuraminidases A and B both have essential roles during infection of the respiratory tract and sepsis. Infect. Immun. 74, 4014–4020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Harty D. W., Chen Y., Simpson C. L., Berg T., Cook S. L., Mayo J. A., Hunter N., and Jacques N. A. (2004) Characterisation of a novel homodimeric N-acetyl-β-d-glucosaminidase from Streptococcus gordonii. Biochem. Biophys. Res. Commun. 319, 439–447 [DOI] [PubMed] [Google Scholar]
- 30. Langley D. B., Harty D. W., Jacques N. A., Hunter N., Guss J. M., and Collyer C. A (2008) Structure of N-acetyl-β-d-glucosaminidase (GcnA) from the endocarditis pathogen Streptococcus gordonii and its complex with the mechanism-based inhibitor NAG-thiazoline. J. Mol. Biol. 377, 104–116 [DOI] [PubMed] [Google Scholar]
- 31. Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R. D., and Bairoch A. (2003) ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Collaborative Computational Project No. 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 33. Emsley P., and Cowtan K. (2004) Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 34. Murshudov G. N., Vagin A. A., and Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 35. Brünger A. T. (1992) Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355, 472–475 [DOI] [PubMed] [Google Scholar]
- 36. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., Snoeyink J., Richardson J. S., and Richardson D. C. (2007) MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, 375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schell M. A., Karmirantzou M., Snel B., Vilanova D., Berger B., Pessi G., Zwahlen M.-C., Desiere F., Bork P., Delley M., Pridmore R. D., and Arigoni F. (2002) The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. U.S.A. 99, 14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bricker A. L., and Camilli A. (1999) Transformation of a type 4 encapsulated strain of Streptococcus pneumoniae. FEMS Microbiol. Lett. 172, 131–135 [DOI] [PubMed] [Google Scholar]
- 39. Chan P. F., O'Dwyer K. M., Palmer L. M., Ambrad J. D., Ingraham K. A., So C., Lonetto M. A., Biswas S., Rosenberg M., Holmes D. J., and Zalacain M. (2003) Characterization of a novel fucose-regulated promoter (PfcsK) suitable for gene essentiality and antibacterial mode-of-action studies in Streptococcus pneumoniae. J. Bacteriol. 185, 2051–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lacks S. (1966) Integration efficiency and genetic recombination in pneumococcal transformation. Genetics 53, 207–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pérez-Dorado I., Galan-Bartual S., and Hermoso J. A (2012) Pneumococcal surface proteins: when the whole is greater than the sum of its parts. Mol. Oral Microbiol. 27, 221–245 [DOI] [PubMed] [Google Scholar]
- 42. Henrissat B., and Davies G. (1997) Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 7, 637–644 [DOI] [PubMed] [Google Scholar]
- 43. Val-Cid C., Biarnés X., Faijes M., and Planas A. (2015) Structural-functional analysis reveals a specific domain organization in family GH20 hexosaminidases. PLoS ONE 10, e0128075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ramasubbu N., Thomas L. M., Ragunath C., and Kaplan J. B. (2005) Structural analysis of dispersin B, a biofilm-releasing glycoside hydrolase from the periodontopathogen Actinobacillus actinomycetemcomitans. J. Mol. Biol. 349, 475–486 [DOI] [PubMed] [Google Scholar]
- 45. Tews I., Perrakis A., Oppenheim A., Dauter Z., Wilson K. S., and Vorgias C. E. (1996) Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease. Nat. Struct. Biol. 3, 638–648 [DOI] [PubMed] [Google Scholar]
- 46. Horsch M., Hoesch L., Vasella A., and Rast D. M. (1991) N-Acetylglucosaminono-1,5-lactone oxime and the corresponding (phenylcarbamoyl)oxime. Novel and potent inhibitors of β-N-acetylglucosaminidase. Eur. J. Biochem. 197, 815–818 [DOI] [PubMed] [Google Scholar]
- 47. Mark B. L., Vocadlo D. J., Knapp S., Triggs-Raine B. L., Withers S. G., and James M. N. (2001) Crystallographic evidence for substrate-assisted catalysis in a bacterial β-hexosaminidase. J. Biol. Chem. 276, 10330–10337 [DOI] [PubMed] [Google Scholar]
- 48. Liu T., Zhang H., Liu F., Wu Q., Shen X., and Yang Q. (2011) Structural determinants of an insect β-N-acetyl-d-hexosaminidase specialized as a chitinolytic enzyme. J. Biol. Chem. 286, 4049–4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bidossi A., Mulas L., Decorosi F., Colomba L., Ricci S., Pozzi G., Deutscher J., Viti C., and Oggioni M. R. (2012) A functional genomics approach to establish the complement of carbohydrate transporters in Streptococcus pneumoniae. PLoS ONE 7, e33320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jeong J. K., Kwon O., Lee Y. M., Oh D.-B., Lee J. M., Kim S., Kim E.-H., Le T. N., Rhee D.-K., and Kang H. A. (2009) Characterization of the Streptococcus pneumoniae BgaC protein as a novel surface β-galactosidase with specific hydrolysis activity for the Galβ1–3GlcNAc moiety of oligosaccharides. J. Bacteriol. 191, 3011–3023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Singh A. K., Pluvinage B., Higgins M. A., Dalia A. B., Woodiga S. A., Flynn M., Lloyd A. R., Weiser J. N., Stubbs K. A., Boraston A. B., and King S. J. (2014) Unravelling the multiple functions of the architecturally intricate Streptococcus pneumoniae β-galactosidase, BgaA. PLoS Pathog. 10, e1004364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Barik S. (1996) Site-directed mutagenesis in vitro by megaprimer PCR. Methods Mol. Biol. 57, 203–215 5 [DOI] [PubMed] [Google Scholar]