Abstract
Regulation of hyaluronan (HA) synthesis and degradation is essential to maintenance of extracellular matrix homeostasis. We recently reported that HYBID (HYaluronan-Binding protein Involved in hyaluronan Depolymerization), also called KIAA1199, plays a key role in HA depolymerization in skin and arthritic synovial fibroblasts. However, regulation of HA metabolism mediated by HYBID and HA synthases (HASs) under stimulation with growth factors remains obscure. Here we report that TGF-β1, basic FGF, EGF, and PDGF-BB commonly enhance total amount of HA in skin fibroblasts through up-regulation of HAS expression, but molecular size of newly produced HA is dependent on HYBID expression levels. Stimulation of HAS1/2 expression and suppression of HYBID expression by TGF-β1 were abrogated by blockade of the MAPK and/or Smad signaling and the PI3K-Akt signaling, respectively. In normal human skin, expression of the TGF-β1 receptors correlated positively with HAS2 expression and inversely with HYBID expression. On the other hand, TGF-β1 up-regulated HAS1/2 expression but exerted only a slight suppressive effect on HYBID expression in synovial fibroblasts from the patients with osteoarthritis or rheumatoid arthritis, resulting in the production of lower molecular weight HA compared with normal skin and synovial fibroblasts. These data demonstrate that although TGF-β1, basic FGF, EGF, and PDGF-BB enhance HA production in skin fibroblasts, TGF-β1 most efficiently contributes to production of high molecular weight HA by HAS up-regulation and HYBID down-regulation and suggests that inefficient down-regulation of HYBID by TGF-β1 in arthritic synovial fibroblasts may be linked to accumulation of depolymerized HA in synovial fluids in arthritis patients.
Keywords: arthritis, cell signaling, fibroblast, growth factor, hyaluronan, skin, transforming growth factor beta (TGF-B), KIAA1199/HYBID, catabolism, hyaluronan synthase
Introduction
Hyaluronan (HA)3 is a nonsulfated linear glycosaminoglycan composed of repeating disaccharide units of β-(1,3)-linked-d-glucuronic acid and β-(1,4)-linked-N-acetyl-d-glucosamine. HA is ubiquitously present as a major component in vertebrate connective tissues such as skin and synovial membrane and contributes to space filling, lubrication, and cell proliferation and migration (1). The turnover of HA in most tissues is extraordinarily rapid; the half-life of HA in the skin, which contains about half of all HA in the body, is 1–1.5 days (2). Thus, the tight control of HA synthesis and degradation is necessary for this turnover and seems to finely balance the amounts of high molecular mass HA (1,000–10,000 kDa) within tissues (1, 2). On the other hand, an imbalance of synthesis and degradation causes the accumulation of HA with different molecular weights, which is commonly observed in diseases such as arthritis and cancers (3–5). Synovial fluids from patients with osteoarthritis (OA) or rheumatoid arthritis (RA) contain lower molecular mass HA (as low as 200 kDa) than that from normal subjects, leading to decreased synovial viscosity and increased inflammatory responses (6). HA is synthesized by HA synthases (Has1, Has2, and Has3) at the cell membrane, and the expression of Has enzymes is dependent on cell types (7, 8). We recently reported that KIAA1199, a novel HA-binding protein, plays a key role in HA degradation in normal human skin fibroblasts independently of the two hyaluronidases HYAL1 and HYAL2 and the cell surface HA receptor CD44 (9–11). This molecule is also involved in HA degradation in synovial fibroblasts and is overexpressed in OA and RA synovial fibroblasts, showing correlation with enhanced HA-degrading activity (9). In the present study, because of the involvement in HA depolymerization, we propose to designate KIAA1199 as “HYBID” (HYaluronan-Binding protein Involvedin hyaluronan Depolymerization).
Growth factors play key roles in extracellular matrix homeostasis and tissue remodeling under physiological and pathological conditions such as development and wound healing, which are commonly associated with increased HA production. Transforming growth factor-β1 (TGF-β1), basic fibroblast growth factor (bFGF), epidermal growth factor (EGF), and platelet-derived growth factor-BB (PDGF-BB) have been reported to up-regulate HAS genes expression, leading to HA overproduction in human skin fibroblasts (12–15). Among them, TGF-β1 and PDGF-BB have been well characterized to stimulate HA synthesis via distinctive mechanisms. The Smad3 signaling pathway is known to take part in TGF-β1-induced HAS1 expression (16), whereas the extracellular signal-regulated kinase (ERK) and phosphatidylinositol 3-kinase (PI3K)-Akt signaling pathways are important for PDGF-BB-stimulated HA production (15). However, regulation mechanisms about the HYBID-mediated HA degradation by growth factors in skin fibroblasts remain elusive. Although TGF-β1 is present at high levels in synovial fluids of patients with OA or RA (17) and is considered to act as a major stimulator of HA synthesis in arthritic synovial fibroblasts (18, 19), no information is available for the expression of HYBID and HAS genes or HA species synthesized under stimulation with TGF-β1.
In the present study we showed that TGF-β1, bFGF, EGF, and PDGF-BB commonly up-regulate HA production by increasing HAS-mediated HA synthesis in skin fibroblasts. Although all of them generally suppressed the expression of HYBID, the degree of the suppressive effect was different among the growth factors. Polydisperse HA species containing intermediate-sized HA derived from growth factor-stimulated fibroblasts were dependent on the HYBID expression. Of these, TGF-β1 conferred the most potent effect on production of high molecular weight HA by up-regulation of HAS expression and down-regulation of HYBID expression via distinct signaling pathways. Arthritic synovial fibroblasts showed similar profiles of the expression of HAS genes and HA production in response to TGF-β1. However, TGF-β1 only modestly down-regulated the HYBID expression and caused the accumulation of a large amount of intermediate-sized HA. Our data provide, to the best of our knowledge, the first evidence that sizes of newly produced HA under stimulation with growth factors are determined by the expression levels of HYBID, and down-regulation of the HYBID expression by TGF-β1 is cell type-specific.
Experimental Procedures
Cell Cultures
Normal human skin fibroblasts including Detroit 551 (American Type Culture Collection), HS27 (American Type Culture Collection), and NHDF-Ad (Takara Bio) cells were cultured in Eagle's minimum essential medium (MP Biomedicals) supplemented with nonessential amino acids, 1 mm sodium pyruvate, and 10% (v/v) fetal bovine serum (FBS) (JRH Biosciences). Human synovial fibroblasts from a normal subject (60-year-old male), a patient with OA (61-year-old male), and a patient with RA (51-year-old male) were purchased from Toyobo. Synovial fibroblasts were maintained in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% (v/v) FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C in a humidified atmosphere containing 5% CO2.
Assay for HA Production
Culture media of skin and synovial fibroblasts were obtained at 24 or 48 h after culturing in the presence or absence of one of the following growth factors (1 or 10 ng/ml): TGF-β1, bFGF, EGF, and PDGF-BB (R&D Systems). The HA content in the media was determined using a QnE hyaluronic acid (HA) ELISA Assay (Biotech Trading Partners, LLC) according to the manufacturer's protocol.
Quantitative Real-time PCR
After stimulation of skin and synovial fibroblasts with or without 1 or 10 ng/ml TGF-β1, bFGF, EGF, or PDGF-BB for 8 or 24 h, total RNA was isolated by an RNeasy Mini Kit (Qiagen), and cDNA synthesis was performed using a High Capacity cDNA Archive kit (Applied Biosystems). Expression of the target mRNAs was quantitatively analyzed using the cDNA templates in a TaqMan real-time PCR assay (Applied Biosystems; StepOnePlusTM Real-Time PCR System) according to the manufacturer's protocol. The relative quantification values of HYBID, HAS1, HAS2, and HAS3 were normalized to endogenous glyceraldehyde-3-phosphate dehydrogenase (GAPDH) unless otherwise specified. In the Akt inhibitor, activator, and knockdown experiments, they were normalized to β-actin, as the level of Akt expression is known to affect the expression of GAPDH (20).
RNA Interference
Knockdown experiments were performed using 25-nucleotide siRNA duplexes chemically synthesized for each gene by Invitrogen. The siRNA oligonucleotide sequences were as follows: HYBID-1, 5′-AAACAUUGAAAUAUUCGCCAUGCUC-3′; HYBID-2, 5′-UUGACAAGGAGGCCAAGACAGUGGU-3′;SMAD2-1, 5′-CAACAACCAGGAAUUUGCUGCUCUU-3′; SMAD2-2, 5′-GCCACAUGUUAUAUAUUGCCGAUUA-3′; SMAD3-1, 5′-CCACCAGGAUGCAACCUGAAGAUCU-3′; SMAD3-2, 5′-GAGAAACCAGUGACCACCAGAUGAA-3′; AKT1-1, 5′-GACGUGGCUAUUGUGAAGGAGGGUU-3′; AKT1-2, 5′-UGCAGCAUCGCUUCUUUGCCGGUAU-3′; AKT2-1, 5′-GGCUCCUUCAUUGGGUACAAGGAGA-3′; AKT2-2, 5′-UCAUCCUCAUGGAAGAGAUCCGCUU-3′; AKT3-1, 5′-GGCUCUUGAUAAAGGAUCCAAAUAA-3′; and AKT3-2, 5′-ACCUCAAGAUGUGGAUUUACCUUAU-3′. These siRNAs were transfected into cells using Lipofectamine RNAiMAX (Invitrogen), and the knocked-down cells were used for the experiments at 48 h after transfection.
Inhibitors and Activator
The pharmacologic inhibitors including p38-MAPK (mitogen-activated protein kinase) inhibitor SB203580, MEK (MAPK/ERK kinase) inhibitor PD98059, JNK (c-Jun N-terminal kinase) inhibitor II, and Akt inhibitor V were purchased from Merck. Ranolazine dihydrochloride was purchased from Santa Cruz Biotechnology.
Assay for Cellular [3H]HA Depolymerization
High molecular mass [3H]HA of >1000 kDa was prepared as described previously (9). Cellular HA depolymerization was assayed by culturing confluent cells in medium containing [3H]HA (40,000 dpm/ml) and applying the media to a Sepharose CL-2B (GE Healthcare) column (1 × 60 cm) equilibrated with 0.5 m NaCl in distilled water. The flow rate was 0.65 ml/min, and fractions of 2.5 ml were collected. The radioactivity of each fraction was measured by a scintillation counter (Aloka; LSC-6100). The column was calibrated with the fluoresceinamine-labeled HA species: H1 (1760 kDa, peak top kDa), M1 (907 kDa), L1 (197 kDa), S1 (56 kDa), T1 (28 kDa), and U1 (9.8 kDa) (PG Research). An excitation wavelength of 490 nm and an emission wavelength of 525 nm were used for the detection of fluoresceinamine.
Antibodies
A rat monoclonal antibody against HYBID was previously developed using a peptide of CARYSPHQDADPLKPRE, which corresponds to the amino acid residues Ala762 to Glu777 of KIAA1199 (GenBankTM accession number NM_018689) (9). An antibody against GAPDH was purchased from Santa Cruz Biotechnology. Antibodies against β-actin, Smad2, phosphorylated Smad2 (p-Smad2), Smad3, phosphorylated Smad3 (p-Smad3), MEK1/2, phosphorylated MEK1/2 (p-MEK1/2), ERK1/2, phosphorylated ERK1/2 (p-ERK1/2), p38-MAPK, phosphorylated p38-MAPK (p-p38-MAPK), JNK, phosphorylated JNK (p-JNK), Akt, and phosphorylated Akt (p-Akt) were purchased from Cell Signaling.
Immunoblotting
Cell homogenate supernatants were separated by electrophoresis on NuPAGE 4–12% Bis-Tris gels (Invitrogen), and proteins were transferred onto polyvinylidene difluoride membranes. The membranes were reacted with antibodies and then incubated with the following horseradish peroxidase-conjugated secondary antibodies: donkey anti-rat IgG antibody for HYBID (Jackson ImmunoResearch) and goat anti-rabbit IgG antibody for Smad2, p-Smad2, Smad3, p-Smad3, MEK1/2, p-MEK1/2, ERK1/2, p-ERK1/2, p38-MAPK, p-p38-MAPK, JNK, p-JNK, Akt, p-Akt, β-actin, and GAPDH (DAKO). Immunoreactive bands were detected using the SuperSignal West Pico Chemiluminescent Substrate System (Thermo Scientific).
Molecular Size Distribution of Synthesized HA
Detroit 551 cells or synovial fibroblasts were cultured for 48 h in the culture media containing 10 μCi/ml d-[1,6-3H(N)]glucosamine hydrochloride (American Radiolabeled Chemicals) in the presence or absence of the indicated growth factors (1 or 10 ng/ml). The harvested culture media were treated with 30 μg/ml Pronase (Merck) and then divided into two parts; one was digested with 0.1 units/ml Streptococcus dysgalactiae hyaluronidase SD (Seikagaku Corp.) in 0.5 m MES buffer, pH 6.0, containing 0.15 m sodium acetate at 37 °C overnight, whereas the other was left undigested by the enzyme. They were applied to a PD-10 desalting column equilibrated with 0.5 m NaCl in distilled water to remove unincorporated radiolabel, and each sample was fractionated in a Sepharose CL-2B (GE Healthcare) column. The chromatography profiles depicted only the hyaluronidase-sensitive activity in each fraction plotted against the fraction number by subtracting the hyaluronidase-resistant counts from the radioactivity of the hyaluronidase-undigested samples.
Human Tissue Samples
Normal human skin tissues were taken with a punch biopsy from the faces of 24 female volunteers (age range: 60–69 years) at Stephens & Associates, Inc. (Dallas, Colorado). The skin tissue collection was approved by the Institutional Review Board of Stephens & Associates, Inc., and informed consent was obtained from all of the volunteers before surgery.
Microarray Gene Analysis
Total RNA was isolated from the normal human skin tissues using RNeasy Mini Kit (Qiagen) and assessed for quality by 2100 Bioanalyzer (Agilent Technologies). The total RNA was labeled with Low Input Quick Amp Labeling kit (Agilent Technologies) and subsequently hybridized to SurePrint G3 Human GE 8 × 60K Microarray (Agilent Technologies). The data were normalized using Agilent GeneSpring GX11 software (normalized by a 75 percentile shift for each chip; normalized to median across all samples for each gene).
Statistics
Statistical significance was assessed by Student's t test, analysis of variance, and Dunnett's test, and all correlations were examined by Pearson's correlation coefficient analysis using EXSUS Version 8.0.0 (CAC EXICARE Corp.). A probability of p < 0.05 was considered significant.
Results
Regulation of HAS-mediated Synthesis and HYBID-mediated Depolymerization of HA by Growth Factors in Normal Human Skin Fibroblasts
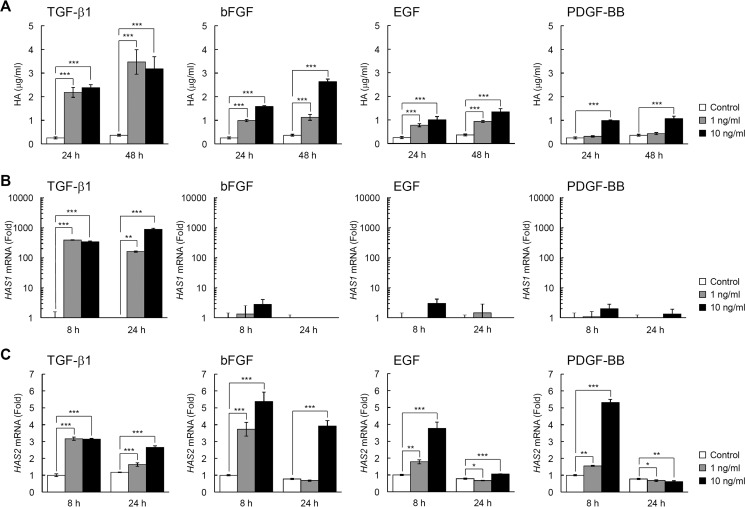
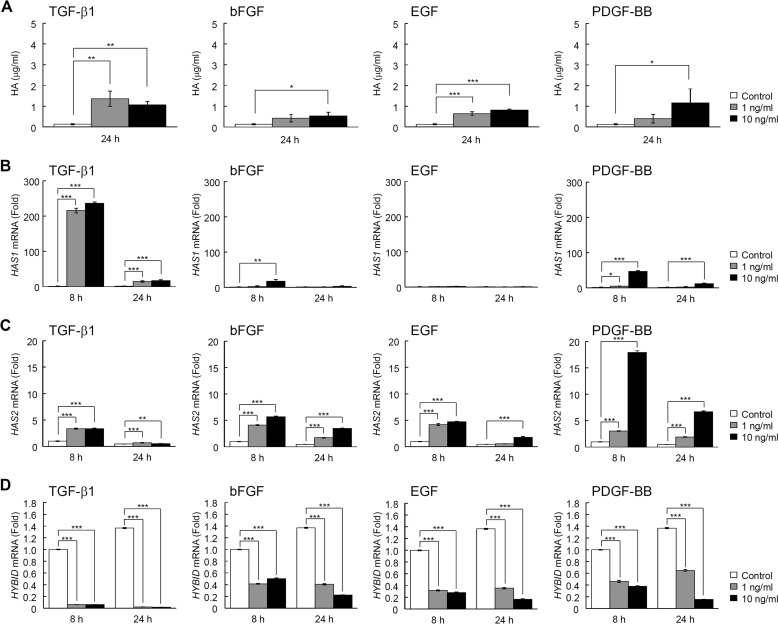
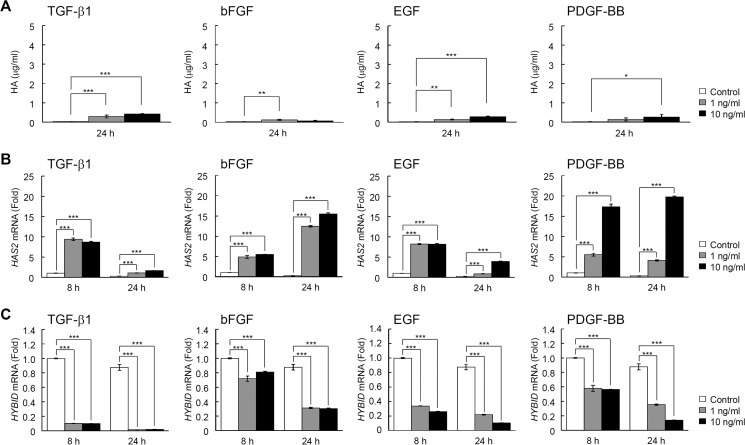
When the effects of TGF-β1, bFGF, EGF, and PDGF-BB on HAS-mediated HA synthesis were examined in normal human embryonic skin fibroblast Detroit 551 cells, all these growth factors significantly enhanced the amounts of HA in the culture media (Fig. 1A) and HAS1 and HAS2 expression (Fig. 1, B and C), confirming the data of previous studies on skin fibroblasts from various origins (12–15). Of note, TGF-β1 exerted maximal stimulatory effects on HA production via up-regulation of both the HAS1 and HAS2 genes, whereas bFGF, EGF, and PDGF-BB up-regulated only the HAS2 gene (Fig. 1, A–C). The basal mRNA expression level of HAS2 was ∼1000-fold higher than that of HAS1 mRNA, and HAS3 mRNA expression was negligible under both basal and growth factor-stimulated conditions (data not shown). Then we examined the effects of TGF-β1, bFGF, EGF, and PDGF-BB on HYBID-mediated HA depolymerization in Detroit 551 cells. As shown in Fig. 2, A–C, TGF-β1 almost completely suppressed HA depolymerization and markedly down-regulated the mRNA and protein expression of HYBID to levels <10% of the original. Although bFGF and EGF moderately suppressed HA depolymerization and HYBID expression (Fig. 2, A–C), PDGF-BB had only a mild or negligible suppressive effect (Fig. 2, A–C). Similar profiles of effects of TGF-β1, bFGF, EGF, and PDGF-BB on the HA production and the mRNA expression of HAS and HYBID were observed with other human skin fibroblasts including NHDF-Ad (adult skin fibroblasts) (Fig. 3, A–D) and HS27 (neonatal skin fibroblasts) (Fig. 4, A–C), although both cells produced a relatively lower amount of HA compared with Detroit 551 cells. Among the growth factors, TGF-β1 was the most effective regulator of HA production in these cells. On the other hand, the effect of PDGF-BB appeared to be dependent on fibroblasts, as the degree of the effect was comparatively high in NHDF-Ad and HS27 cells compared with Detroit 551 cells. These data suggest that TGF-β1, bFGF, EGF, and PDGF-BB enhance HA production not only by up-regulation of HAS-mediated HA synthesis but also down-regulation of HYBID-mediated HA degradation in skin fibroblasts.
FIGURE 1.
Up-regulation of HAS-mediated HA synthesis by TGF-β1, bFGF, EGF, and PDGF-BB in normal human skin fibroblasts. A, HA content in the culture media of Detroit 551 skin fibroblasts stimulated with growth factors. Cells were cultured in the absence (Control) or presence of 1 or 10 ng/ml TGF-β1, bFGF, EGF, or PDGF-BB for 24 or 48 h. The HA concentrations in the media were quantified by ELISA. Values represent the mean ± S.D. (n = 3). Dunnett's test was used for statistical analysis. ***, p < 0.001. B and C, the expression levels of HAS1 (B) and HAS2 (C) mRNAs in cells treated without (Control) or with 1 or 10 ng/ml TGF-β1, bFGF, EGF, or PDGF-BB for 8 or 24 h. Levels of mRNAs were measured by quantitative real-time PCR. Values are expressed as the mean ± S.D. (n = 3) and shown as -fold increases in mRNA expression relative to 8 h control cells. Dunnett's test was used for statistical analysis. ***, p < 0.001; **, p < 0.01; *, p < 0.05.
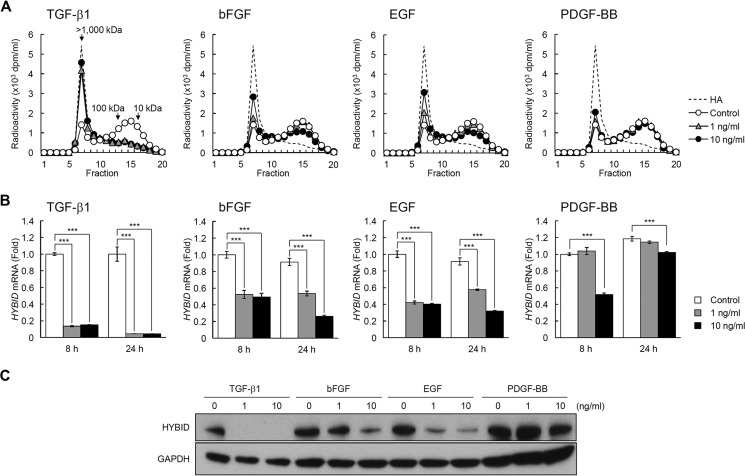
FIGURE 2.
Down-regulation of HYBID-mediated HA depolymerization by TGF-β1, bFGF, EGF, and PDGF-BB. A, HA depolymerization by Detroit 551 cells treated with growth factors. Cells treated without (Control; open circle) or with 1 (gray triangle) or 10 ng/ml (closed circle) TGF-β1, bFGF, EGF, or PDGF-BB were cultured with [3H]HA for 48 h, and HA fragments in the culture media were examined by size-exclusion chromatography. B and C, the expression levels of HYBID mRNA (B) and protein (C) in cells treated without (Control) or with 1 or 10 ng/ml TGF-β1, bFGF, EGF, or PDGF-BB. The mRNA levels at 8 and 24 h and the protein expression at 24 h after the treatment were measured by quantitative real-time PCR and immunoblotting, respectively. Values are expressed as the mean ± S.D. (n = 3) and shown as -fold increases in mRNA expression relative to 8 h control cells. Dunnett's test was used for statistical analysis. ***, p < 0.001.
FIGURE 3.
Up-regulation of HAS-mediated HA synthesis and down-regulation of HYBID mRNA expression by TGF-β1, bFGF, EGF, and PDGF-BB in NHDF-Ad cells. A, HA content in the culture media of NHDF-Ad skin fibroblasts stimulated with growth factors. Cells were cultured in the absence (Control) or presence of 1 or 10 ng/ml TGF-β1, bFGF, EGF, or PDGF-BB for 24 h. The HA concentrations in the conditioned media were quantified by ELISA. Values represent the mean ± S.D. (n = 3). Dunnett's test was used for statistical analysis. ***, p < 0.001; **, p < 0.01; *, p < 0.05. B–D, the expression levels of HAS1 (B), HAS2 (C), and HYBID (D) mRNAs in cells treated without (Control) or with 1 or 10 ng/ml TGF-β1, bFGF, EGF, or PDGF-BB for 8 or 24 h. Levels of mRNAs were measured by quantitative real-time PCR. Values are expressed as the mean ± S.D. (n = 3) and shown as -fold increases in mRNA expression relative to 8 h control cells. Dunnett's test was used for statistical analysis. ***, p < 0.001; **, p < 0.01; *, p < 0.05.
FIGURE 4.
Up-regulation of HAS-mediated HA synthesis and down-regulation of HYBID mRNA expression by TGF-β1, bFGF, EGF, and PDGF-BB in HS27 cells. A, HA content in the culture media of HS27 skin fibroblasts stimulated with growth factors. Cells were cultured in the absence (Control) or presence of 1 or 10 ng/ml TGF-β1, bFGF, EGF, or PDGF-BB for 24 h. The HA concentrations in the conditioned media were quantified by ELISA. Values represent the mean ± S.D. (n = 3). Dunnett's test was used for statistical analysis. ***, p < 0.001; **, p < 0.01; *, p < 0.05. B and C, the expression levels of HAS2 (B) and HYBID (C) mRNAs in cells treated without (Control) or with 1 or 10 ng/ml TGF-β1, bFGF, EGF, or PDGF-BB for 8 or 24 h. Levels of mRNAs were measured by quantitative real-time PCR. HAS1 mRNA expression was negligible under either basal or stimulated conditions with the growth factors tested. Values are expressed as the mean ± S.D. (n = 3) and shown as -fold increases in mRNA expression relative to 8 h control cells. Dunnett's test was used for statistical analysis. ***, p < 0.001.
Effects of Growth Factors on Molecular Size of HA Synthesized in Detroit 551 Cells
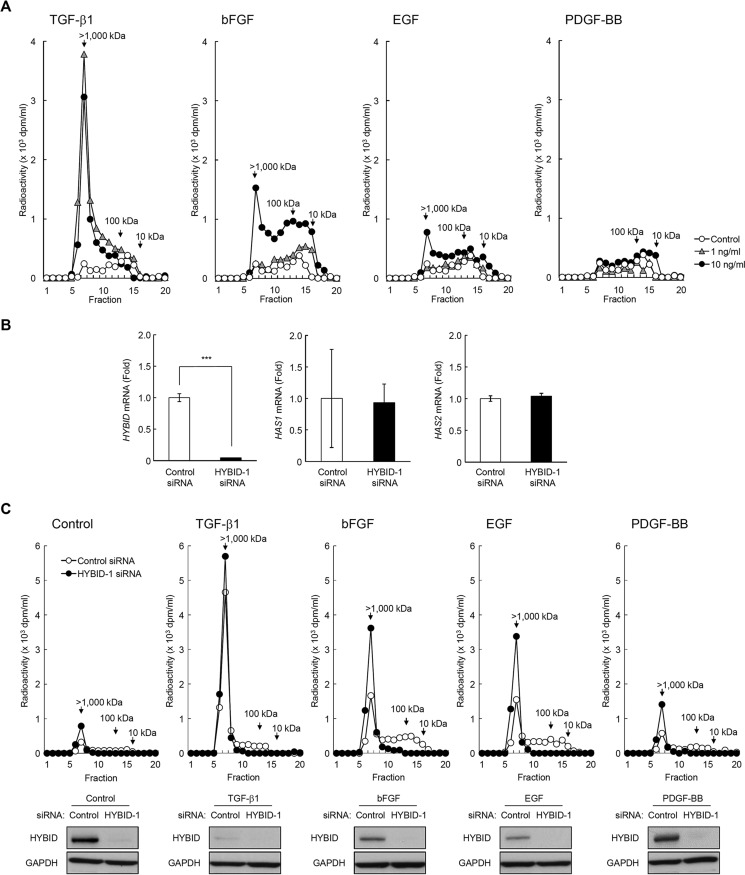
We then analyzed molecular weight distribution of newly synthesized HA in Detroit 551 cells under stimulation with TGF-β1, bFGF, EGF, or PDGF-BB. The cells were incubated with [3H]glucosamine in the presence and absence of TGF-β1, bFGF, EGF, or PDGF-BB, and the gel filtration chromatography profiles of metabolically labeled HA were examined. HA synthesized by non-stimulated cells was polydisperse but composed of two main fractions, i.e. high molecular mass [3H]HA of >1000 kDa and intermediate-sized [3H]HA of 10–100 kDa (Fig. 5A). Because Has1 and Has2 proteins synthesize only high molecular weight HA (7), the intermediate-sized [3H]HA was considered to be a degradation product from the high molecular weight [3H]HA. As shown in Fig. 5A, treatment with TGF-β1 remarkably increased the synthesis of high molecular weight [3H]HA, consistent with the data of almost complete suppression of HYBID expression (Fig. 2, B and C). On the other hand, bFGF and EGF, which showed moderate suppressive effects on HYBID expression (Fig. 2, B and C), enhanced production of both high molecular weight [3H]HA and intermediate-sized [3H]HA (Fig. 5A). Treatment with PDGF-BB, which exerted little effect on HYBID expression (Fig. 2, B and C), resulted in no or negligible changes in HA fractions, showing mainly intermediate-sized [3H]HA (Fig. 5A). To further study the direct involvement of HYBID in newly produced HA species, we knocked down HYBID expression by siRNA. As shown in Fig. 5B, siRNA successfully knocked down the HYBID expression without changing HAS1/2 expression in Detroit 551 cells. Accordingly, HYBID knocked-down cells produced only high molecular weight [3H]HA under both control and TGF-β1, bFGF, EGF, or PDGF-BB-stimulated conditions (Fig. 5C). These data demonstrate that the expression level of HYBID is a determinant for size distribution of newly synthesized HA in Detroit 551 cells under stimulation with the growth factors.
FIGURE 5.
Effects of TGF-β1, bFGF, EGF, and PDGF-BB, and knockdown of HYBID on the size distribution of newly produced HA. A, Detroit 551 cells were metabolically labeled with 10 μCi/ml [3H]glucosamine in the absence (Control; open circle) or presence of 1 (gray triangle) or 10 (closed circle) ng/ml TGF-β1, bFGF, EGF, or PDGF-BB for 48 h. The radiolabeled HA was isolated from the conditioned media and subjected to size-exclusion chromatography. B, knockdown efficiency and specificity for HYBID was evaluated by quantitative real-time PCR. The expression levels of HYBID, HAS1, and HAS2 mRNAs in HYBID knocked-down cells at 24 h after treatment with siRNA were measured by quantitative real-time PCR. As for controls, the cells were transfected with control non-silencing siRNA (Control siRNA). Values are expressed as the mean ± S.D. (n = 3) and shown as -fold increases in mRNA expression relative to control siRNA-treated cells. Student's t test was used for statistical analysis. ***, p < 0.001. Representative data for two siRNAs are shown. C, control (open circle) or HYBID (closed circle) siRNA-treated Detroit 551 cells were metabolically labeled with 10 μCi/ml [3H]glucosamine in the absence (Control) or presence of 10 ng/ml TGF-β1, bFGF, EGF, or PDGF-BB for 48 h. The radiolabeled HA was isolated from the conditioned media and subjected to size-exclusion chromatography. Lower, immunoblotting for HYBID and GAPDH (a loading control). The expression levels of HYBID protein in control or HYBID siRNA-treated cells stimulated without (Control) or with 10 ng/ml TGF-β1, bFGF, EGF, or PDGF-BB at 24 h after the stimulation.
Differential Regulation of HAS1/2 and HYBID Gene Expression by TGF-β1 through the Smad3, MAPK, or PI3K-Akt Pathways in Detroit 551 Cells
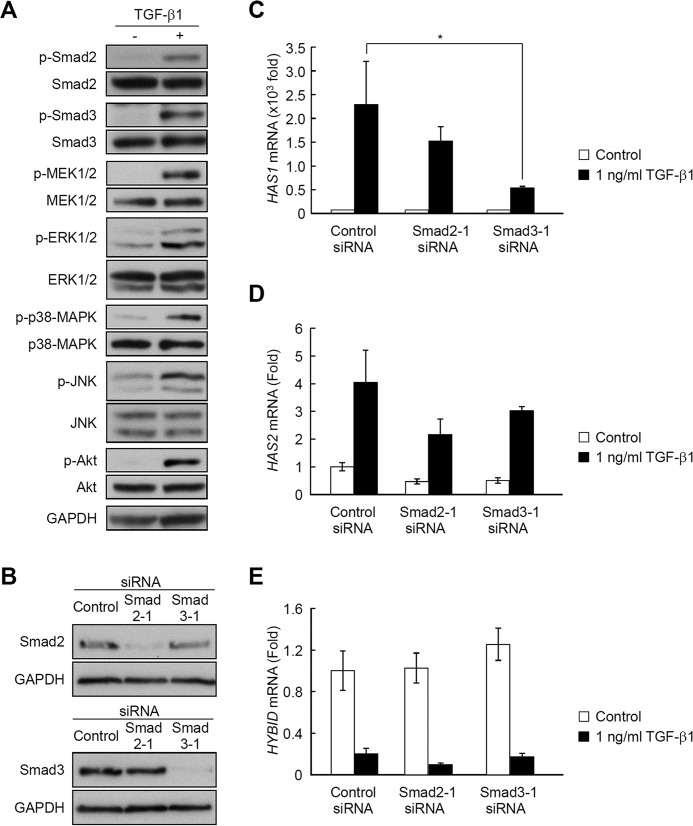
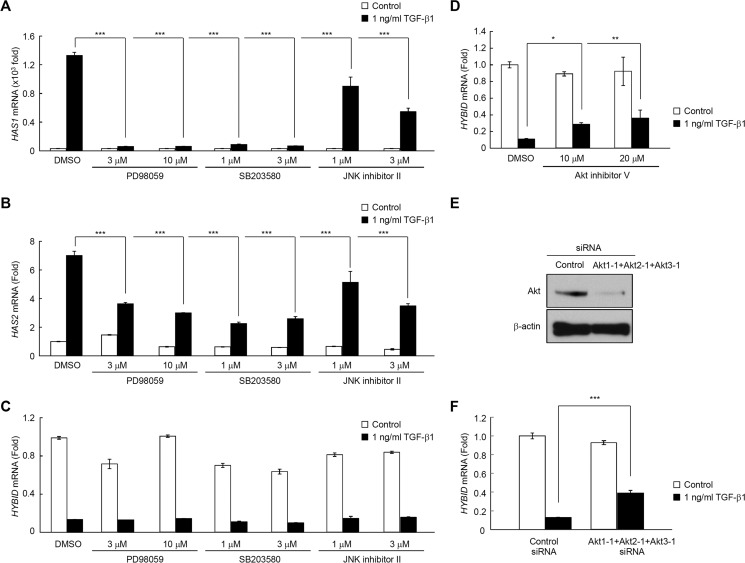
Because TGF-β1 was found to be a key inducer of high molecular weight HA synthesis by inversely regulating HAS1/2 and HYBID expression in Detroit 551 cells, we then investigated the signaling pathways for TGF-β1-mediated HAS1/2 and HYBID expression. As shown in Fig. 6A, we observed phosphorylation of Smad2 and Smad3 (the Smad2/3 signaling), MEK1/2, ERK1/2, p38-MAPK, and JNK (the MAPK signaling), and Akt (the PI3K-Akt signaling) in response to TGF-β1. When Smad2 or Smad3 expression was knocked down using siRNAs (Fig. 6B), TGF-β1-stimulated expression of HAS1 was partially, but significantly, reduced by Smad3 knockdown (Fig. 6C), but there was no significant effect on HAS2 (Fig. 6D) or HYBID (Fig. 6E) expression. We also examined involvement of the MAPK and the PI3K-Akt signaling pathways using pharmacological inhibitors. MEK inhibition (PD98059) and p38-MAPK inhibition (SB203580) greatly inhibited the TGF-β1-mediated up-regulation of HAS1 (Fig. 7A) and HAS2 (Fig. 7B), and JNK inhibitor (JNK inhibitor II) also showed a slight effect (Fig. 7, A and B). None of these inhibitors, however, exhibited apparent effects on the TGF-β1-mediated down-regulation of HYBID (Fig. 7C). On the other hand, Akt inhibition (Akt inhibitor V) partially attenuated the down-regulation of HYBID by TGF-β1 (Fig. 7D), and similar observations were obtained with knockdown experiments of Akt using siRNAs (Fig. 7, E and F). These results suggest that the TGF-β1-mediated gene expression of HAS1/2 and HYBID in Detroit 551 cells is regulated by distinct signaling pathways; the up-regulation of HAS1/2 expression is largely mediated via the Smad3, ERK1/2, and/or p38-MAPK pathways, and the PI3K-Akt signaling pathway is, at least in part, involved in the down-regulation of HYBID.
FIGURE 6.
TGF-β1-mediated phosphorylation of Smad2, Smad3, MEK1/2, ERK1/2, p38-MAPK, JNK, and Akt and contribution of the Smad signaling pathways to the regulation of TGF-β1-mediated HAS1, HAS2, and HYBID mRNA expression. A, phosphorylation of Smad2, Smad3, MEK1/2, ERK1/2, p38-MAPK, JNK, and Akt in response to TGF-β1. Detroit 551 cells were cultured in the absence or presence of 1 or 10 ng/ml TGF-β1 for 10 min. Levels of p-Smad2, total Smad2, p-Smad3, total Smad3, p-MEK1/2, total MEK1/2, p-ERK1/2, total ERK1/2, p-p38-MAPK, total p38-MAPK, p-JNK, total JNK, p-Akt, and total Akt were evaluated by immunoblotting. GAPDH was used as a loading control. Representative data obtained from the incubation with 10 ng/ml TGF-β1 are shown. B, knockdown efficiency for Smad2 and Smad3 was evaluated by immunoblotting. Smad2 and Smad3 were knocked down by treating Detroit 551 cells with siRNAs to Smad2 or Smad3. As for controls, the cells were transfected with control non-silencing siRNA (Control siRNA). GAPDH was used as a loading control. Representative data for two siRNAs are shown. C–E, effects of siRNA knockdown of Smad2 or Smad3 on HAS1 (C), HAS2 (D), and HYBID (E) mRNA expression in response to TGF-β1. The expression levels of HAS1, HAS2, and HYBID mRNAs in Smad2 or Smad3 knocked-down cells treated without (Control) or with 1 ng/ml TGF-β1 for 8 h were measured by quantitative real-time PCR. Values are expressed as the mean ± S.D. (n = 3) and shown as -fold increases in mRNA expression relative to control siRNA-treated cells in the absence of TGF-β1. Dunnett's test was used for statistical analysis. Representative data for two siRNAs are shown. *, p < 0.05.
FIGURE 7.
Contribution of the MAPK and PI3K-Akt signaling pathways to the regulation of TGF-β1-mediated HAS1, HAS2, and HYBID mRNA expression. A–C, effects of inhibitors to MEK, p38-MAPK, and JNK on HAS1, HAS2, or HYBID mRNA expression in response to TGF-β1. Detroit 551 cells were incubated for 1 h in the absence (DMSO) or presence of 3 or 10 μm PD98059 (MEK inhibitor), 1 or 3 μm SB203580 (p38-MAPK inhibitor), and 1 or 3 μm JNK inhibitor II followed by additional incubation for 8 h without (Control) or with 1 ng/ml TGF-β1. The expression levels of HAS1, HAS2, and HYBID mRNAs were measured by quantitative real-time PCR. Values are expressed as the mean ± S.D. (n = 3) and shown as -fold increases in mRNA expression relative to DMSO-treated cells in the absence of TGF-β1. Dunnett's test was used for statistical analysis. ***, p < 0.001. D, effect of Akt inhibitor V on HYBID mRNA expression. Cells were treated for 1 h with 10 or 20 μm Akt inhibitor V followed by additional incubation for 8 h without (Control) or with 1 ng/ml TGF-β1, and the HYBID mRNA expression was measured as described above. **, p < 0.01; *, p < 0.05. E, knockdown efficiency for Akt. Akt was knocked down by treating Detroit 551 cells with siRNAs to Akt1, Akt2, and Akt3. For controls, the cells were transfected with control non-silencing siRNA (control siRNA). β-Actin was used as a loading control. F, effect of siRNA knockdown of Akt on HYBID mRNA expression in response to TGF-β1. The expression levels of HYBID mRNA in Akt knocked-down cells treated without (Control) or with 1 ng/ml TGF-β1 for 8 h were measured by quantitative real-time PCR. Values are expressed as the mean ± S.D. (n = 3) and shown as -fold increases in mRNA expression relative to control siRNA-treated cells in the absence of TGF-β1. Student's t test was used for statistical analysis. ***, p < 0.001.
Correlation of TGF-β Receptor type I and II Expression with HAS2 and HYBID Expression in Normal Human Skin
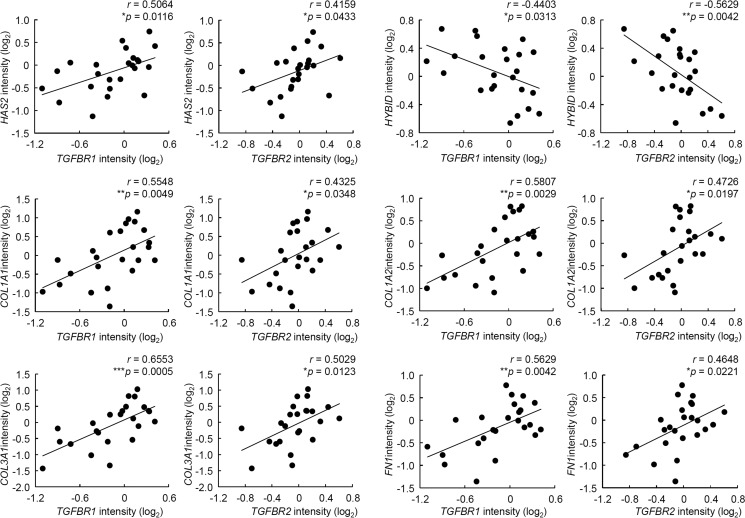
Because the levels of TGF-β1 receptors determine the response of cells to TGF-β1 (21, 22), we further assessed whether the expression levels of the HAS1/2 and HYBID genes correlate with expression of the receptors for TGF-β1 (TGFBR1 and TGFBR2) in human skin. By a microarray analysis of biopsy samples of normal skin (n = 24), the expression of TGFBR1 and TGFBR2 positively correlated with the HAS2 expression (r = 0.5064, p = 0.0116 and r = 0.4159, p = 0.0433, respectively) and inversely with the HYBID expression (r = −0.4403, p = 0.0313 and r = −0.5629, p = 0.0042, respectively) (Fig. 8). On the other hand, analysis of the correlation between the expression of HAS1 and TGFBR1 or TGFBR2 was impossible because of the low level expression of HAS1. Positive correlations of TGFBR1 and TGFBR2 were also obtained for COL1A1 (collagen type I, α-1) (r = 0.5548, p = 0.0049 and r = 0.4325, p = 0.0348, respectively), COL1A2 (collagen type I, α-2) (r = 0.5807, p = 0.0029 and r = 0.4726, p = 0.0197, respectively), COL3A1 (collagen type III, α-1) (r = 0.6553, p = 0.0005 and r = 0.5029, p = 0.0123, respectively), and FN1 (fibronectin 1) (r = 0.5629, p = 0.0042 and r = 0.4648, p = 0.0221, respectively) genes (Fig. 8), all of which are known to be up-regulated in cultured human skin fibroblasts in response to TGF-β1 (23).
FIGURE 8.
Correlation of TGFBR1 and TGFBR2 gene expression levels with HAS2, HYBID, COL1A1, COL1A2, COL3A1, and FN1 gene expression levels in normal human skin. Levels of TGFBR1, TGFBR2, HAS2, HYBID, COL1A1, COL1A2, COL3A1, and FN1 mRNAs in the normal human skin tissues (n = 24) were measured by microarray gene analysis. Correlations were examined by Pearson's correlation coefficient analysis. A probability of p < 0.05 was considered significant. r = correlation coefficient; **, p < 0.01; *, p < 0.05.
Production of Lower Molecular Weight HA by OA and RA Synovial Fibroblasts under Stimulation with TGF-β1
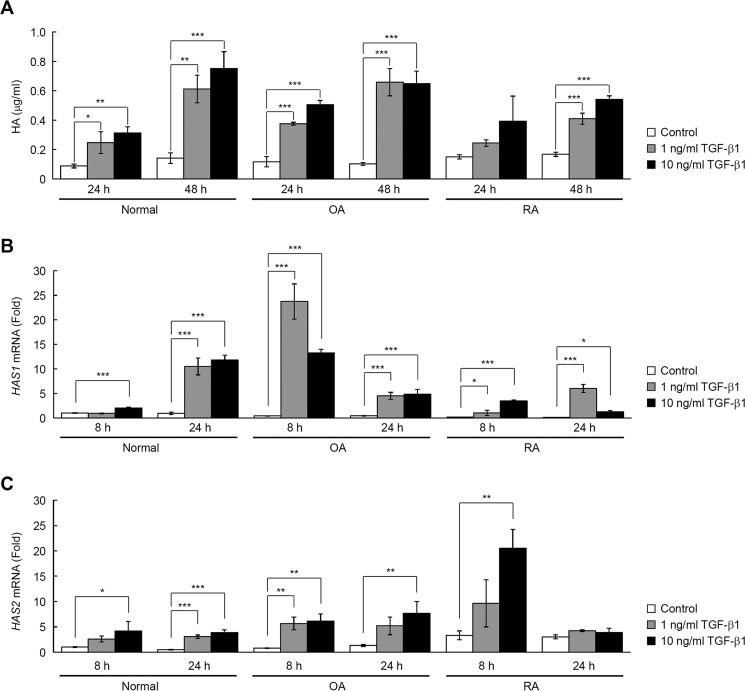
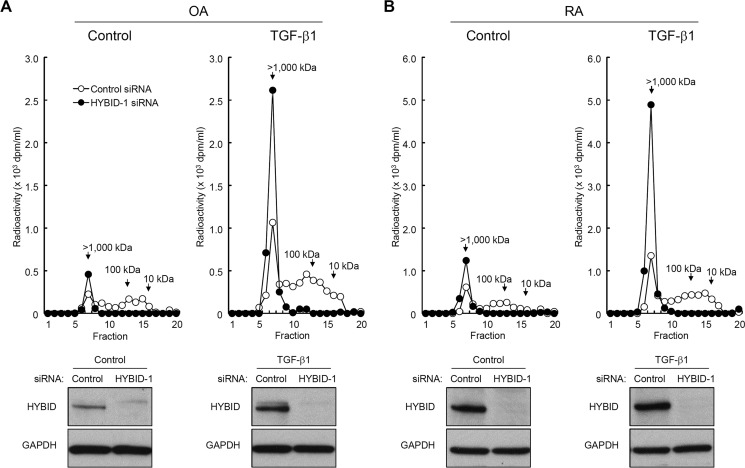
To study the possible role of TGF-β1 in HA metabolism by synovial fibroblasts, we examined production and degradation of HA and expression of the HAS and HYBID genes by treating normal and arthritic (OA and RA) synovial fibroblasts with TGF-β1. In agreement with previous findings (17–19), TGF-β1 increased the amount of HA in the culture media of normal, OA, and RA synovial fibroblasts (Fig. 9A). This was associated with up-regulation of HAS1 and HAS2 expression (Fig. 9, B and C), although HAS3 expression was negligible under this condition (data not shown). On the other hand, TGF-β1 exerted little effect on HA depolymerization in normal, OA, and RA synovial fibroblasts (Fig. 10A). The HYBID expression levels appeared to be higher in OA and RA synovial fibroblasts compared with normal synovial fibroblasts (Fig. 10, B and C), and TGF-β1-induced suppression of HYBID expression in synovial fibroblasts was minimal, i.e. up to the levels of 60% of the original (Fig. 10, B and C). Unlike skin fibroblasts, Akt was not phosphorylated in response to TGF-β1 in RA synovial fibroblasts (Fig. 10D), and ranolazine, which is known to activate PI3K-Akt (24), significantly enhanced TGF-β1-induced suppression of HYBID expression in RA synovial fibroblasts (Fig. 10E). Accordingly, newly produced HA was composed of high molecular weight HA in normal synovial fibroblasts treated with TGF-β1, but OA and RA synovial fibroblasts produced large amounts of lower molecular weight HA in response to TGF-β1 (Fig. 10F). To more directly show the involvement of HYBID in HA degradation during production of HA by synovial fibroblasts, we knocked down HYBID expression in OA and RA synovial fibroblasts by siRNA. As shown in Fig. 11, knockdown of HYBID resulted in the production of high molecular weight [3H]HA in the absence (Fig. 11, A and B, Control) or presence of TGF-β1. These results strongly suggest that TGF-β1 stimulation leads to the accumulation of intermediate-sized HA by increasing the expression of HAS1/2 with only a slight decrease in HYBID expression in arthritic synovial fibroblasts.
FIGURE 9.
Up-regulation of HAS-mediated HA synthesis by TGF-β1 in synovial fibroblasts from a normal subject and OA and RA patients. A, HA content in the culture media of normal, OA, and RA synovial fibroblasts stimulated with TGF-β1. Cells were cultured in the absence (Control) or presence of 1 or 10 ng/ml TGF-β1 for 24 or 48 h. HA concentrations in conditioned media were quantified by ELISA. Values represent the mean ± S.D. (n = 3). Dunnett's test was used for statistical analysis. ***, p < 0.001; **, p < 0.01; *, p < 0.05. B and C, the expression levels of HAS1 (B) and HAS2 (C) mRNAs in cells treated without (Control) or with 1 or 10 ng/ml TGF-β1 for 8 or 24 h. mRNA levels were measured by quantitative real-time PCR. Values are expressed as the mean ± S.D. (n = 3) and shown as -fold increases in mRNA expression relative to normal 8 h control cells. Dunnett's test was used for statistical analysis. ***, p < 0.001; **, p < 0.01; *, p < 0.05.
FIGURE 10.
Effect of TGF-β1 on HYBID-mediated HA depolymerization and the size distribution of HA synthesized by synovial fibroblasts from a normal subject, and OA and RA patients. A, HA depolymerization by synovial fibroblasts treated with TGF-β1. Cells treated without (Control; open circle) or with 1 (gray triangle) or 10 (closed circle) ng/ml TGF-β1 were cultured with [3H]HA for 48 h, and HA fragments in the culture media were examined by size-exclusion chromatography. B and C, the expression levels of HYBID mRNA (B) and protein (C) in cells treated without (Control) or with 1 or 10 ng/ml TGF-β1. mRNA levels at 8 and 24 h and protein expression at 24 h after the treatment were measured by quantitative real-time PCR and immunoblotting, respectively. Values are expressed as the mean ± S.D. (n = 3) and shown as -fold increases in mRNA expression relative to normal 8 h control cells. Dunnett's test was used for statistical analysis. ***, p < 0.001. D, phosphorylation of Akt in response to TGF-β1. Detroit 551 skin fibroblasts and RA synovial fibroblasts were incubated in the absence or presence of 1 or 10 ng/ml TGF-β1 for 10 min. Levels of p-Akt and total Akt were evaluated by immunoblotting. Representative data obtained from the incubation with 10 ng/ml TGF-β1 are shown. E, effect of PI3K-Akt activation on HYBID mRNA expression in response to TGF-β1. RA synovial fibroblasts were incubated for 1 h in the absence or presence of 200 μm ranolazine dihydrochloride followed by additional incubation for 8 h without (Control) or with 1 ng/ml TGF-β1. The expression level of HYBID mRNA was measured by quantitative real-time PCR. Values are expressed as the mean ± S.D. (n = 3) and shown as -fold increases in mRNA expression relative to control cells in the absence of TGF-β1. Student's t test was used for statistical analysis. ***, p < 0.001. F, newly produced HA. Cells were metabolically labeled with 10 μCi/ml [3H]glucosamine in the absence (Control; open circle) or presence of 1 (gray triangle) or 10 (closed circle) ng/ml TGF-β1 for 48 h. The radiolabeled HA was isolated from the conditioned media and subjected to size-exclusion chromatography.
FIGURE 11.
Size distribution of newly produced HA by HYBID knocked-down OA and RA synovial fibroblasts stimulated with TGF-β1. A and B, control (open circle) or HYBID (closed circle) siRNA-treated OA (A) and RA (B) synovial fibroblasts were metabolically labeled with 10 μCi/ml [3H]glucosamine in the absence or 10 ng/ml TGF-β1 for 48 h. The radiolabeled HA was isolated from the conditioned media and subjected to size-exclusion chromatography. Bottom, immunoblotting for HYBID and GAPDH (a loading control). Shown are expression levels of HYBID protein in control or HYBID siRNA-treated cells stimulated without (Control) or with 10 ng/ml TGF-β1 at 24 h after the stimulation.
Discussion
In the present study to the best of our knowledge we have demonstrated for the first time that although TGF-β1, bFGF, EGF, and PDGF-BB enhance total amount of HA by stimulation of HAS-mediated HA synthesis in normal human skin fibroblasts, distribution patterns of molecular sizes of newly synthesized HA are different depending on the growth factors used for treatment. Among them, TGF-β1 was the most effective stimulator for high molecular weight HA production by up-regulation of HAS1/2 genes and down-regulation of HYBID gene. We also found that the effect of TGF-β1 on the HYBID down-regulation differs between normal skin fibroblasts and arthritic synovial fibroblasts.
TGF-β1, bFGF, EGF, and PDGF-BB are reported to promote HA synthesis through enhanced HAS expression in human skin fibroblasts (12–15). In the present study we confirmed the data by showing that they increase HA production by overexpression of HAS1 and/or HAS2 genes in three different normal skin fibroblasts (Detroit 551, HS27, and NHDF-Ad cells) and further disclosed that in contrast to TGF-β1-mediated production of high molecular mass HA (>1000 kDa), bFGF, EGF, or PDGF-BB treatment results in accumulation of mainly intermediate-sized HA (10–100 kDa) in Detroit 551 cells. Stimulation of HAS1 and HAS2 gene expression and almost complete inhibition of HYBID expression by TGF-β1 accorded well with accumulation of high molecular weight HA in Detroit 551 cells. On the other hand, bFGF, EGF, and PDGF-BB enhanced HAS2 gene expression, but the suppression of HYBID expression was modest by bFGF and EGF and only minimum by PDGF-BB. In addition, we demonstrated that siRNA-mediated knockdown of HYBID in Detroit 551 cells results in production of only high molecular weight HA in the absence and the presence of TGF-β1, bFGF, EGF, or PDGF-BB. Accordingly, it is likely that the expression levels of HYBID determine the molecular sizes of newly produced HA under stimulation with these growth factors.
All these growth factors are implicated in skin wound healing (25, 26), which is composed of continuous and overlapping processes of inflammatory, proliferative, and remodeling phases (27). PDGF-BB, which is secreted from platelets present within blood coagula formed in the fresh wound, acts as a stimulator for recruitment of dermal fibroblasts and leukocytes into the wound site in the early stage of the inflammatory phase (25, 26). In the proliferative phase, bFGF and EGF play key roles in angiogenesis in the dermal granulation tissue and re-epithelialization by stimulating proliferation and migration of endothelial cells and epidermal keratinocytes, respectively (25, 26). Because biological functions of HA are dependent on the molecular sizes (28, 29) and smaller-sized HA is known to promote inflammatory and angiogenic reactions and migration of dermal fibroblasts and keratinocytes (28–32), accumulation of intermediate-sized HA elaborated by the actions of PDGF-BB, bFGF, and EGF in wound tissue may be a condition facilitating the inflammatory reactions, granulation tissue formation, and re-epithelialization, all of which occur in these phases. On the other hand, TGF-β1, one of the major growth factors in the proliferative and remodeling phases, promotes proliferation of fibroblasts, their differentiation into myofibroblasts, and extracellular matrix production (25–27). TGF-β1-mediated production of high molecular weight HA may suppress angiogenesis (29) and provide more solid extracellular matrix structures composed of high molecular weight HA and collagens, the tissue microenvironment suitable for remodeling phase.
Previous studies suggested that the Smad3, ERK, and p38-MAPK pathways are implicated in HAS1 induction by TGF-β1 in fibroblasts (16, 19). Our data have demonstrated that the TGF-β1-stimulated HAS1 expression is via the Smad3, ERK1/2, and p38-MAPK pathways in Detroit 551 cells and also that the HAS2 expression by TGF-β1 is dependent on the ERK1/2 and p38-MAPK pathways. These data indicate that TGF-β1-mediated induction of HAS1 and HAS2 expression shares the ERK1/2 and p38-MAPK pathways. In contrast, the TGF-β1-mediated down-regulation of HYBID was not canceled by knockdown of Smad2/3 or inhibition of ERK1/2, p38-MAPK, and JNK but partly recovered by the Akt inhibitor or siRNAs for Akt1/2/3. Thus, the TGF-β1-mediated expression of the HAS1/2 and HYBID genes appeared to be controlled through different intracellular signaling pathways, i.e. the Smad3 and/or MAPK pathways for HAS1/2 and the PI3K-Akt pathway for HYBID. Importantly, the present study on the normal human skin tissues demonstrated that the expression levels of the TGF-β1 receptors (TGFBR1 and TGFBR2) are positively and negatively correlated with those of HAS2 and HYBID, respectively. These data seem to reflect the results of the inverse regulation of HAS2 and HYBID expression by TGF-β1 obtained by the experiments using cultured skin fibroblasts. Altogether, the TGF-β1/TGF-β1 receptor axis may function for maintenance of extracellular matrix homeostasis through the two different pathways in human normal skin.
One of the interesting findings in the present study is that TGF-β1 did not efficiently down-regulate the HYBID expression in arthritic synovial fibroblasts, resulting in accumulation of intermediate-sized HA. This contrasts to the reaction of normal skin fibroblasts to TGF-β1. It is possible to speculate that insufficient effect of TGF-β1 on HYBID down-regulation in arthritic synovial fibroblasts is due to different expression levels of TGF-β1 receptors in skin and synovial fibroblasts. However, another plausible explanation might be epigenetic regulation of the HYBID gene expression. In human breast cancers, a link between hypomethylation of the regulatory region of the HYBID gene and up-regulation of HYBID has been reported (33). Genomic hypomethylation is known to contribute to RA, and the expression of matrix-degrading proteinases including matrix metalloproteinases-1 and -14 is increased by hypomethylation in RA synovial fibroblasts (34). Thus, the epigenetic modifications of OA and RA synovial fibroblasts may be involved in the overexpression of HYBID and inefficient down-regulation by TGF-β1, although future studies are definitely needed to demonstrate the hypothesis.
High molecular weight HA is essential to maintain the viscoelasticity and lubrication of the synovial fluid in joints, but molecular sizes of HA are decreased in synovial fluids from OA and RA patients (3, 4). TGF-β1 is present at high levels in synovial fluids of OA and RA patients (17) and expected to act as an anabolic factor for HA production. However, our data provide evidence that the stimulation of arthritic synovial fibroblasts by TGF-β1 fails in generation and accumulation of high molecular weight HA because of insufficient down-regulation of HYBID. These findings suggest that the strategies to modulate TGF-β1 activity and/or expression have limitations to maintain high molecular weight HA in arthritic conditions. Therefore, to better control the quality of HA in synovial fluids, new remedies to efficiently down-regulate HYBID expression or inhibit the HYBID-mediated HA degradation should be developed by future studies.
In summary, we have investigated the regulation of HAS-mediated synthesis and HYBID-mediated degradation of HA in normal skin fibroblasts under stimulation with TGF-β1, bFGF, EGF, and PDGF-BB and provided evidence that distribution patterns of molecular sizes of newly produced HA, but not amounts of produced HA, are substantially different depending on the HYBID expression levels modulated by the growth factors. In addition, TGF-β1-mediated down-regulation of HYBID was cell type-specific. The different molecular sizes of newly synthesized HA under stimulation with the growth factors may be related to molecular size-dependent HA functions under physiological and pathological conditions.
Author Contributions
A. N. and H. Y. designed, performed, and analyzed most of the experiments and wrote the paper. S. N. and T. M. in part performed and analyzed the experiments in Figs. 6–8. K. K., M. K., and S. S. contributed to human tissue sampling and in part performed and analyzed experiments in Fig. 8. Y. O. wrote the paper and reviewed and edited the manuscript. Y. T. and S. I. coordinated the study and reviewed the manuscript. All authors read and approved the final manuscript.
Acknowledgment
We thank Mika Aoki for skillful technical assistance.
The authors declare that they have no conflicts of interest with the contents of this article.
- HA
- hyaluronan
- HYBID
- hyaluronan binding protein involved in hyaluronan depolymerization
- HAS
- hyaluronan synthase
- OA
- osteoarthritis
- RA
- rheumatoid arthritis
- bFGF
- basic fibroblast growth factor
- EGF
- epidermal growth factor
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
References
- 1. Laurent T. C., and Fraser J. R. (1992) Hyaluronan. Hyaluronan. FASEB J. 6, 2397–2404 [PubMed] [Google Scholar]
- 2. Pandey M. S., Harris E. N., Weigel J. A., and Weigel P. H. (2008) The cytoplasmic domain of the hyaluronan receptor for endocytosis (HARE) contains multiple endocytic motifs targeting coated pit-mediated internalization. J. Biol. Chem. 283, 21453–21461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ghosh P. (1994) The role of hyaluronic acid (hyaluronan) in health and disease: interactions with cells, cartilage, and components of synovial fluid. Clin. Exp. Rheumatol. 12, 75–82 [PubMed] [Google Scholar]
- 4. Yoshida M., Sai S., Marumo K., Tanaka T., Itano N., Kimata K., and Fujii K. (2004) Expression analysis of three isoforms of hyaluronan synthase and hyaluronidase in the synovium of knees in osteoarthritis and rheumatoid arthritis by quantitative real-time reverse transcriptase polymerase chain reaction. Arthritis Res. Ther. 6, R514–R520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sugahara K. N., Murai T., Nishinakamura H., Kawashima H., Saya H., and Miyasaka M. (2003) Hyaluronan oligosaccharides induce CD44 cleavage and promote cell migration in CD44-expressing tumor cells. J. Biol. Chem. 278, 32259–32265 [DOI] [PubMed] [Google Scholar]
- 6. Vuorio E., Einola S., Hakkarainen S., and Penttinen R. (1982) Synthesis of underpolymerized hyaluronic acid by fibroblasts cultured from rheumatoid and non-rheumatoid synovitis. Rheumatol. Int. 2, 97–102 [DOI] [PubMed] [Google Scholar]
- 7. Itano N., and Kimata K. (2002) Mammalian hyaluronan synthases. IUBMB Life 54, 195–199 [DOI] [PubMed] [Google Scholar]
- 8. Törrönen K., Nikunen K., Kärnä R., Tammi M., Tammi R., and Rilla K. (2014) Tissue distribution and subcellular localization of hyaluronan synthase isoenzymes. Histochem. Cell Biol. 141, 17–31 [DOI] [PubMed] [Google Scholar]
- 9. Yoshida H., Nagaoka A., Kusaka-Kikushima A., Tobiishi M., Kawabata K., Sayo T., Sakai S., Sugiyama Y., Enomoto H., Okada Y., and Inoue S. (2013) KIAA1199, a deafness gene of unknown function, is a new hyaluronan binding protein involved in hyaluronan depolymerization. Proc. Natl. Acad. Sci. U.S.A. 110, 5612–5617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yoshida H., Nagaoka A., Nakamura S., Sugiyama Y., Okada Y., and Inoue S. (2013) Murine homologue of the human KIAA1199 is implicated in hyaluronan binding and depolymerization. FEBS Open Bio. 3, 352–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yoshida H., Nagaoka A., Nakamura S., Tobiishi M., Sugiyama Y., and Inoue S. (2014) N-terminal signal sequence is required for cellular trafficking and hyaluronan-depolymerization of KIAA1199. FEBS Lett. 588, 111–116 [DOI] [PubMed] [Google Scholar]
- 12. Sugiyama Y., Shimada A., Sayo T., Sakai S., and Inoue S. (1998) Putative hyaluronan synthase mRNA are expressed in mouse skin and TGF-β up-regulates their expression in cultured human skin cells. J. Invest. Dermatol. 110, 116–121 [DOI] [PubMed] [Google Scholar]
- 13. Ellis I., Banyard J., and Schor S. L. (1997) Differential response of fetal and adult fibroblasts to cytokines: cell migration and hyaluronan synthesis. Development 124, 1593–1600 [DOI] [PubMed] [Google Scholar]
- 14. Yamada Y., Itano N., Hata K., Ueda M., and Kimata K. (2004) Differential regulation by IL-1β and EGF of expression of three different hyaluronan synthases in oral mucosal epithelial cells and fibroblasts and dermal fibroblasts: quantitative analysis using real-time RT-PCR. J. Invest. Dermatol. 122, 631–639 [DOI] [PubMed] [Google Scholar]
- 15. Li L., Asteriou T., Bernert B., Heldin C. H., and Heldin P. (2007) Growth factor regulation of hyaluronan synthesis and degradation in human dermal fibroblasts: importance of hyaluronan for the mitogenic response of PDGF-BB. Biochem. J. 404, 327–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen L., Neville R. D., Michael D. R., Martin J., Luo D. D., Thomas D. W., Phillips A. O., and Bowen T. (2012) Identification and analysis of the human hyaluronan synthase 1 gene promoter reveals Smad3- and Sp3-mediated transcriptional induction. Matrix Biol. 31, 373–379 [DOI] [PubMed] [Google Scholar]
- 17. Tanimoto K., Suzuki A., Ohno S., Honda K., Tanaka N., Doi T., Yoneno K., Ohno-Nakahara M., Nakatani Y., Ueki M., and Tanne K. (2004) Effects of TGF-β on hyaluronan anabolism in fibroblasts derived from the synovial membrane of the rabbit temporomandibular joint. J. Dent. Res. 83, 40–44 [DOI] [PubMed] [Google Scholar]
- 18. Oguchi T., and Ishiguro N. (2004) Differential stimulation of three forms of hyaluronan synthase by TGF-β, IL-1β, and TNF-α. Connect. Tissue Res. 45, 197–205 [DOI] [PubMed] [Google Scholar]
- 19. Stuhlmeier K. M., and Pollaschek C. (2004) Differential effect of transforming growth factor β (TGF-β) on the genes encoding hyaluronan synthases and utilization of the p38 MAPK pathway in TGF-β-induced hyaluronan synthase 1 activation. J. Biol. Chem. 279, 8753–8760 [DOI] [PubMed] [Google Scholar]
- 20. Shen W., Brown N. S., Finn P. F., Dice J. F., and Franch H. A. (2006) Akt and mammalian target of rapamycin regulate separate systems of proteolysis in renal tubular cells. J. Am. Soc. Nephrol. 17, 2414–2423 [DOI] [PubMed] [Google Scholar]
- 21. Kawakami T., Ihn H., Xu W., Smith E., LeRoy C., and Trojanowska M. (1998) Increased expression of TGF-β receptors by scleroderma fibroblasts: evidence for contribution of autocrine TGF-β signaling to scleroderma phenotype. J. Invest. Dermatol. 110, 47–51 [DOI] [PubMed] [Google Scholar]
- 22. Rojas A., Padidam M., Cress D., and Grady W. M. (2009) TGF-β receptor levels regulate the specificity of signaling pathway activation and biological effects of TGF-β. Biochim. Biophys. Acta 1793, 1165–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Varga J., Rosenbloom J., and Jimenez S. A. (1987) Transforming growth factor β (TGF β) causes a persistent increase in steady-state amounts of type I and type III collagen and fibronectin mRNAs in normal human dermal fibroblasts. Biochem. J. 247, 597–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Polytarchou K., and Manolis A. S. (2015) Ranolazine and its antiarrhythmic actions. Cardiovasc. Hematol. Agents Med. Chem. 13, 31–39 [DOI] [PubMed] [Google Scholar]
- 25. Greaves N. S., Ashcroft K. J., Baguneid M., and Bayat A. (2013) Current understanding of molecular and cellular mechanisms in fibroplasia and angiogenesis during acute wound healing. J. Dermatol. Sci. 72, 206–217 [DOI] [PubMed] [Google Scholar]
- 26. Barrientos S., Stojadinovic O., Golinko M. S., Brem H., and Tomic-Canic M. (2008) Growth factors and cytokines in wound healing. Wound Repair Regen. 16, 585–601 [DOI] [PubMed] [Google Scholar]
- 27. Hattori N., Mochizuki S., Kishi K., Nakajima T., Takaishi H., D'Armiento J., and Okada Y. (2009) MMP-13 plays a role in keratinocyte migration, angiogenesis, and contraction in mouse skin wound healing. Am. J. Pathol. 175, 533–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Noble P. W. (2002) Hyaluronan and its catabolic products in tissue injury and repair. Matrix Biol. 21, 25–29 [DOI] [PubMed] [Google Scholar]
- 29. Stern R., Asari A. A., and Sugahara K. N. (2006) Hyaluronan fragments: an information-rich system. Eur. J. Cell Biol. 85, 699–715 [DOI] [PubMed] [Google Scholar]
- 30. Bourguignon L. Y. (2014) Matrix hyaluronan-activated CD44 signaling promotes keratinocyte activities and improves abnormal epidermal functions. Am. J. Pathol. 184, 1912–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bourguignon L. Y., Wong G., Xia W., Man M. Q., Holleran W. M., and Elias P. M. (2013) Selective matrix (hyaluronan) interaction with CD44 and RhoGTPase signaling promotes keratinocyte functions and overcomes age-related epidermal dysfunction. J. Dermatol. Sci. 72, 32–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Frenkel J. S. (2014) The role of hyaluronan in wound healing. Int. Wound J. 11, 159–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kuscu C., Evensen N., Kim D., Hu Y. J., Zucker S., and Cao J. (2012) Transcriptional and epigenetic regulation of KIAA1199 gene expression in human breast cancer. PLoS ONE 7, e44661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Karouzakis E., Gay R. E., Michel B. A., Gay S., and Neidhart M. (2009) DNA hypomethylation in rheumatoid arthritis synovial fibroblasts. Arthritis Rheum. 60, 3613–3622 [DOI] [PubMed] [Google Scholar]