Abstract
The HIV-1 transactivator protein Tat is implicated in the neuronal damage that contributes to neurocognitive impairment affecting people living with HIV/AIDS. Aberrant splicing of TAU exon 10 results in tauopathies characterized by alterations in the proportion of TAU isoforms containing three (3R) or four (4R) microtubule-binding repeats. The splicing factor SC35/SRSF2 binds to nuclear RNA and facilitates the incorporation of exon 10 in the TAU molecule. Here, we utilized clinical samples, an animal model, and neuronal cell cultures and found that Tat promotes TAU 3R up-regulation through increased levels of phosphorylated SC35, which is retained in nuclear speckles. This mechanism involved Tat-mediated increased expression of DYRK1A and was prevented by DYRK1A silencing. In addition, we found that Tat associates with TAU RNA, further demonstrating that Tat interferes with host RNA metabolism in the absence of viral infection. Altogether, our data unravel a novel mechanism of Tat-mediated neuronal toxicity through dysregulation of the SC35-dependent alternative splicing of TAU exon 10. Furthermore, the increased immunostaining of DYRK1A in HIV+ brains without pathology points at dysregulation of DYRK1A as an early event in the neuronal complications of HIV infection.
Keywords: DYRK1A, human immunodeficiency virus (HIV), phosphorylation, RNA splicing, Tau protein (Tau)
Introduction
Neuro-inflammation and cognitive impairment caused by HIV-1 infection persist despite the use of antiretroviral therapies (1, 2), and importantly, these therapies do not limit the amount of HIV Tat in the brain (3, 4) where it contributes to chronic immune activation (4). Therefore, knowledge of how Tat contributes to neuronal dysfunction, ultimately leading to cognitive impairment, may have implications both for understanding the pathophysiology of HIV-associated neurocognitive disorders and for devising better-targeted therapies.
The microtubule-associated protein TAU is highly involved in stabilizing neuronal cytoskeletal structures to promote axonal and synaptic maintenance and to ensure physiological vesicular and protein transport. A phenotypic trademark of TAU is its differential ability to polymerize and stabilize microtubules (5), which depends on the availability of different TAU isoforms produced as a result of alternative splicing (6, 7). The human TAU gene undergoes alternative splicing of exons 2, 3, and 10, forming six different isoforms, in which exon 10 is responsible for providing one of the four microtubule-binding domains (MTBDs)2 at the C terminus. Therefore, inclusion of exon 10 generates TAU proteins with four MTBDs (Tau 4R), although its exclusion results in three MTBDs (TAU 3R) (8–10). In this respect, the major form of TAU in fetal human and mouse brains is a TAU 3R isoform lacking all three alternative exons, called fetal TAU, whereas TAU 3R and Tau 4R are thought to be equally represented in the normal adult brain (9). There are multiple different neurodegenerative diseases in which TAU abnormalities have been reported. For example, abundant TAU aggregates are trademarks of Alzheimer disease (11, 12). In addition, prominent Tau inclusions with altered TAU 3R:4R ratios have been detected in corticobasal degeneration, frontotemporal dementia with parkinsonism-type 17 (FTDP-17), Down syndrome, Pick disease, progressive supranuclear palsy, and Niemann-Pick disease (7, 8, 13–15).
The SC35 protein belongs to the family of splicing factors that have a serine/arginine (SR)-rich C-terminal domain that undergoes dynamic phosphorylation changes. Dephosphorylation and phosphorylation cycles are thought to determine the subcellular localization of SR proteins and mediate protein-protein interactions required for the assembly of the spliceosome, RNA splicing, and mRNA export (16–18). Importantly, SC35 has been shown to bind TAU exon 10, stabilize Tau mRNA, and promote the expression of TAU 4R (19, 20), inhibiting the formation of exon 10-spliced 3R isoforms. Altered TAU 3R:4R ratios can result from either silent mutations on cis-elements (e.g. FTDP-17) or by aberrant phosphorylation of splicing factors, such as SC35, resulting from the activity of kinases including the dual-specificity tyrosine phosphorylation-regulated kinase 1A (DYRK1A) (21–24) and glycogen synthase kinase-3β (GSK-3β) (25–30). The dyrk1a gene is located on the Down syndrome critical region of chromosome 21 (21q22.2) (31). Because YRK1A is constitutively activated by autophosphorylation during translation, the activity of this protein is dependent on the dosage. Increased expression of DYRK1A has been implicated in learning deficits in Down syndrome and other neurological disorders, as well as in impaired synaptic plasticity (32–38).
Although aberrant splicing of TAU has been involved in multiple neurodegenerative disorders (7), to the best of our knowledge there are no reports of TAU exon 10 aberrant splicing in HIV-associated neurocognitive disorders. In this study, we investigated whether Tat alters the normal structure and organization of SC35 nuclear speckle domains, thereby affecting TAU exon 10 alternative splicing. We report increased phosphorylation of SC35 and altered Tau 3R:4R ratios in brain tissues from individuals with HIV-encephalitis, in an inducible Tat-transgenic mouse model and in neuronal cell cultures. In vitro studies further confirmed the ability of Tat to impair SC35-dependent TAU exon 10 inclusion through a mechanism that involves up-regulation of DYRK1A and association of Tat with TAU RNA. Finally, we found increased levels of DYRK1A in HIV+ cases without brain pathology, indicating that up-regulation of this kinase could be an early event in the neurological dysfunction associated with HIV infection.
Experimental Procedures
Primary Cells and Cell Lines
Mouse cortical neurons were isolated from CD1 mice (Charles River Laboratories) at the embryonic day 17 and cultured as described previously (39, 40). SH-SY5Y and HEK-293T (293T) cells were obtained from American Type Culture Collection (ATCC, Manassas, VA) and maintained under standard growth conditions. All cells were incubated at 37 °C (5% CO2).
Doxycycline-inducible Tat Transgenic Mouse Model
The adult transgenic mouse model, in which a tetracycline “on” bi-transgenic system permits expression of Tat(1–86) (IIIB) in astrocytes, has been previously described (41, 42). In brief, doxycycline was administered to both Tat+ and Tat− mice through a specially formulated chow (Harlan Labs, Indianapolis, IN, 6 mg/kg) for 8 days, followed by the extraction of specified brain structures and storage at −80 °C. Administration of doxycycline to the Tat mouse (which express the reverse tetracycline transactivator RTTA but not the tat gene) controls for any nonspecific actions of doxycycline ingestion (41). Experiments involving the Tat-tg mice were performed at the Virginia Commonwealth University, and snap-frozen brain samples were shipped to Louisiana State University Health Sciences Center for further analysis.
Plasmids, siRNAs, and Recombinant Tat
PCI/SI9-SI10 containing TAU exons 9–21, and part of introns 9 and 10, was a kind gift from Dr. Fei Liu (New York State Institute for Basic Research in Developmental Disabilities, Staten Island, NY). Plasmids containing full-length Tat (Tat101) were previously described (39, 43) and used at a 0.2 μg/ml concentration. Human wild-type SC35 sequence (NM_003016.4) was cloned into pcDNA 3.1+ vector by PCR amplification from a human cDNA library to generate pcDNA3.1/SC35. The primers for the PCR contained KpnI and EcoRI restriction sites (shown with underlines) and were as follows: forward 5′-GGGGTACCACTCAGAGCTATGAGCTACGG-3′ and reverse 5′-GGAATTCTTAAGAGGACACCGCTCCT-3′. A pool of human DYRK1A siRNAs and scrambled siRNAs was obtained from Santa Cruz Biotechnology (Santa Cruz, CA) and used at a concentration of 50 nm. Recombinant HIV-Tat-101 protein was purchased from ImmunoDX, LLC (Woburn, MA), and used at 100 nm concentrations. Recombinant Tat was diluted in a saline/phosphate buffer (150 mm NaCl, 50 mm sodium phosphate, pH 6.5) as recommended by ImmunoDX immediately before the treatment of the cells. The same amount of saline solution served as control.
For transfection, 293T and SH-SY5Y cells were seeded at a density of 5 × 104 cells/cm2 and 1 × 105 cells/cm2, respectively. Cells were transfected using Lipofectamine 2000 (Life Technologies, Inc.) according to the manufacturer's instructions.
Western Blots and Antibodies
Cultured cells were collected by gently scraping in the presence of cold PBS containing 1 mm of PMSF (Sigma) and 1 mm sodium orthovanadate, followed by centrifugation and disruption of the cell pellet in a lysis buffer (50 mm Hepes, pH 7.5, 150 mm NaCl, 1.5 mm MgCl2, 1 mm EGTA, pH 8.4, 10% glycerol, 1% Triton X-100) containing 1 mm protease inhibitor mixture (Sigma), 1 mm PMSF (Sigma), 1 mm phosphatase inhibitor (Sigma), and 1 mm sodium orthovanadate. Whole-cell lysates (50 μg) were denatured in 1× Laemmli buffer, boiled, and electrophoresed on a 4–15% SDS-polyacrylamide gel (Bio-Rad) before being transferred to a 0.2-μm nitrocellulose membrane (Bio-Rad) using the Trans-Blot TURBOTM apparatus (Bio-Rad). Mouse anti-SC35 (1SC-4F11), mouse anti-TAU RD3 (8E6/C11), and mouse anti-TAU RD4 (1E1/A6) were obtained from Millipore (Temecula, CA). Rabbit anti-Tat antibodies were obtained from ImmunoDX, LLC (Woburn, MA). Anti-Myc tag (71D10), anti-phospho-GSK-3α/β (Ser-21/9), anti-total GSK-3β, and anti-DYRK1A antibodies were obtained from Cell Signaling Technology (Beverly, MA), and the anti-14-3-3ζ was purchased from Santa Cruz Biotechnology (Dallas, TX). All antibodies were used at a 1:1000 dilution except anti-TAU RD4 (1:2000). Recombinant TAU protein ladder from Sigma was utilized as a marker for TAU isoforms. Quantification of the bands detected by Western blots was performed using ImageJ software (National Institutes of Health, Bethesda, MD).
TAU Exon 10 Quantification by RT-PCR
RNA was isolated using the RNeasy mini kit from Qiagen (Valencia, CA). One μg of total RNA was subjected to first-strand cDNA synthesis using the high capacity cDNA reverse transcription kit (Life Technologies, Inc.). Multiplex RT-PCR was performed using PCR master mix 2× (Thermo Scientific, Pittsburgh, PA) to amplify the minigene transcripts (E10+ and E10−) from cells transfected with pCI/SI9-SI10. The same PCR mix contained primers specific for the hypoxanthine-guanine phosphoribosyltransferase (HPRT) for semi-quantification purposes. The primers for PCR were as follows: TAU forward 5′-GGTGTCCACTCCCAGTTCAA-3′ and TAU reverse 5′-CCCTGGTTTATGATGGATGTTGCCTAATGAG-3′; HPRT forward5′-TCGAGATGTGATGAAGGAGATGGG-3′ and HPRT reverse 5′-GATGTAATCCAGCAGGTCAGCAAAG-3′. PCR cycling conditions were set as follows: denaturation for 2 min at 95 °C, followed by 25 cycles of denaturation for 30 s at 95 °C, annealing for 30 s at 55 °C, and polymerization for 45 s at 72 °C, and a final extension for 5 min at 72 °C. TAU and HPRT transcripts were resolved on 1.8% agarose gel, visualized using G:Box (Syngene, Frederick, MD), and quantified with ImageJ software (National Institutes of Health).
Quantitative RT-PCR to Detect Levels of REST and DYRK1A
Total RNA was isolated as described previously. RNA was reverse-transcribed using the High Capacity cDNA reverse transcription kit (Applied Biosystems, Carlsbad, CA). Quantitative real time PCR was performed in duplicate using a LightCycler 480 Real Time PCR System (Roche Applied Science). Human and mouse primer sets specific for REST, DYRK1A, and β-actin were purchased from Life Technologies, Inc. Each sample was normalized using β-actin (ΔCt), and relative quantification of gene expression was calculated using the comparative Ct method (2−ΔΔCt).
In Vitro Dephosphorylation of SC35 by Alkaline Phosphatase
Cellular lysates were collected as described previously with the exception that the lysis buffer did not contain phosphatase inhibitors and sodium orthovanadate. Lysates (30 μg) were incubated with or without calf intestinal alkaline phosphatase at the concentration of 1 unit/μg of protein in CutSmart buffer, both obtained from New England Biolabs (Ipswich, MA). The reaction was supplemented with 1 mm protease inhibitor mixture and PMSF and incubated for 30 min at 37 °C. The samples were denatured in 1× Laemmli buffer and processed for Western blot analysis as described above.
Immunohistochemistry
Samples from four cases each of formalin-fixed, paraffin-embedded (FFPE) HIV-encephalopathy, HIV-negative, and controls were collected from the HIV Manhattan Brain Bank at Mount Sinai Medical School. FFPE tissue samples were sectioned at 4–6-μm thickness and placed on positively charged glass slides. Hematoxylin and eosin staining was performed for routine histological evaluation. Immunohistochemistry was performed using the avidin-biotin-peroxidase complex system Vectastain (Vector Laboratories, Burlingame, CA). Our modified protocol includes paraffin melting at 58 °C for 40 min, deparaffination in xylene, and rehydration through descending grades of ethanol (100, 85, and 70%). For non-enzymatic antigen retrieval, sections were heated in 0.01 m sodium citrate buffer, pH 6.0, at 95 °C for 30 min in a vacuum oven. After cooling, the slides were rinsed in PBS and then incubated for 20 min in 6% H2O2 in methanol to quench endogenous peroxidases. Sections were then washed with PBS and blocked with 5% normal horse serum (for mouse monoclonal antibodies) in 0.1% PBS/BSA for 2 h at room temperature. Consecutive sections were incubated overnight at room temperature in a humidified chamber with either anti-Tau RD3 (1:200 dilution) or anti-Tau RD4 (1:200) (Millipore, Temecula, CA). Anti-DYRK1A (1:200) was from Santa Cruz Biotechnology. The next day, slides were rinsed with PBS and then incubated with biotinylated secondary anti-mouse in 0.1% PBS/BSA for 1 h at room temperature (1:200 dilution). Sections were then incubated with avidin-biotin-peroxidase complexes for 60 min at room temperature, rinsed with PBS, and developed with diaminobenzidine (Sigma) for 3–5 min. Slides were then counterstained with hematoxylin and mounted with Permount (Fisher).
Indirect Immunofluorescence
Cells were washed twice with PBS and fixed with 10% ultrapure EM grade formaldehyde (Polysciences, Warrington, PA) for 15 min at room temperature. After two washes with PBS, the slides were incubated in −20 °C acetone for 5 min and subsequently washed with PBS. The slides were blocked in 5% normal goat serum (Pierce) in PBS, followed by primary antibody incubation diluted in 5% normal goat serum in PBS overnight at 4 °C. The next day, the cells were rinsed in PBS and incubated with the appropriate Alexa Fluor secondary antibody conjugated to a fluorophore (1:1000 dilution) in 1% normal goat serum. Finally, the slides were rinsed with PBS twice and mounted with ProLong Gold Antifade Reagent with DAPI (Invitrogen). Phospho-SC35 antibody (clone: αSC35; 1:400 dilution) was obtained from BD Biosciences (San Jose, CA). MAP2 antibody (1:400) was from GeneTex, San Antonio, TX. Anti-DYRK1A (1:200 dilution) and anti-Myc tag (1:200) antibodies were obtained from Cell Signaling Technology (Beverly, MA). For immunofluorescence on FFPE tissues, sections were prepared according to immunohistochemistry protocol. After blocking with 5% normal goat serum in PBS for 1 h at room temperature, immunofluorescence was carried out as described above with phospho-SC35 (1:200 dilution) and Tat (ImmunoDX, Woburn, MA; 1:200 dilution) antibodies. Images were acquired using an Olympus FLUOVIEW confocal laser scanning microscope (FV 1000) equipped with FV10-ASW software (version 4.0, Olympus). For subsequent three-dimensional rendering and mask/quantification analysis, SlideBook5 acquisition/deconvolution software (Intelligent Imaging Innovations, Inc., Denver, CO) was utilized. For purposes of three-dimensional representations, images were stacked and surface-rendered. Quantification of fluorescent filters, both in the nucleus and cytoplasm, were performed by utilizing Mask analysis included in SlideBook5 software according to the manufacturer's recommendation and presented as area (pixels) per nucleus.
RNA-binding Protein Immunoprecipitation
The RNA-binding protein immunoprecipitation Tat-IP was performed essentially as described previously (44). Briefly, cells were lysed in a lysis buffer consisting of 150 mm KCl, 25 mm Tris-HCl, pH 7.4, 5 mm EDTA, 0.5% Nonidet P-40, and 5 mm DTT and supplemented with 10 mm protease inhibitor mixture, 10 mm PMSF, 10 mm phosphatase inhibitor, 10 mm Na3VO4, and 100 units/ml RNase inhibitor (Applied Biosystems, Carlsbad, CA). For the immunoprecipitation, 30 μl of protein A/G magnetic beads (Pierce) were washed three times with a blocking solution (0.5% BSA dissolved in DPBS+/+) and incubated for 2 h at 4 °C with 3 μg of anti-Tat mouse monoclonal IgG1κ (ImmunoDX, LLC) or with isotype IgG1κ control antibodies (Millipore, Temecula, CA). The immune complexes were washed three times with blocking solution before adding 500 μg of proteins diluted in 500 μl of IP buffer (150 mm KCl, 25 mm Tris-HCl, pH 7.4, 5 mm EDTA, 0.5% Nonidet P-40, and 5 mm DTT) supplemented with protein, phosphatase, and RNase inhibitors as outlined previously. Immunoprecipitations with specific lysates were carried out at 4 °C overnight. The next day, the protein-immune complexes were washed three times with an IP buffer supplemented with the inhibitors. After the last wash, the protein-immune complexes were resuspended in 20 μl of IP buffer, of which 10 μl were used for the RNA extraction and 10 μl for Western blot analysis. RNA extraction from the beads was followed by reverse transcription and RT-PCR, as described above.
Statistical Analysis
Data are presented as mean ± S.D. Comparison between two experimental groups was performed using the Student's t test. One-way analysis of variance was used to compare three or more groups. p values ≤0.05 were considered statistically significant. In all figures showing densitometric analysis, experiments were performed three times, and representative images are shown.
Ethics Statement
All of the procedures described that involved isolation and culture of embryonic primary neurons from experimental animals were performed under a protocol approved by the Louisiana State University Health Sciences Center-New Orleans Institutional Animal Care and Use Committee (IACUC), in accordance with the Animal Welfare Act and all other federal and state regulations. Procedures that involved the Tat-tg mice were performed under the Virginia Commonwealth University Institutional Animal Care and Use Committee.
The archived adult human brain tissues used in this study (four cases each of controls, HIV+, and HIVE) were collected from the HIV Manhattan Brain Bank at Mount Sinai Medical School and have been previously described (39, 45).
Results
TAU 3R Isoforms Are Increased and SC35 Is Dysregulated in Human Frontal Cortex with HIV-Encephalitis
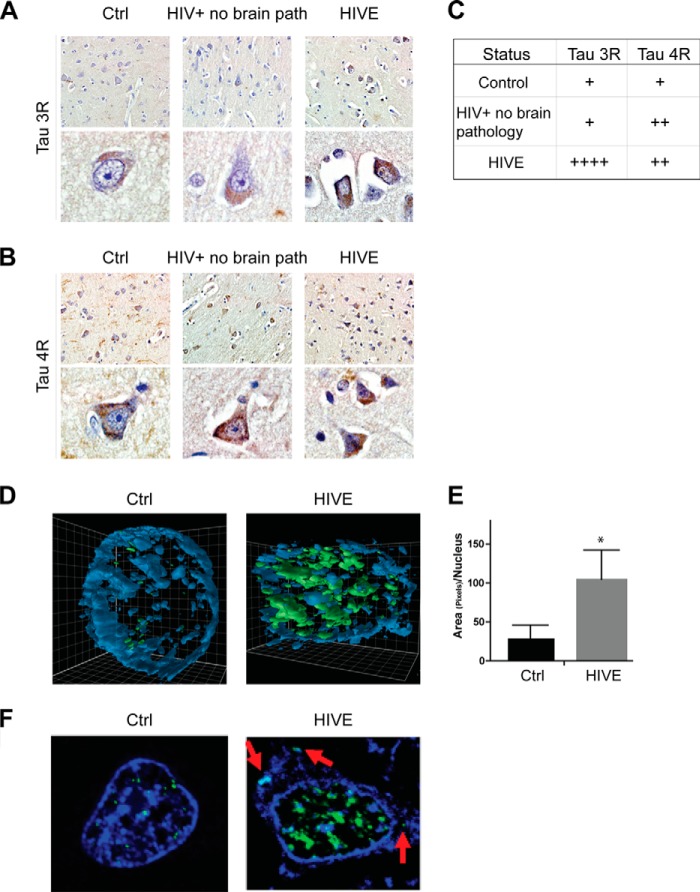
Because dysregulation of the TAU 3R:4R ratios is highly correlative with many neurodegenerative diseases (9), we investigated the expression of TAU 3R and 4R in brain samples from HIV+ patients with encephalitis (HIVE, n = 4) and controls (HIV− and HIV+ patients without brain pathology; n = 4 each group). Immunohistochemical analysis of FFPE sections labeled with anti-TAU RD3 (3R)- and anti-TAU RD4 (4R)-specific antibodies (Fig. 1, A and B, respectively) revealed TAU 3R to be preferentially increased in HIV-encephalitis (HIVE) cases in respect to controls, although the increase in 4R was less apparent because controls demonstrated relatively high levels of this splicing variant. The TAU 3R and 4R histology grading is reported in Fig. 1C, suggesting possible differences in the expression of 3R and 4R, which may derive from alteration in TAU exon 10 splicing, a process that involves the activity of the SR splicing factor SC35 (19, 20). We next determined the expression pattern of SC35 in the same brain samples, the results of which are shown in Fig. 1D. Representative three-dimensional confocal images (Fig. 1D) indicate a strong up-regulation in phospho-SC35 (green) in neurons from HIVE cases as compared with neurons from normal brains. The red arrows in Fig. 1F indicate an abnormal cytoplasmic localization of phospho-SC35, which can be observed sporadically in HIVE cases and is not present in controls. It is noteworthy that available SC35 antibody, which is used for immunofluorescence, recognizes a nuclear speckle-specific phosphorylated form of this protein. Quantitatively, phospho-SC35 was 3.5-fold higher in the neuronal nuclei of HIVE cases compared with the control (Fig. 1E). Altogether, these data indicate increased levels of phospho-SC35 and increased TAU 3R isoforms in HIVE brain tissues as compared with controls.
FIGURE 1.
Expression pattern of Tau 3R and 4R and phosphorylation status of SC35 are altered in HIV brain tissues. A and B, immunohistochemistry analyses to detect changes in TAU 3R (RD3) and TAU 4R (RD4) isoforms in archived brain tissues of HIV+ without brain pathology, HIVE, and controls (n = 4/group). C, table indicates the intensity of TAU 3R (A) and 4R (B) immunostaining grading, using a scale in which 1+ and 4+ represent the lowest and the highest intensity, respectively. D, three-dimensional imaging of neuronal nuclei (blue) from FFPE human brain samples to detect phospho-SC35 (green). E, quantification of phospho-SC35, measured as area of pixels per nucleus, obtained from five different fields in control (Ctrl) and HIVE cases. The asterisk indicates p ≤ 0.05. F, confocal images showing P-SC35 (green) neuronal nuclei in control and HIVE brain tissue. Red arrows indicate cytoplasmic P-SC35.
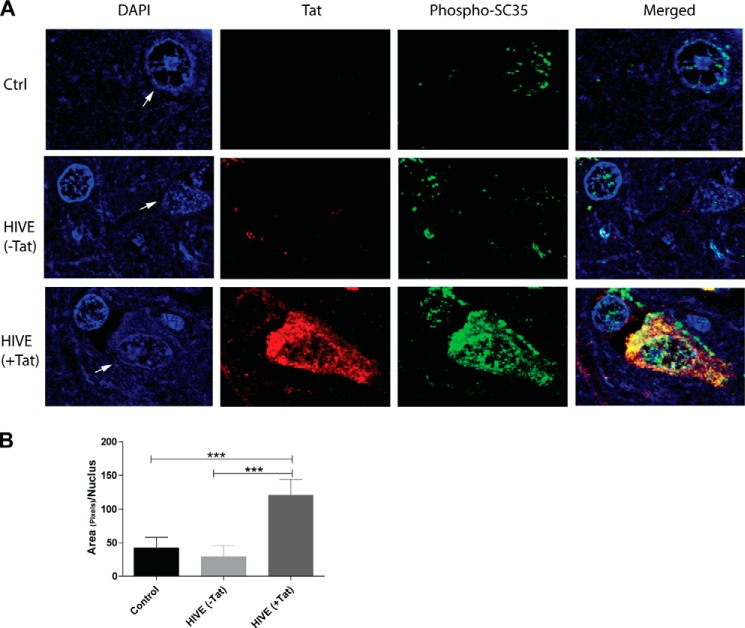
Because Tat immunohistochemistry data demonstrated high accumulation of Tat in areas of inflammation in HIVE cases compared with areas without inflammation or HIV-negative brain tissues (39), we asked whether increased phosphorylation of SC35 was associated with the presence of Tat. Fig. 2 shows representative confocal images (×100) of consecutive sections of brain tissue samples used in Fig. 1 that were subjected to immunofluorescence using anti-Tat and anti-phospho-SC35 antibodies. In general, Tat was not detectable in HIV+ brain tissues in the absence of inflammation and/or brain pathology; however, in areas of inflammation, the cells most robustly labeled with anti-Tat antibodies (in red) were neurons compared with astrocytes and microglia (Fig. 1), as reported previously (39, 46). Conversely, phospho-SC35 (in green) was detected in all cell types. In controls and in HIVE (−Tat) the pattern of phospho-SC35 appeared punctate and normal. Strikingly, neurons with the strongest Tat labeling (HIVE (+Tat)) also had the greatest amount of phospho-SC35 immunoreactivity. In addition to losing the typical nucleus-speckled pattern, phospho-SC35 appeared in the cytoplasm of Tat-positive cells. Quantification of nuclear phospho-SC35 further confirmed a 3-fold increase (p ≤ 0.05) in neurons positive for Tat (Fig. 2B).
FIGURE 2.
Co-expression of Tat and SC35 in HIVE. A, representative images to detect Tat (red) and phospho-SC35 antibodies (green) in FFPE tissue samples from controls (Ctrl) and HIVE cases. Areas with intense (+Tat) or background (−Tat) Tat immunolabeling can be found in HIVE cases and parallel a strong or normal phospho-SC35 immunolabeling pattern, respectively. Arrows indicate neuronal nuclei. B, diagram showing quantification analysis of nuclear phospho-SC35 in the indicated samples obtained from three experiments. The three asterisks show statistical significance between groups (p ≤ 0.001).
Tat Dysregulates SC35 in Primary Neuronal Cultures
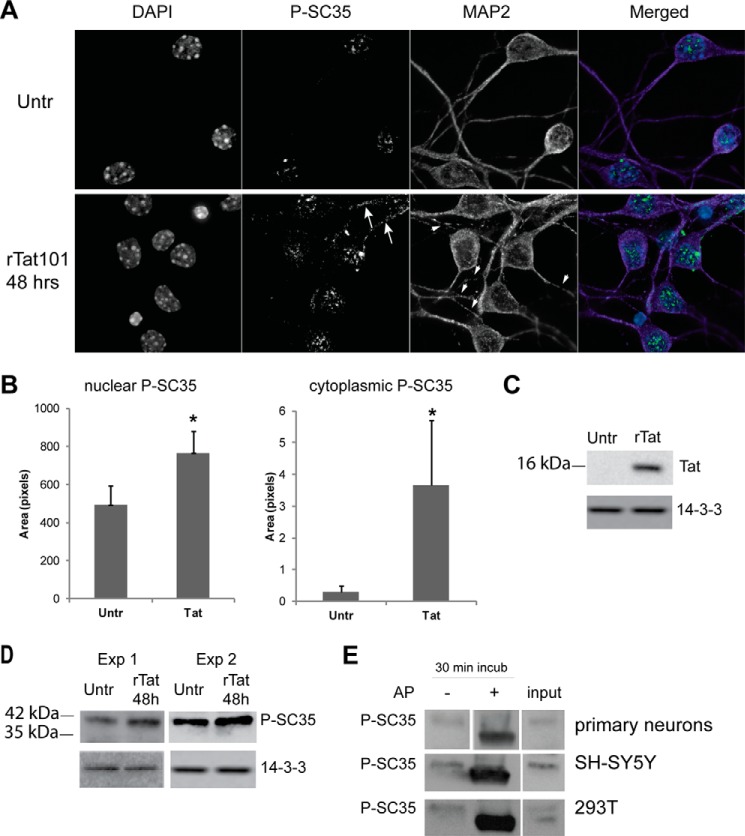
To investigate the molecular mechanisms by which Tat affects the SC35 splicing factor, we utilized several cellular models. First, we utilized embryonic neurons isolated from E17 mice (“Experimental Procedures”). After 5 days in vitro (DIV5), neurons were treated with 100 nm recombinant Tat (rTat101, 101 amino acids) for 48 h, and immunofluorescent microscopy was performed to characterize the distribution of phospho-SC35. Double labeling with anti-MAP-2 (microtubule-associated protein 2) served as a marker for both neuronal processes and for Tat-mediated neuronal toxicity, as we reported previously (39). In fact, neurons treated with Tat show a discontinuous pattern of MAP-2 labeling as compared with healthy neurons (Fig. 3A, arrowheads). Results in Fig. 3A show a punctate nuclear pattern of SC35 distribution in untreated neurons, as reported previously in multiple cell types, and in neurons treated with Tat, the pattern appears less organized and/or levels of phospho-SC35 are increased. To our surprise, we also observed the cytoplasmic presence of phospho-SC35 in response to Tat (Fig. 3A, white arrows), a pattern usually associated with cell division (47). Densitometric analysis of the nuclear and cytoplasmic areas labeled with anti-SC35 antibody was performed on five fields from two independent experiments and confirmed the increased staining in Tat-treated cells (Fig. 3B). Efficient Tat uptake was also evaluated after 24 h of Tat treatment (Fig. 3C) and confirmed our previous reports (39, 43). Using the same experimental conditions, we have evaluated levels of SC35 protein by Western blotting on lysates obtained from two experiments, which confirmed an increase in phospho-SC35 in response to Tat treatment (Fig. 3B). Interestingly, the SC35 band was detected at the level of 42 kDa instead of the predicted 35 kDa. This discrepancy in the migration pattern could be explained by the high content of serine residues, which, when phosphorylated, are known to cause retardation in gel mobility. To test this interpretation of the obtained results, we treated neuronal lysates (extracted without addition of phosphatase inhibitors in the lysis buffer) with calf-intestinal alkaline phosphatase for 30 min. We found that the immunoreactivity associated with the 42-kDa band was lost after treatment with alkaline phosphatase as compared with lysates not treated with alkaline phosphatase (Fig. 3C, top panel). We have repeated these experiments in SH-SY5Y and 293T cells transfected with Tat, and we obtained similar results (Fig. 3E, middle and bottom panels, respectively). Our results indicate that Tat affects SC35 subcellular localization and increases SC35 phosphorylation.
FIGURE 3.
Treatment of mouse embryonic cortical neurons with Tat results in increased phosphorylation of SC35 protein. A, confocal microscopy images showing phospho-SC35 (green) immunolabeling of DIV7 mouse primary neurons performed 48 h after treatment with recombinant Tat. DAPI staining (blue) identifies the nucleus, and MAP-2 (purple) labels neuronal processes. The arrows in the bottom panel indicate cytoplasmic P-SC35, and arrowheads indicate discontinuous pattern of MAP-2 in neuronal processes. Untr, untreated. B, quantification analysis of nuclear and cytoplasmic P-SC35 labeling was performed on five fields from each condition from two experiments. Asterisks indicate p < 0.05. C, representative Western blot showing efficient uptake of recombinant Tat in mouse primary neurons. D, Western blots showing increased phospho-SC35 in two experiments of Tat-treated neurons. E, dephosphorylation by alkaline phosphatase (AP) confirmed that the SC35 detected in Tat-treated neurons, Tat-transfected SH-SY-5Y and 293T cells is the phosphorylated form. Anti-14-3-3 is used as a loading control.
Tat Promotes Tau 3R Expression in Tat-tg Mouse Model and in Cultured Neuronal Cells
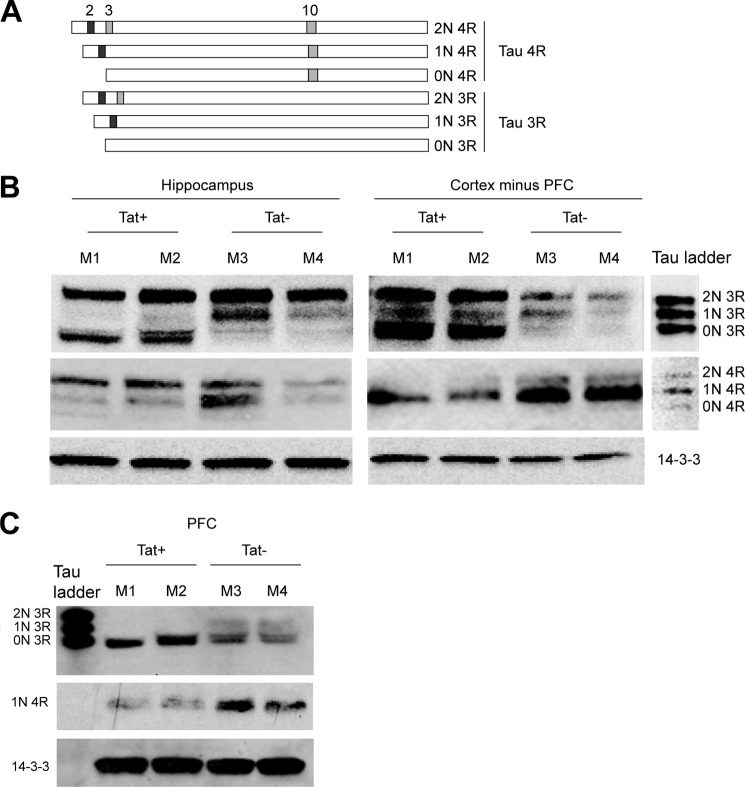
Because SC35 participates in the TAU exon 10 splicing (9, 19, 20), we asked whether Tat-mediated dysregulation of phospho-SC35 would affect Tau exon 10 splicing using a Tat-inducible transgenic mouse model (41, 48, 49). First, we determined the expression levels of Tau 3R and 4R isoforms in adult mice in which Tat expression is induced in the brain. To induce Tat expression in astroglial cells, doxycycline was administered as described under “Experimental Procedures.” We utilized frozen tissues from seven different areas of the brain (hippocampus, striatum, prefrontal cortex (PFC), cortex minus PFC, cerebellum, forebrain, and brainstem) taken from control Tat (Tat−) (n = 7) and Tat+ mice (n = 7). Alternative splicing of Tau exons 2, 3, and 10 results in six combinatorial isoforms, which can be further divided into two categories depending on the exclusion or inclusion of exon 10 (Tau 3R and 4R, respectively), as represented in the schematic picture (Fig. 4A). Western blot analysis was performed with protein extracts isolated from all the brain areas at the indicated conditions. In general, different areas of the brain expressed different levels of Tau-splicing variants and the effects of Tat varied from modest to severe. Fig. 4B shows representative results from the hippocampus and cortical areas minus PFC, as determined in two mice per group, Tat+ (M1 and M2) and Tat− (M3 and M4). The Tau ladder was used to identify all six Tau isoforms. The most significant response to Tat is observed for the Tau 0N 3R (fetal Tau), which is highly increased in Tat+ mice in lieu of either the 1N 3R (hippocampus), or the Tau 4R (cortex minus PFC) isoforms. This latter effect is particularly evident in the cortex minus PFC, in which expression of all three Tau 3R proteins is up-regulated by Tat and parallels a significant down-regulation of Tau 4R. Of interest, in the prefrontal cortex, 0N 3R and 1N 4R were the predominant Tau isoforms detected, although some of the other six Tau variants were undetectable (Fig. 4C). Nevertheless, there was an increase in Tau 3R with a concomitant down-regulation of Tau 4R in PFC areas expressing Tat. The Tat-mediated increase in Tau 3R was additionally observed in the striatum, cerebellum, and forebrain structures (data not shown).
FIGURE 4.
Altered Tau 3R and 4R expression in Tat-transgenic mice. A, diagram showing the six Tau isoforms originated from the alternative splicing of exons 2, 3, and 10 (modified from Ref. 7). Inclusion of exon 10 results in a Tau that possesses four tubulin-binding domains (4R), whereas exclusion results in three tubulin-binding domains (3R). The fully spliced 0N 3R is also called fetal Tau. B and C, expression of Tau 3R and 4R in the hippocampus, prefrontal cortex, and cortex without PFC of brains from Tat-tg mice in which expression of Tat was induced (Tat+) or in mice lacking the Tat transgene (Tat−).
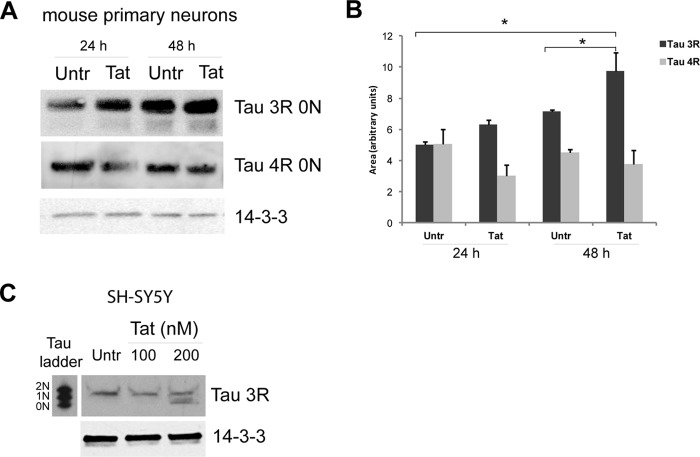
Next, we determined expression levels of Tau 3R and 4R isoforms in mouse embryonic primary neurons. DIV7 neuronal cultures were incubated with 100 nm recombinant Tat for 24 and 48 h, and expression levels of Tau 3R and 4R isoforms were determined by Western blotting (Fig. 5A). The levels of Tau 3R increased over time in control untreated cells. In Tat-treated neuronal cultures, Tau 3R isoforms were significantly higher in comparison with the control at 24 h, and this Tat-induced increase in Tau 3R splicing variant continued at 48 h. In contrast, we observed a slight decrease in the 4R isoform in the Tat-treated sample at 24 h and no difference at 48 h (Fig. 5A, middle panel). These highly significant changes in Tau 3R levels and only small changes in 4R contributed to the significant change of the Tau 3R:4R ratio induced by Tat. It is important to emphasize that human and mouse embryonic neurons express mainly the Tau 0N 3R isoform, which is also called fetal Tau. However, once the neurons are allowed to differentiate in vitro, the Tau 4R may also be detectable by Western blot by loading at least 70 μg of protein extracts (Fig. 5A). We have also confirmed Tat-induced Tau 3R expression in undifferentiated human SH-SY5Y cells treated with two different concentrations of recombinant Tat for 48 h. The reason for choosing undifferentiated cells for this experiment is because the treatments that induce differentiation in SH-SY5Y cells (retinoic acid followed by BDNF) robustly increase expression of Tau 3R (50, 51), preventing the detection of a further increase eventually due to Tat. Similarly to mouse primary neurons and Tat-tg mice, Tat treatment in SH-SY5Y cells resulted in the up-regulation of Tau 3R 0N (Fig. 5C).
FIGURE 5.
Tat increases Tau 3R expression in cultured neuronal cells. A, representative Western blot analysis to detect Tau 3R and 4R isoforms in DIV7 mouse primary neurons treated with Tat for 24 and 48 h and controls. Note that three times more protein lysates were loaded into the gels to detect Tau 4R compared with the amount used to detect Tau 3R. B, diagram showing densitometric quantification of Tau 3R and 4R expression relative to loading controls obtained from three independent experiments. Asterisk indicates p ≤ 0.05. C, Western blotting showing a dose response of Tau 3R isoforms in SH-SY5Y cells transfected with increasing concentrations of Tat. In all panels anti-14-3-3 was used as a loading control. Untr, untreated.
Tat Promotes Aberrant Splicing of TAU Exon 10
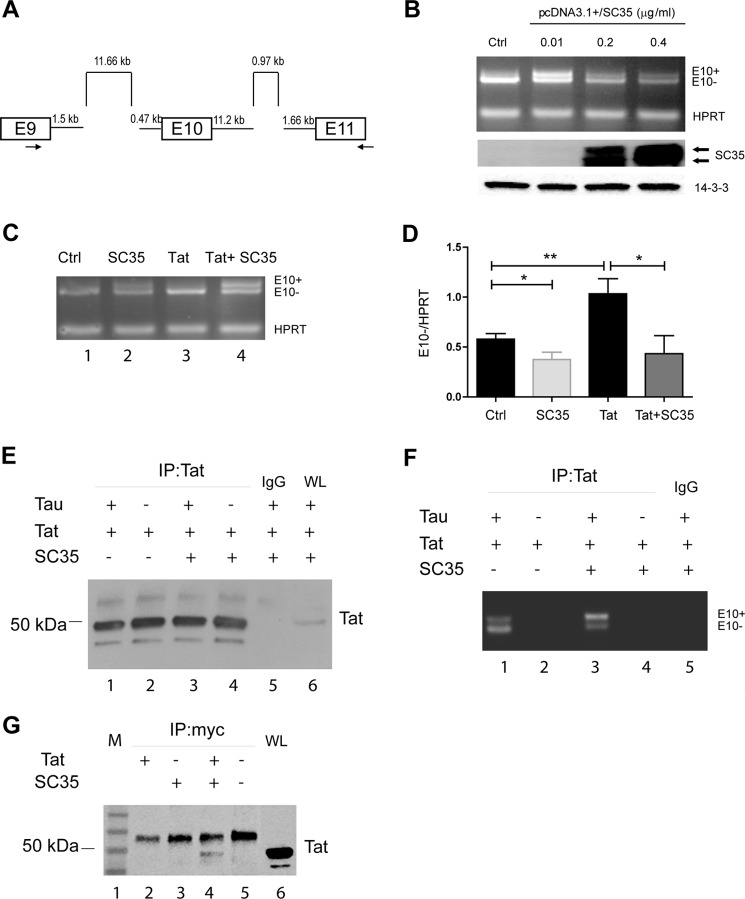
To elucidate the role of Tat in the splicing of TAU exon 10, we used a TAU minigene, pCI/SI9-SI10 (20, 52). The TAU minigene consists of exons 9–11 and their corresponding partial introns that contain the necessary elements required for correct splicing (Fig. 6A). Inclusion (E10+) and exclusion (E10−) of exon 10 in the pre-mRNA transcript was RT-PCR-amplified and detected on a 1.8% agarose gel as two bands of 513 and 420 bp, respectively. First, to check the function of SC35 on the TAU exon 10 splicing, 293T cells were co-transfected with the TAU minigene, and increasing concentrations of pcDNA3.1-SC35 (pSC35) and total RNA were used to set up a multiplex RT-PCR to detect TAU exon 10 inclusion/exclusion (primers are indicated as arrows in Fig. 6A) and HPRT for normalization. The results in Fig. 6B show that in control cells the band corresponding to the exclusion of exon 10 (E10−) is more visible than the one corresponding to exon 10 inclusion (E10+). This is in line with previous reports (19, 20) and with the fact that TAU exon 10 is naturally spliced out unless it is retained by alternative splicing. The ectopic expression of SC35 promoted inclusion of exon 10 in our system when the level of SC35 expression was relatively low (0.01 μg/ml). Gross overexpression of SC35 was achieved in the presence of 0.2 and 0.4 μg/ml plasmid DNA, and although it did not affect the E10+/E10− ratio, it partially attenuated both transcripts, as reported previously (20). For the next experiments, we utilized 0.01 μg/ml pSC35 plasmid, an amount in which both the inclusion and exclusion of exon 10 can be easily visualized. The results in Fig. 6C demonstrate that transfection of Tat resulted in an increased E10 exclusion (lane 3), although co-transfection with pSC35 prevented Tat-induced exon 10 splicing by promoting E10 inclusion (lane 4). The effects of Tat and SC35 on the exclusion of TAU exon 10 could be further appreciated after quantification and statistical analysis, as shown in Fig. 6D. These results confirm that Tat indeed promotes aberrant splicing of TAU through dysregulation of SC35.
FIGURE 6.
Direct evidence that Tat promotes exon 10 exclusion. A, scheme of TAU minigene as reported previously (20, 52). B, titration of SC35 overexpression in 293T cells to determine the concentration of pSC35 at which E10+ and E10− TAU mRNA are equally produced (0.01 μg/ml) in the cells transfected for 36 h with the TAU minigene. HPRT was PCR-multiplexed and used as an internal control (Ctrl) and for normalization. Levels of SC35 upon its expression were detected by Western blots (middle panel). Anti-14-3-3 was used as a loading control. C, multiplex RT-PCR to detect E10+ and E10− TAU RNAs in controls, cells transfected with SC35, Tat, or both. HPRT was used for normalization. D, quantitatively, the TAU E10 exclusion is increased when Tat is present, and overexpression of SC35 prevents this up-regulation. One and two asterisks represent the significance between groups (p ≤ 0.05 and p ≤ 0.01, respectively) resulting from three independent experiments. E, Western blotting to detect Tat in cells transfected with pEYFP-Tat, Myc-SC35, and TAU minigene in the indicated combinations after immunoprecipitation (IP) with anti-Tat antibody or IgG control. WL stands for whole lysates and was used as a positive control for the expression of Tat. Note that EYFP-Tat is about 50 kDa. F, image of a representative agarose gel showing RT-PCR-amplified E10+ and E10− Tau RNAs in the indicated samples as in E. G, representative Western blots (n = 3) to detect Tat in 293T cells transfected with pEYFP-Tat101 (lane 2), Myc-SC35 (lane 3), co-transfected with Myc-SC35 and pEYFP-Tat101 (lane 4), and control mock-transfected (lane 5). M and WL stand for marker and whole lysate (lanes 1 and 6, respectively).
In addition to its role in splicing, SC35 plays an active role in transcription elongation (53), and in releasing paused RNA polymerase II through binding to promoter-associated nascent RNA (54). Interestingly, the role of SC35 in gene activation has been suggested to be analogous to the mechanism by which Tat activates transcription. Furthermore, Tat has been shown to associate with cellular RNAs (55), suggesting that Tat and SC35 may compete for the same substrate, TAU exon 10, limiting the function of SC35. In addition, Tat may directly bind to SC35, limiting the function of this protein. To examine this possibility, we performed immunoprecipitation analysis and found that SC35 can pull down Tat in 293T cells transfected with Myc-SC35 and pEYFP-Tat101 (Fig. 6G). To test whether Tat associates with TAU RNA, we performed Tat-RNA immunoprecipitation following our protocol (44). In this experiment, pEYFP-Tat (Tat), Myc-SC35 (SC35), and TAU minigene (Tau) were transfected into 293T cells with the following combinations: Tat + TAU (sample 1); Tat alone (sample 2); SC35 +Tat + TAU (sample 3); and Tat + SC35 (sample 4). Efficient immunoprecipitation of Tat in all tested conditions was evaluated by Western blots and is shown in Fig. 6E. The amount of RNA extracted from samples immunoprecipitated with anti-Tat or IgG was about 25 and 1 ng/μl, respectively, confirming that Tat can associate with cellular RNAs, as recently reported (55). Semiquantitative RT-PCR revealed the presence of both E10 included (top band) and E10 excluded (lower band) in the samples in which Tat and TAU were present (Fig. 6F, lanes 1 and 3) but not in the controls without Tau (lanes 2 and 4) or without Tat antibody (IgG, lane 5). Of note, similar to the results in Fig. 6C, Tat promoted E10 exclusion, and this effect was prevented by the addition of SC35 (compare lanes 1 and 3). It should be noted that the PCR primers (with the reverse primer spanning through the plasmid sequence) are designed to detect E10 splicing only and not the TAU pre-mRNA.
Tat-mediated Increase in Phospho-SC35 Is Associated with Cytoplasmic SC35
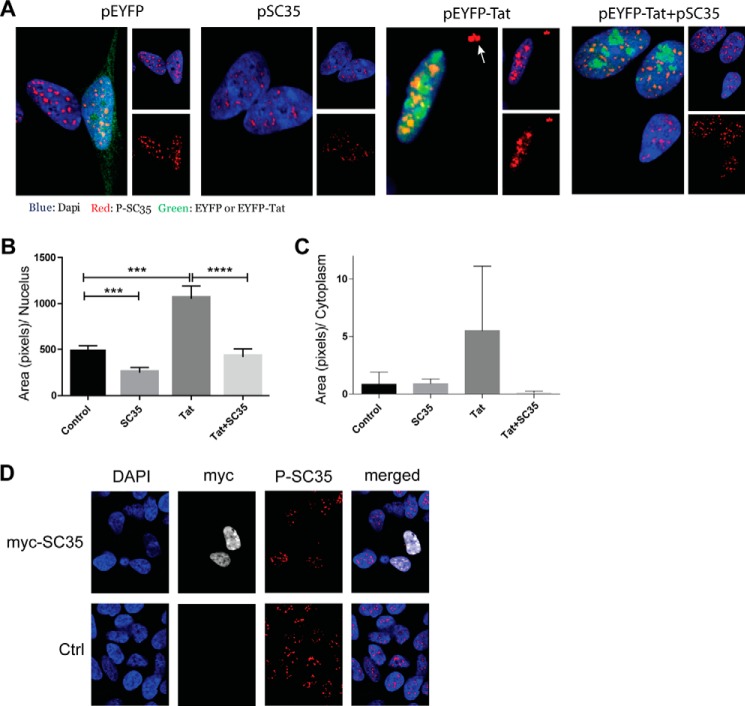
Because the release of SC35 into the nucleoplasm is associated with increased TAU 4R (20, 56), we hypothesized that Tat-mediated increases in exon 10 exclusion may be due to retention of phospho-SC35 in the nuclear speckles. SH-SY5Y cells were transfected with pEYFP, pSC35, or pEYFP-Tat or co-transfected with pEYFP-Tat and pSC35 for 24 h and subjected to immunofluorescence. Representative images from each group show the effect of Tat on phospho-SC35 (Fig. 7A). In control cells (pEYFP, green), phospho-SC35 (red) was detected in nuclear speckles, a pattern of expression also observed in primary neurons (Fig. 3A). In Tat-transfected cells (pEYFP-Tat), phospho-SC35 was dramatically increased in nuclear speckles with an increase in its cytoplasmic distribution in large clusters (arrow in Fig. 7A) as compared with the control. The Tat-mediated cytoplasmic and nuclear SC35 speckles were rescued when SC35 was co-transfected with pEYFP-Tat (Fig. 7A, right panel). The results in Fig. 7B demonstrate that in comparison with the control, comparison to s demonstrated an over 2-fold increase in nuclear phospho-SC35 (p ≤ 0.001) and that this phenotype was prevented by the addition of exogenous SC35 (p ≤ 0.0001). We also observed increased cytoplasmic phospho-SC35 in Tat-expressing cells compared with controls, although this increase was not statistically significant (Fig. 7C). In addition, we noted that the overexpression of SC35 alone reduced the fluorescent intensity of phospho-SC35 in our system (Fig. 7, A and B, p ≤ 0.001). This putative paradoxical effect observed during overexpression of SC35 is most likely mediated by the specificity of the anti-SC35 antibody used for immunofluorescence. This antibody (known also as a nuclear-speckle marker) has a high affinity for phospho-SC35 localized in the nuclear speckles. However, dephosphorylated or differentially phosphorylated SC35, which leaves the nuclear speckles to promote TAU exon 10 inclusion (20, 56), is most likely not detected or weakly detected by this antibody. To confirm this hypothesis, we overexpressed SC35 using a plasmid in which SC35 was cloned in-frame with a Myc tag (Fig. 7D). Results showed a very high expression of SC35 throughout the nucleus, as detected by the anti-Myc antibody. As anticipated, the pattern of phospho-SC35 immunolabeling did not overlap with anti-Myc immunoreactivity, indicating the specificity of the anti-phospho-SC35 antibody in detecting SC35 contained in nuclear speckles.
FIGURE 7.
Effects of Tat on SC35 phosphorylation in human SH-SY5Y cells. A, representative confocal microscopy images of SH-SY5Y cells transfected with control (pEYFP), pSC35, pEYFP-Tat, or pEYFP-Tat and pSC35. Immunofluorescence was performed 24 h post-transfection. B and C, quantification of phospho-SC35, represented as area in pixels per nucleus, was performed on the acquired images. Three and four asterisks represent the significance between groups (p ≤ 0.005 and p ≤ 0.0001, respectively) resulting from three independent experiments. D, representative images showing the distribution of SC35 in SH-SY5Y cells transfected with pcDNA3-Myc-SC35, using anti-Myc (white) and the nuclear speckle marker P-SC35 (red) antibodies. Nuclei are stained with DAPI (blue). Ctrl, control.
Involvement of GSK-3β and DYRK1A Kinases in the Tat-induced SC35 Dysfunction and Tau Exon 10 Splicing
Glycogen synthase kinase 3 (GSK-3) and DYRK1A have emerged as key kinases of SC35. Previous studies have shown that inactivation of GSK-3β leads to increased TAU 4R (8), suggesting that activation of this kinase may lead to increased TAU 3R. Additionally, another study found that overexpression of DYRK1A results in increased TAU 3R (20). Thus, the increased phosphorylation of SC35 and increased TAU 3R levels observed in our clinical samples, as well as in mouse and cellular models, prompted us to examine whether GSK-3β and/or DYRK1A was/were activated or up-regulated by Tat.
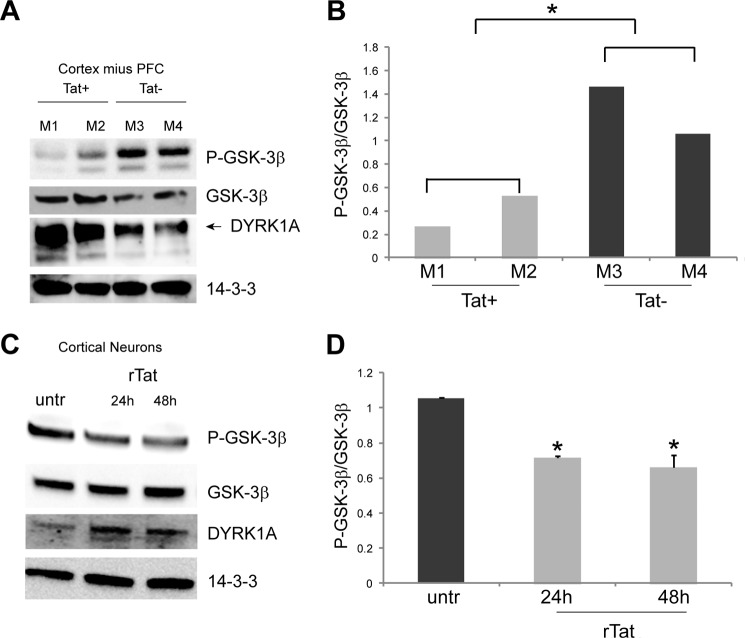
Fig. 8A shows the down-regulation (therefore activation) of phospho-GSK-3β (P-GSK-3β) in the cortex minus PFC of Tat+ mice compared with controls despite increased levels of total GSK-3β. Even more prominent was the up-regulation of DYRK1A in the same brain tissue samples. Similar results were obtained in DIV7 primary neurons treated with recombinant Tat (100 nm) at 24 and 48 h, in which GSK-3β was activated, and DYRK1A was increased (Fig. 8C). The Tat-mediated down-regulation of P-GSK-3β could be further appreciated in Fig. 8, B and D, in which levels of expression of P-GSK-3β and total GSK-3β were quantified and normalized with the loading control and expressed as the ratio P-GSK-3β/GSK-3β. Statistical analysis in Fig. 8B was performed on the average values between the two groups of mice (Tat+ and Tat−).
FIGURE 8.
Tat mediates up-regulation of DYRK1A and activation of GSK-3β in vivo and in cell culture. Western blots showing expression of P-GSK-3β (inactive form), total GSK-3β, and DYRK1A in Tat-Tg mouse brain tissues (A) and cultured mouse primary neurons (C). B, quantification of P-GSK-3β expression levels compared with total GSK-3β. Statistical analysis was performed on the average intensities between the groups of mice (Tat+ and Tat−). D, diagram showing densitometric quantification of the bands relative to P-GSK-3β and total GSK-3β from three independent experiments and expressed as a ratio P-GSK-3β/GSK-3β after normalization with 14-3-3 loading controls. Asterisks indicate p ≤ 0.05.
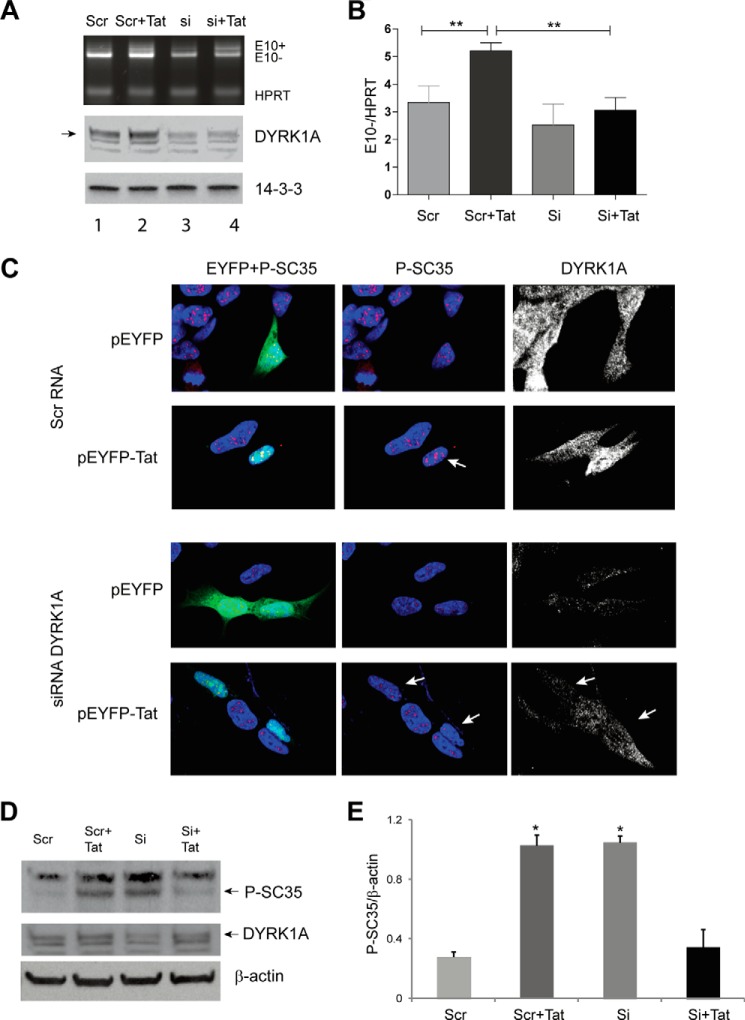
Because DYRK1A is a priming kinase for GSK-3β (57), we reasoned that down-regulation of DYRK1A would be sufficient to prevent the Tat-mediated increased expression of TAU 3R. This hypothesis was tested using the Tau minigene. The results in Fig. 9A show that the down-regulation of DYRK1A fully prevented the Tat-mediated Tau exon 10 splicing compared with all controls (Scr, Scr + Tat, si), as further documented by gel densitometry in which E10 exclusion was quantified and normalized with HPRT (Fig. 9B). siRNA-mediated down-regulation of DYRK1A was confirmed by Western blot analysis (Fig. 9A, middle panel).
FIGURE 9.
Silencing DYRK1A inhibits Tat-mediated increase in P-SC35 and promotes TAU 10 exclusion. A, agarose gel of a representative multiplex RT-PCR to determine changes in Tau E10 inclusion (E10+) and exclusion (E10−) in cells co-transfected with Tat and scrambled siRNA (Scr) or siRNA to DYRK1A (si) at 24 h post-transfection. HPRT was used as a loading control. Levels of DYRK1A upon its down-regulation by siRNAs were detected by Western blots. Anti-14-3-3 was used as a loading control. B, quantification analysis of Tau E10 exclusion in the indicated experimental conditions. Statistical significance is indicated by the asterisks (p ≤ 0.01) and results from three independent experiments. C, representative confocal immunofluorescent microscopy images of pEYFP and pEYFP-Tat-transfected SH-SY5Y cells (green cells) in the presence of scrambled siRNA (top two panels) or DYRK1A siRNA (bottom two panels). Phospho-SC35 (P-SC35) is identified in red and DYRK1A in white. Nuclei are stained with DAPI (blue). White arrows indicate differences as detailed in the text. D, Western blots showing levels of P-SC35 and DYRK1A in the experimental setting as in C. β-Actin was used as loading control. E, densitometric analysis of the bands relative to P-SC35 compared with β-actin from two experiments. Asterisks indicate p ≤ 0.05 when compared with values corresponding to either Scr or Si +Tat samples.
We next determined whether down-regulation of DYRK1A would restore baseline levels of SC35 phosphorylation in Tat-expressing cells by performing confocal immunofluorescent microscopy. SH-SY5Y cells were co-transfected with pEYFP or pEYFP-Tat and either the scramble control (Fig. 9C, top two panels) or DYRK1A siRNAs (Fig. 9C, lower panels). Cells expressing Tat again showed increased levels of phospho-SC35 (Fig. 9C, white arrow) and DYRK1A compared with untransfected cells in the same field or pEYFP-transfected cells; co-transfection with the control siRNA did not alter the immunolabeling pattern of SC35 in all conditions. Silencing DYRK1A in control pEYFP-transfected cells resulted in lower levels of phospho-SC35 in nuclear speckles, confirming previous studies (20). Importantly, the down-regulation of DYRK1A in Tat-expressing cells (Fig. 9C, bottom panels, arrows) was sufficient to prevent increased levels of phosphorylated SC35, and the punctate pattern of the nuclear speckles was similar to the control (Scr RNA). In the same experimental setting, we evaluated levels of SC35 by Western blots, and results are shown in Fig. 9, D and E. In the presence of the scrambled RNA, transfection of Tat increased the levels of SC35 compared with the control transfected only with the scrambled RNA (Fig. 9D, 1st two lanes), although silencing DYRK1A efficiently prevented this increase (Si + Tat). Interestingly, down-regulation of DYRK1A determined an increase in P-SC35 (Fig. 9D, Si). This effect might be due to the phosphorylation of SC35 from other kinases after SC35 leaves the nuclear speckles, although specific experiments would be required to evaluate this hypothesis. Altogether, these results indicate that up-regulation of DYRK1A is required for Tat to alter the SC35-dependent TAU exon 10 splicing.
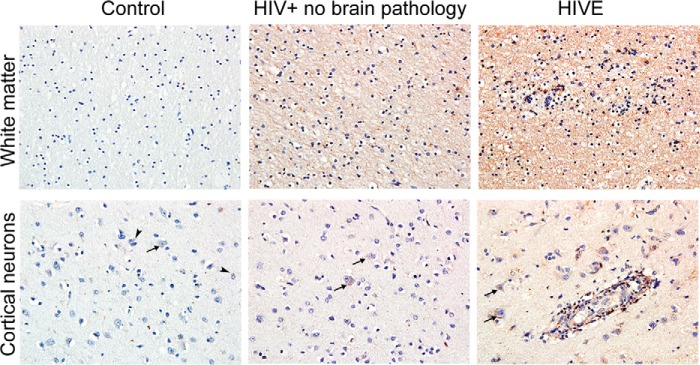
Finally, immunohistochemistry analysis of brain autopsies demonstrated low levels of DYRK1A in neurons (arrows) and glia (arrowheads) of control brains (Fig. 10). In contrast, HIVE brains showed a different cellular distribution with the increased presence of DYRK1A in the neuronal soma and in axons. Interestingly, HIV+ cases without detected encephalitis demonstrated an intermediate degree of labeling.
FIGURE 10.
DYRK1A expression in clinical samples. Representative images of DYRK1A immunolabeling in the white matter (original magnification ×20) and prefrontal cortex (original magnification ×40) of control brains, HIV+ without brain pathology, and HIVE (n = 4/group). A modest labeling of DYRK1A was observed in normal white matter and frontal cortex. Strikingly, intense labeling of axons and neuronal soma were observed in HIV+ without brain pathology, and this pattern was even stronger in HIVE cases. Arrows and arrowheads indicate neurons and glial cells, respectively.
Discussion
The HIV-1 transactivator Tat has been shown to interact with a variety of cellular factors altering their normal function in both infected and uninfected cells (58–63). In respect to RNA metabolism, Tat directly participates in the splicing of viral RNA (64); however, the effect(s) of Tat on mechanisms involved in processing cellular RNA in the absence of the viral genome is/are largely unexplored. Intuitively, Tat should still be able to bind and alter the function of transcriptional/translational complexes in uninfected cells even in the absence of other viral factors that usually mediate those interactions, such as the LTR or other viral proteins. In addition, recent reports indicating that cellular factors such as SC35 can function in a manner similar to Tat (53, 54) further support the possibility that Tat itself may compete with cellular proteins or with host RNA species to affect RNA metabolism. This possibility was confirmed by RNA immunoprecipitation coupled with microarray analysis, which demonstrated that Tat can interact with cellular RNAs in T cells (55). Although in that report TAU RNA was not detected, likely due to the low abundance of TAU in T cells, there is a surprising similarity between Tau exon 10 and TAR LTR stem-loop structures, suggesting the possibility of a Tat-TAU RNA interaction. Indeed, our data in which Tat pulled down TAU RNA strongly indicate that such a possibility may exist (Fig. 6).
Neurodegenerative tauopathies have been reported to be associated with an aberrant splicing of TAU (7, 8, 13, 14); however, to the best of our knowledge TAU aberrant splicing has never been investigated in the setting of HIV. Because our immunohistochemistry data demonstrate both alterations in TAU 3R and 4R isoforms and apparent changes in the levels of SC35 phosphorylation in HIVE cases compared with controls (Figs. 1 and 2), we sought to investigate molecular mechanisms involved in the SC35-mediated TAU exon 10 splicing. With respect to SC35, incubation of primary mouse cortical neurons with recombinant Tat affected both the phosphorylation levels and gel mobility of SC35 (Fig. 3). Our data showing increased intensity in the band relative to dephosphorylated SC35 (Fig. 3D) are in line with a previous report suggesting that this phenomenon is due to the presence of a domain in SC35 that confers a remarkable resistance to phosphatases (65).
Increased phosphorylation of SC35 paralleled the shift toward TAU 3R isoform in primary neurons treated with Tat (Fig. 5). Because of the presence of four microtubule-binding domains, TAU 4R isoforms bind to microtubules much more efficiently than the 3R isoforms (6, 7). Previous studies have shown that the Tau 3R is the most abundant isoform in fetal and newborn mice, although in the adult mouse Tau 4R is the predominant isoform (66–69). However, regional and cell-specific variations in the expression of Tau isoforms have been identified, which likely reflect unique regional differences in the function of Tau isoforms (69). Regional variations in TAU isoforms were also demonstrated here. For instance, our data show more Tau 3R than 4R in the mouse hippocampus (Fig. 4A), which may reflect higher neuronal plasticity of these parts of the adult brain. In contrast, other cortical regions demonstrate Tau 4R as the most prevalent isoform (Fig. 4, B and C). Although our controls are not wild-type mice but mice transgenic for the RTTA gene (see under “Experimental Procedures”), our data are in line with previous reports showing Tau 4R as the most prevalent isoform in the adult mouse brain (69). In the hippocampus the switch from Tau 3R to 4R occurs later than in other brain regions, and expression of Tau 3R is maintained in specific areas (69); therefore, it may not be surprising that we still detect more Tau 3R than 4R in the hippocampus of our RTTA control mice.
Increased expression of SC35 has been observed in HIV-infected cells (70, 71) and correlated with the expression of Tat (70). However, there are no reports on the effect of Tat on SC35 expression in non-infected cells. In vitro experiments using the TAU minigene demonstrated that in the presence of Tat, TAU exon 10 (E10) is spliced out more efficiently and that SC35 co-transfection can counteract this effect, leading to more balanced E10 inclusion (Fig. 6, C and D). Interestingly, similar E10 inclusion/exclusion ratios were obtained when Tat-RNA immunoprecipitation was performed (Fig. 6F). Importantly, these results also demonstrated that Tat can associate with TAU RNA. Because both alternatively spliced forms of TAU RNAs (E10 excluded and included) were pulled down by Tat, it is possible that Tat-TAU RNA association is mediated by another molecule that, in addition to Tat, also interacts with SC35. The positive transcription elongation factor b (P-TEFb) would indeed fit in this model because SC35 and Tat both have the ability to associate with this complex (53, 54, 72–74). In line with this model, Tat and SC35 could compete for the association of the P-TEFb complex to the nascent TAU mRNA in a manner similar to Tat and another member of the SR family of splicing factors, SF2/ASF, as recently demonstrated for the association of the HIV transcript (75). Our experiments further indicate that Tat could inhibit SC35 function through binding (Fig. 6G). In addition to affecting the function of SC35, the expression of Tat could result in the down-regulation and/or dysfunction of other molecules that stabilize the TAU exon 10 RNA stem loop and promote its inclusion, such as the DEAD-box p68 RNA helicase (DDX5) (76).
Tat-mediated increases in phospho-SC35 and the enlargement of SC35-containing nuclear speckles was reversed by co-transfection with pSC35 (Fig. 7, A–C). The fact that overexpression of SC35 results in lower P-SC35 detected in the nuclear speckles (Fig. 7), despite the increase in the overall amount of P-SC35 (Fig. 6B, top band), may also explain the increased levels of P-SC35 when DYRK1A is down-regulated (Fig. 9, C and D) and confirms the phosphorylation of SC35 by other kinases.
Earlier studies have shown that SC35 promotes TAU exon 10 inclusion by binding to the SC35-like enhancer region on the TAU pre-mRNA and that DYRK1A-mediated SC35 phosphorylation inhibits exon 10 inclusion (20). DYRK1A maps to chromosome 21 (77) and is overexpressed in Down syndrome (78, 79), Alzheimer disease, and Pick disease (79). We also found robust increase of DYRK1A in HIVE autopsies compared with controls (Fig. 10). Interestingly, HIV+ cases without encephalitis showed intermediate levels of immunolabeling, which may point to DYRK1A dysregulation as an early event in the neuronal dysfunction associated with HIV infection.
Previous reports have shown that inhibition of GSK-3β results in TAU exon 10 inclusion (8) and that DYRK1A is a priming kinase for GSK-3β (57). In line with these data, we found a substantial increase in DYRK1A expression and GSK-3β activation in Tat-tg mice and in cell cultures. Importantly, we found that the Tat-mediated up-regulation of phospho-SC35, its retention in the nuclear speckles, and up-regulation of TAU exon 10 exclusion were dependent on the increased expression of DYRK1A, because siRNA-mediated DYRK1A reduction prevented these effects (Figs. 8 and 9).
Among the known mechanisms that determine an increase in DYRK1A, the RE1 silencing transcription factor has been shown to regulate DYRK1A transcription in a negative feedback loop (80). In an attempt to identify the mechanism(s) by which Tat may promote increased expression of DYRK1A, we have evaluated expression levels of DYRK1A and REST mRNAs in Tat-Tg mice as well as in SH-SY5Y cells transfected with Tat. However, no significant changes in DYRK1A and REST mRNAs were observed in any of the experimental conditions (data not shown), suggesting that Tat could promote up-regulation of DYRK1A through protein stabilization. Indeed, Tat has been shown to block the catalytic activity of the proteasome through direct binding with several subunits of the 26S (81) and to switch the proteasome to a non-proteolytic mode through dissociation of the proteasome into 19S and 20S particles (82). In addition, we have previously shown a Tat-mediated subcellular redistribution of the 20S subunits in cultured neurons and in HIVE clinical samples (39). Altogether, our data show that the Tat-induced DYRK1A up-regulation does not involve increased transcription, but previous literature suggests it could likely be due to increased protein stability through dysregulation of the proteasome. Although further experiments are necessary to determine the involvement of the proteasome in the Tat-mediated DYRK1A stability, our data unravel a novel mechanism in which Tat can dysregulate the DYRK1A/SC35 axis and promote TAU exon 10 exclusion.
Author Contributions
F. K. performed most of the experiments; A. W., M. P., D. J., and A. L. performed the rest of the experiments; A. P. S. and L. D. V. performed and interpreted immunohistochemistry analyses; K. F. H. and P. E. K. provided mouse brain tissues; C. P. contributed to discussion and interpretation of data; and F. P. designed the study and wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Michelle Holt for providing editorial assistance. We thank Dr. F. Liu for the Tau minigene.
This work was supported in part by the National Institutes of Health NIGMS Grant 1U54 GM104940 (to F. P.), which funds the Louisiana Clinical and Translational Science Center, by the National Institutes of Health Grants P20 GM103501 (to F. P. and C. P.), R01 DA034231 (to P. E. K.), and K02 DA027374 (to K. F. H.) and by the Louisiana Cancer Research Center and the Stanley S. Scott Cancer Research Center at Louisiana State University Health Sciences Center. The authors declare that they have no conflicts of interest with the content of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- MTBD
- microtubule-binding domain
- FFPE
- formalin-fixed, paraffin-embedded
- HPRT
- hypoxanthine-guanine phosphoribosyltransferase
- PFC
- prefrontal cortex
- HIVE
- HIV-encephalitis
- DIV
- days in vitro
- REST
- RE1 silencing transcription factor.
References
- 1. Heaton R. K., Clifford D. B., Franklin D. R. Jr., Woods S. P., Ake C., Vaida F., Ellis R. J., Letendre S. L., Marcotte T. D., Atkinson J. H., Rivera-Mindt M., Vigil O. R., Taylor M. J., Collier A. C., Marra C. M., et al. (2010) HIV-associated neurocognitive disorders persist in the era of potent antiretroviral therapy: CHARTER study. Neurology 75, 2087–2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McArthur J. C., Steiner J., Sacktor N., and Nath A. (2010) Human immunodeficiency virus-associated neurocognitive disorders: mind the gap. Ann. Neurol. 67, 699–714 [DOI] [PubMed] [Google Scholar]
- 3. Bachani M., Sacktor N., McArthur J. C., Nath A., and Rumbaugh J. (2013) Detection of anti-tat antibodies in CSF of individuals with HIV-associated neurocognitive disorders. J. Neurovirol. 19, 82–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Johnson T. P., Patel K., Johnson K. R., Maric D., Calabresi P. A., Hasbun R., and Nath A. (2013) Induction of IL-17 and nonclassical T-cell activation by HIV-Tat protein. Proc. Natl. Acad. Sci. U.S.A. 110, 13588–13593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Panda D., Samuel J. C., Massie M., Feinstein S. C., and Wilson L. (2003) Differential regulation of microtubule dynamics by three- and four-repeat tau: implications for the onset of neurodegenerative disease. Proc. Natl. Acad. Sci. U.S.A. 100, 9548–9553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Avila J., Lucas J. J., Perez M., and Hernandez F. (2004) Role of tau protein in both physiological and pathological conditions. Physiol. Rev. 84, 361–384 [DOI] [PubMed] [Google Scholar]
- 7. Buée L., Bussière T., Buée-Scherrer V., Delacourte A., and Hof P. R. (2000) Tau protein isoforms, phosphorylation and role in neurodegenerative disorders. Brain Res. Brain Res. Rev. 33, 95–130 [DOI] [PubMed] [Google Scholar]
- 8. Hernández F., Pérez M., Lucas J. J., Mata A. M., Bhat R., and Avila J. (2004) Glycogen synthase kinase-3 plays a crucial role in tau exon 10 splicing and intranuclear distribution of SC35. Implications for Alzheimer's disease. J. Biol. Chem. 279, 3801–3806 [DOI] [PubMed] [Google Scholar]
- 9. Liu F., and Gong C. X. (2008) Tau exon 10 alternative splicing and tauopathies. Mol. Neurodegener. 3, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yin X., Jin N., Gu J., Shi J., Zhou J., Gong C. X., Iqbal K., Grundke-Iqbal I., and Liu F. (2012) Dual-specificity tyrosine phosphorylation-regulated kinase 1A (Dyrk1A) modulates serine/arginine-rich protein 55 (SRp55)-promoted Tau exon 10 inclusion. J. Biol. Chem. 287, 30497–30506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Espinoza M., de Silva R., Dickson D. W., and Davies P. (2008) Differential incorporation of tau isoforms in Alzheimer's disease. J. Alzheimers Dis. 14, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shi J., Qian W., Yin X., Iqbal K., Grundke-Iqbal I., Gu X., Ding F., Gong C. X., and Liu F. (2011) Cyclic AMP-dependent protein kinase regulates the alternative splicing of tau exon 10: a mechanism involved in tau pathology of Alzheimer disease. J. Biol. Chem. 286, 14639–14648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dawson H. N., Cantillana V., Chen L., and Vitek M. P. (2007) The tau N279K exon 10 splicing mutation recapitulates frontotemporal dementia and parkinsonism linked to chromosome 17 tauopathy in a mouse model. J. Neurosci. 27, 9155–9168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wegiel J., Kaczmarski W., Barua M., Kuchna I., Nowicki K., Wang K. C., Wegiel J., Yang S. M., Frackowiak J., Mazur-Kolecka B., Silverman W. P., Reisberg B., Monteiro I., de Leon M., Wisniewski T., et al. (2011) Link between DYRK1A overexpression and several-fold enhancement of neurofibrillary degeneration with 3-repeat tau protein in Down syndrome. J. Neuropathol. Exp. Neurol. 70, 36–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Foster N. L., Wilhelmsen K., Sima A. A., Jones M. Z., D'Amato C. J., and Gilman S. (1997) Frontotemporal dementia and parkinsonism linked to chromosome 17: a consensus conference. Conference participants. Ann. Neurol. 41, 706–715 [DOI] [PubMed] [Google Scholar]
- 16. Bourgeois C. F., Lejeune F., and Stévenin J. (2004) Broad specificity of SR (serine/arginine) proteins in the regulation of alternative splicing of pre-messenger RNA. Prog. Nucleic Acid Res. Mol. Biol. 78, 37–88 [DOI] [PubMed] [Google Scholar]
- 17. Huang Y., Yario T. A., and Steitz J. A. (2004) A molecular link between SR protein dephosphorylation and mRNA export. Proc. Natl. Acad. Sci. U.S.A. 101, 9666–9670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ma X., and He F. (2003) Advances in the study of SR protein family. Genomics Proteomics Bioinformatics 1, 2–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Qian W., Iqbal K., Grundke-Iqbal I., Gong C. X., and Liu F. (2011) Splicing factor SC35 promotes tau expression through stabilization of its mRNA. FEBS Lett. 585, 875–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Qian W., Liang H., Shi J., Jin N., Grundke-Iqbal I., Iqbal K., Gong C. X., and Liu F. (2011) Regulation of the alternative splicing of tau exon 10 by SC35 and Dyrk1A. Nucleic Acids Res. 39, 6161–6171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ghosh G., and Adams J. A. (2011) Phosphorylation mechanism and structure of serine-arginine protein kinases. FEBS J. 278, 587–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lamond A. I., and Spector D. L. (2003) Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 4, 605–612 [DOI] [PubMed] [Google Scholar]
- 23. Spector D. L., and Lamond A. I. (2011) Nuclear speckles. Cold Spring Harb. Perspect. Biol. 3, a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhou Z., and Fu X. D. (2013) Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma 122, 191–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Alvarez G., Muñoz-Montaño J. R., Satrústegui J., Avila J., Bogónez E., and Díaz-Nido J. (1999) Lithium protects cultured neurons against β-amyloid-induced neurodegeneration. FEBS Lett. 453, 260–264 [DOI] [PubMed] [Google Scholar]
- 26. Dewhurst S., Maggirwar S. B., Schifitto G., Gendelman H. E., and Gelbard H. A. (2007) Glycogen synthase kinase 3 β (GSK-3 β) as a therapeutic target in neuroAIDS. J. Neuroimmune Pharmacol. 2, 93–96 [DOI] [PubMed] [Google Scholar]
- 27. Eldar-Finkelman H., and Martinez A. (2011) GSK-3 Inhibitors: preclinical and clinical focus on CNS. Front. Mol. Neurosci. 4, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lovestone S., Davis D. R., Webster M. T., Kaech S., Brion J. P., Matus A., and Anderton B. H. (1999) Lithium reduces tau phosphorylation: effects in living cells and in neurons at therapeutic concentrations. Biol. Psychiatry 45, 995–1003 [DOI] [PubMed] [Google Scholar]
- 29. Medina M., and Wandosell F. (2011) Deconstructing GSK-3: the fine regulation of its activity. Int. J. Alzheimers Dis. 2011, 479249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Plattner F., Angelo M., and Giese K. P. (2006) The roles of cyclin-dependent kinase 5 and glycogen synthase kinase 3 in tau hyperphosphorylation. J. Biol. Chem. 281, 25457–25465 [DOI] [PubMed] [Google Scholar]
- 31. Hämmerle B., Ulin E., Guimera J., Becker W., Guillemot F., and Tejedor F. J. (2011) Transient expression of Mnb/Dyrk1a couples cell cycle exit and differentiation of neuronal precursors by inducing p27KIP1 expression and suppressing NOTCH signaling. Development 138, 2543–2554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wegiel J., Gong C. X., and Hwang Y. W. (2011) The role of DYRK1A in neurodegenerative diseases. FEBS J. 278, 236–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ahn K. J., Jeong H. K., Choi H. S., Ryoo S. R., Kim Y. J., Goo J. S., Choi S. Y., Han J. S., Ha I., and Song W. J. (2006) DYRK1A BAC transgenic mice show altered synaptic plasticity with learning and memory defects. Neurobiol. Dis. 22, 463–472 [DOI] [PubMed] [Google Scholar]
- 34. Altafaj X., Martín E. D., Ortiz-Abalia J., Valderrama A., Lao-Peregrín C., Dierssen M., and Fillat C. (2013) Normalization of Dyrk1A expression by AAV2/1-shDyrk1A attenuates hippocampal-dependent defects in the Ts65Dn mouse model of Down syndrome. Neurobiol. Dis. 52, 117–127 [DOI] [PubMed] [Google Scholar]
- 35. Chen C. K., Bregere C., Paluch J., Lu J. F., Dickman D. K., and Chang K. T. (2014) Activity-dependent facilitation of synaptojanin and synaptic vesicle recycling by the minibrain kinase. Nat. Commun. 5, 4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Grau C., Arató K., Fernández-Fernández J. M., Valderrama A., Sindreu C., Fillat C., Ferrer I., de la Luna S., and Altafaj X. (2014) DYRK1A-mediated phosphorylation of GluN2A at Ser(1048) regulates the surface expression and channel activity of GluN1/GluN2A receptors. Front. Cell. Neurosci. 8, 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Peineau S., Nicolas C. S., Bortolotto Z. A., Bhat R. V., Ryves W. J., Harwood A. J., Dournaud P., Fitzjohn S. M., and Collingridge G. L. (2009) A systematic investigation of the protein kinases involved in NMDA receptor-dependent LTD: evidence for a role of GSK-3 but not other serine/threonine kinases. Mol. Brain 2, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Souchet B., Guedj F., Sahún I., Duchon A., Daubigney F., Badel A., Yanagawa Y., Barallobre M. J., Dierssen M., Yu E., Herault Y., Arbones M., Janel N., Créau N., and Delabar J. M. (2014) Excitation/inhibition balance and learning are modified by Dyrk1a gene dosage. Neurobiol. Dis. 69, 65–75 [DOI] [PubMed] [Google Scholar]
- 39. Aprea S., Del Valle L., Mameli G., Sawaya B. E., Khalili K., and Peruzzi F. (2006) Tubulin-mediated binding of human immunodeficiency virus-1 Tat to the cytoskeleton causes proteasomal-dependent degradation of microtubule-associated protein 2 and neuronal damage. J. Neurosci. 26, 4054–4062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pacifici M., and Peruzzi F. (2012) Isolation and culture of rat embryonic neural cells: a quick protocol. J. Vis. Exp. 63, e3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bruce-Keller A. J., Turchan-Cholewo J., Smart E. J., Geurin T., Chauhan A., Reid R., Xu R., Nath A., Knapp P. E., and Hauser K. F. (2008) Morphine causes rapid increases in glial activation and neuronal injury in the striatum of inducible HIV-1 Tat transgenic mice. Glia 56, 1414–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fitting S., Scoggins K. L., Xu R., Dever S. M., Knapp P. E., Dewey W. L., and Hauser K. F. (2012) Morphine efficacy is altered in conditional HIV-1 Tat transgenic mice. Eur. J. Pharmacol. 689, 96–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Passiatore G., Rom S., Eletto D., and Peruzzi F. (2009) HIV-1 Tat C-terminus is cleaved by calpain 1: implication for Tat-mediated neurotoxicity. Biochim. Biophys. Acta 1793, 378–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jeansonne D., DeLuca M., Marrero L., Lassak A., Pacifici M., Wyczechowska D., Wilk A., Reiss K., and Peruzzi F. (2015) Anti-tumoral Effects of miR-3189–3p in Glioblastoma. J. Biol. Chem. 290, 8067–8080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rom S., Rom I., Passiatore G., Pacifici M., Radhakrishnan S., Del Valle L., Piña-Oviedo S., Khalili K., Eletto D., and Peruzzi F. (2010) CCL8/MCP-2 is a target for mir-146a in HIV-1-infected human microglial cells. FASEB J. 24, 2292–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rom S., Pacifici M., Passiatore G., Aprea S., Waligorska A., Del Valle L., and Peruzzi F. (2011) HIV-1 Tat binds to SH3 domains: cellular and viral outcome of Tat/Grb2 interaction. Biochim. Biophys. Acta 1813, 1836–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Spector D. L., Fu X. D., and Maniatis T. (1991) Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO J. 10, 3467–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fitting S., Ignatowska-Jankowska B. M., Bull C., Skoff R. P., Lichtman A. H., Wise L. E., Fox M. A., Su J., Medina A. E., Krahe T. E., Knapp P. E., Guido W., and Hauser K. F. (2013) Synaptic dysfunction in the hippocampus accompanies learning and memory deficits in human immunodeficiency virus type-1 Tat transgenic mice. Biol. Psychiatry 73, 443–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fitting S., Xu R., Bull C., Buch S. K., El-Hage N., Nath A., Knapp P. E., and Hauser K. F. (2010) Interactive comorbidity between opioid drug abuse and HIV-1 Tat: chronic exposure augments spine loss and sublethal dendritic pathology in striatal neurons. Am. J. Pathol. 177, 1397–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen Q., Zhou Z., Zhang L., Xu S., Chen C., and Yu Z. (2014) The cellular distribution and Ser262 phosphorylation of tau protein are regulated by BDNF in vitro. PLoS One 9, e91793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jämsä A., Hasslund K., Cowburn R. F., Bäckström A., and Vasänge M. (2004) The retinoic acid and brain-derived neurotrophic factor differentiated SH-SY5Y cell line as a model for Alzheimer's disease-like tau phosphorylation. Biochem. Biophys. Res. Commun. 319, 993–1000 [DOI] [PubMed] [Google Scholar]
- 52. Yu Q., Guo J., and Zhou J. (2004) A minimal length between tau exon 10 and 11 is required for correct splicing of exon 10. J. Neurochem. 90, 164–172 [DOI] [PubMed] [Google Scholar]
- 53. Lin S., Coutinho-Mansfield G., Wang D., Pandit S., and Fu X. D. (2008) The splicing factor SC35 has an active role in transcriptional elongation. Nat. Struct. Mol. Biol. 15, 819–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ji X., Zhou Y., Pandit S., Huang J., Li H., Lin C. Y., Xiao R., Burge C. B., and Fu X. D. (2013) SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell 153, 855–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bouwman R. D., Palser A., Parry C. M., Coulter E., Rasaiyaah J., Kellam P., and Jenner R. G. (2014) Human immunodeficiency virus Tat associates with a specific set of cellular RNAs. Retrovirology 11, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Misteli T., Cáceres J. F., Clement J. Q., Krainer A. R., Wilkinson M. F., and Spector D. L. (1998) Serine phosphorylation of SR proteins is required for their recruitment to sites of transcription in vivo. J. Cell Biol. 143, 297–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Aranda S., Laguna A., and de la Luna S. (2011) DYRK family of protein kinases: evolutionary relationships, biochemical properties, and functional roles. FASEB J. 25, 449–462 [DOI] [PubMed] [Google Scholar]
- 58. Brigati C., Giacca M., Noonan D. M., and Albini A. (2003) HIV Tat, its TARgets and the control of viral gene expression. FEMS Microbiol. Lett. 220, 57–65 [DOI] [PubMed] [Google Scholar]
- 59. Johnson T. P., and Nath A. (2014) New insights into immune reconstitution inflammatory syndrome of the central nervous system. Curr. Opin. HIV AIDS 9, 572–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Johri M. K., Mishra R., Chhatbar C., Unni S. K., and Singh S. K. (2011) Tits and bits of HIV Tat protein. Expert Opin. Biol. Ther. 11, 269–283 [DOI] [PubMed] [Google Scholar]
- 61. King J. E., Eugenin E. A., Buckner C. M., and Berman J. W. (2006) HIV tat and neurotoxicity. Microbes Infect. 8, 1347–1357 [DOI] [PubMed] [Google Scholar]
- 62. Peruzzi F. (2006) The multiple functions of HIV-1 Tat: proliferation versus apoptosis. Front. Biosci. 11, 708–717 [DOI] [PubMed] [Google Scholar]
- 63. Del Valle L., Croul S., Morgello S., Amini S., Rappaport J., and Khalili K. (2000) Detection of HIV-1 Tat and JCV capsid protein, VP1, in AIDS brain with progressive multifocal leukoencephalopathy. J. Neurovirol. 6, 221–228 [DOI] [PubMed] [Google Scholar]
- 64. Jablonski J. A., Amelio A. L., Giacca M., and Caputi M. (2010) The transcriptional transactivator Tat selectively regulates viral splicing. Nucleic Acids Res. 38, 1249–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lin S., Xiao R., Sun P., Xu X., and Fu X. D. (2005) Dephosphorylation-dependent sorting of SR splicing factors during mRNP maturation. Mol. Cell 20, 413–425 [DOI] [PubMed] [Google Scholar]
- 66. Janke C., Beck M., Stahl T., Holzer M., Brauer K., Bigl V., and Arendt T. (1999) Phylogenetic diversity of the expression of the microtubule-associated protein tau: implications for neurodegenerative disorders. Brain Res. Mol. Brain Res. 68, 119–128 [DOI] [PubMed] [Google Scholar]
- 67. Kampers T., Pangalos M., Geerts H., Wiech H., and Mandelkow E. (1999) Assembly of paired helical filaments from mouse tau: implications for the neurofibrillary pathology in transgenic mouse models for Alzheimer's disease. FEBS Lett. 451, 39–44 [DOI] [PubMed] [Google Scholar]
- 68. Takuma H., Arawaka S., and Mori H. (2003) Isoforms changes of tau protein during development in various species. Brain Res. Dev. Brain Res. 142, 121–127 [DOI] [PubMed] [Google Scholar]
- 69. McMillan P., Korvatska E., Poorkaj P., Evstafjeva Z., Robinson L., Greenup L., Leverenz J., Schellenberg G. D., and D'Souza I. (2008) Tau isoform regulation is region- and cell-specific in mouse brain. J. Comp. Neurol. 511, 788–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Dowling D., Nasr-Esfahani S., Tan C. H., O'Brien K., Howard J. L., Jans D. A., Purcell D. F., Stoltzfus C. M., and Sonza S. (2008) HIV-1 infection induces changes in expression of cellular splicing factors that regulate alternative viral splicing and virus production in macrophages. Retrovirology 5, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Maldarelli F., Xiang C., Chamoun G., and Zeichner S. L. (1998) The expression of the essential nuclear splicing factor SC35 is altered by human immunodeficiency virus infection. Virus Res. 53, 39–51 [DOI] [PubMed] [Google Scholar]
- 72. Wei P., Garber M. E., Fang S. M., Fischer W. H., and Jones K. A. (1998) A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92, 451–462 [DOI] [PubMed] [Google Scholar]
- 73. Zhu Y., Pe'ery T., Peng J., Ramanathan Y., Marshall N., Marshall T., Amendt B., Mathews M. B., and Price D. H. (1997) Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 11, 2622–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Karn J., and Stoltzfus C. M. (2012) Transcriptional and posttranscriptional regulation of HIV-1 gene expression. Cold Spring Harb. Perspect. Med. 2, a006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Paz S., Krainer A. R., and Caputi M. (2014) HIV-1 transcription is regulated by splicing factor SRSF1. Nucleic Acids Res. 42, 13812–13823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kar A., Fushimi K., Zhou X., Ray P., Shi C., Chen X., Liu Z., Chen S., and Wu J. Y. (2011) RNA helicase p68 (DDX5) regulates tau exon 10 splicing by modulating a stem-loop structure at the 5′ splice site. Mol. Cell. Biol. 31, 1812–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Korenberg J. R., Chen X. N., Schipper R., Sun Z., Gonsky R., Gerwehr S., Carpenter N., Daumer C., Dignan P., and Disteche C. (1994) Down syndrome phenotypes: the consequences of chromosomal imbalance. Proc. Natl. Acad. Sci. U.S.A. 91, 4997–5001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Dowjat W. K., Adayev T., Kuchna I., Nowicki K., Palminiello S., Hwang Y. W., and Wegiel J. (2007) Trisomy-driven overexpression of DYRK1A kinase in the brain of subjects with Down syndrome. Neurosci. Lett. 413, 77–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ferrer I., Barrachina M., Puig B., Martínez de Lagrán M., Martí E., Avila J., and Dierssen M. (2005) Constitutive Dyrk1A is abnormally expressed in Alzheimer disease, Down syndrome, Pick disease, and related transgenic models. Neurobiol. Dis. 20, 392–400 [DOI] [PubMed] [Google Scholar]
- 80. Lu M., Zheng L., Han B., Wang L., Wang P., Liu H., and Sun X. (2011) REST regulates DYRK1A transcription in a negative feedback loop. J. Biol. Chem. 286, 10755–10763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Apcher G. S., Heink S., Zantopf D., Kloetzel P. M., Schmid H. P., Mayer R. J., and Krüger E. (2003) Human immunodeficiency virus-1 Tat protein interacts with distinct proteasomal α and β subunits. FEBS Lett. 553, 200–204 [DOI] [PubMed] [Google Scholar]
- 82. Lassot I., Latreille D., Rousset E., Sourisseau M., Linares L. K., Chable-Bessia C., Coux O., Benkirane M., and Kiernan R. E. (2007) The proteasome regulates HIV-1 transcription by both proteolytic and nonproteolytic mechanisms. Mol. Cell 25, 369–383 [DOI] [PubMed] [Google Scholar]