Graphical abstract
Keywords: NMR, 1H NMR, 13C NMR, Diketopiperazine, Fungi, Secondary metabolites
Introduction
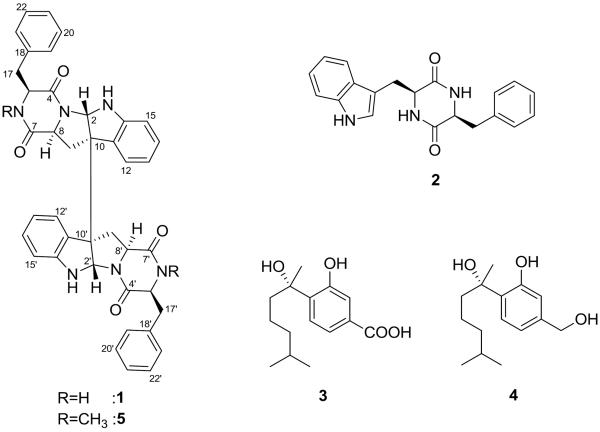
Prior investigations of filamentous fungi in our group have resulted in the isolation of several new and biologically active natural products.[1–3] Continuing these investigations, we have now analyzed the metabolites from a fungal isolate of Aspergillus sydowii (MSX19583) that was obtained from spruce litter collected in 1984 in Colorado, USA. The extracts from solid-substrate fermentation cultures exhibited cytotoxic activity against MDA-MB-435 (human melanoma) cells and were therefore pursued for further analysis. Chemical separation of the CH3CN/CH3OH extract afforded a new diketopiperazine dimer (1) in addition to three known compounds including cyclo-(L-phenylalaninyl-L-tryptophanyl) [2],[4, 5] S-sydonic acid (3), and S-sydonol (4) (Figure 1).[6]
Figure 1.
Structures of compounds 1–5.
Results and Discussion
Compound 1 was assigned the molecular formula C40H36N6O4 (Index of Hydrogen Deficiency of 26) based on the HRESIMS data. The 1H and 13C NMR spectra of 1 displayed signals for only 18 protons and 20 carbons, respectively, suggesting a symmetrical dimeric structure (Table 1). The 1H signals were attributed to nine aromatic protons, a pair of methylene units, three methine protons, and two exchangeable protons (Table 1; Figure S1). In addition to the 13C NMR signals expected for the above structural features, one quaternary (δC 59.1, C-10), five non-protonated sp2-hybridized carbons [three aromatic (δC 126.5, 135.6, and 150.1 for C-11, C-18, and C-16, respectively) and two carbonyl carbons (δC 165.9, C-4 and 168.7, C-7)] were also observed (Table 1; Figure S2). Phenylalanine and tryptophan-derived subunits were readily identified after analysis of HSQC and HMBC NMR data (Figure 2). Presence of phenylalanine was also confirmed by amino acid analysis using Marfey's method.[7] HMBC correlations from H-6 (δH 5.45) to C-4, C-5 (δC 56.3), C-7, and C-8 (δC 58.9) supported the diketopiperazine ring system in 1. A C–N covalent bond between C-2 and N-3 was identified by HMBC correlations from H-2 (δH 5.26) to C-8, C-9 (δC 35.4), C-11, and C-16. Finally, the monomeric units were linked at the only remaining position, C-10, thereby completing the gross structure of 1.
Table 1.
1H (400 MHz) and 13C NMR (100 MHz) data of 1 in CDCl3.
| # | δH (mult., J) | δ C | HMBC (H→ #C) |
|---|---|---|---|
| 1, 1′ | 5.17 (s) | 10, 11 | |
| 2, 2′ | 5.26 (s) | 78.9 | 8, 9, 11, 16 |
| 4, 4′ | 165.9 | ||
| 5, 5′ | 4.15 (br d; 11.3) | 56.3 | 4, 17, 18 |
| 6, 6′ | 5.45 (s) | 4, 5, 7, 8, 17 | |
| 7, 7′ | 168.7 | ||
| 8, 8′ | 3.87 (dd, 11.0, 5.9) | 58.9 | 7, 9 |
| 9a, 9′a | 2.67 (dd, 12.9, 11.0) | 35.4 | 7, 8, 10, 11 |
| 9b, 9′b | 2.56 (dd, 12.9, 5.9) | 2, 8, 10, 11 | |
| 10, 10′ | 59.1 | ||
| 11, 11′ | 126.5 | ||
| 12, 12′ | 7.20 (d, 7.6) | 125.5 | 10, 14, 16 |
| 13, 13′ | 6.79 (t, 7.6) | 119.6 | 11, 15 |
| 14, 14′ | 7.15 (m) | 130.2 | 12, 16 |
| 15, 15′ | 6.65 (d, 7.9) | 110.5 | 11, 13 |
| 16, 16′ | 150.1 | ||
| 17a, 17′a | 3.55 (dd, 14.4, 3.4) | 36.7 | 4, 5, 18, 19/23 |
| 17b, 17′b | 2.70 (dd, 14.4, 11.3) | 4, 5, 18, 19/23 | |
| 18, 18′ | 135.6 | ||
| 19, 19′ | 7.15 (m) | 129.1 | 17, 21, 23 |
| 20, 20′ | 7.32 (m) | 129.6 | 18, 22 |
| 21, 21′ | 7.28 (m) | 127.9 | 19/23 |
| 22, 22′ | 7.32 (m) | 129.6 | 18, 20 |
| 23, 23′ | 7.15 (m) | 129.1 | 17, 19, 21 |
Figure 2.
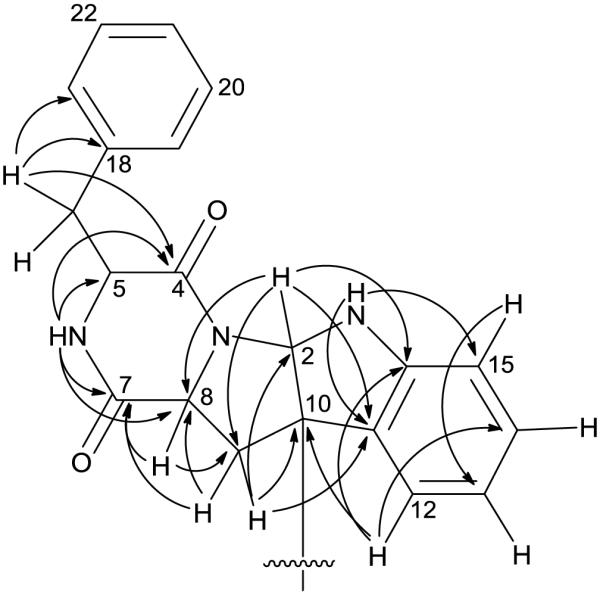
Key HMBC correlations for 1.
A search of the Dictionary of Natural Products[8] identified two compounds with identical molecular formulae, a symmetric diketopiperazine, WIN 64821, and an asymmetric analogue, asperazine.[9, 10] Although the gross structure of 1 was found to be similar to WIN 64821 after analysis of 1H, 13C, HSQC, and HMBC NMR data (Table 1 and Figure 2), the 1H NMR signals for 1 were not fully consistent with those reported in the literature for WIN 64821 or a synthetic analogue, ent-WIN 64821,[11] suggesting differences in configuration. Four additional secondary metabolites with identical molecular formulae and structural skeletons are known in the literature; however, the lack of reported NMR data did not permit comparisons between these and 1,[12, 13] which is a general challenge with this class of compounds.
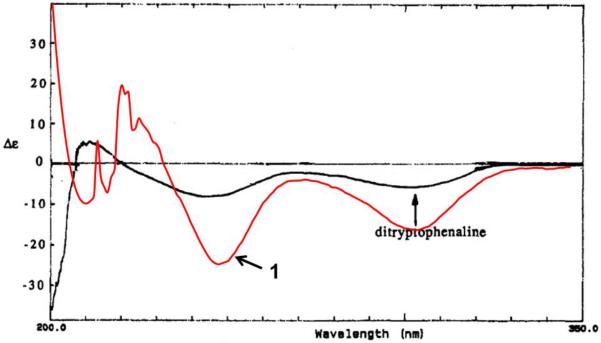
The absolute configuration of 1 was determined using a suite of techniques. Analysis of the NOESY data for 1 did not provide sufficient information for complete assignment of the relative configuration. Marfey's method established the absolute configuration at C-5. Briefly, a sample of 1 was hydrolyzed in 6N HCl (110 °C; 24 h), and Marfey's derivative of the resulting phenylalanine standard sample was prepared using the methods reported previously.[7] HPLC analysis of the product (tR = 5.06 min) and comparison with standards (L-Phe: tR = 5.06 min and D-Phe: tR = 5.92 min) prepared in an analogous manner revealed the presence of L-phenylalanine. By analogy to compound 2, the C-8 stereocenter was presumed to be an S-configuration based on biosynthetic considerations. Due to the limited number of protons, a NOESY correlation to assign the configuration at C-2 relative to other stereocenters could not be observed; however, the absence of a correlation between H-2 and H-8 was consistent with the orientation of these protons on opposite faces of the ring system. More convincingly, the ECD spectrum of 1 resembled that of a closely related analogue, ditryptophenaline (5),[14] where both compounds, each with an anti orientation for H-2 and H-8, exhibited a pair of negative Cotton effects near 245 and 300 nm (Figure 3).[9] In the same literature,[9] WIN 64821 was shown to have an opposite Cotton effect at 245 nm. Thus, the relative orientation of these two protons, anti in 1 and syn in WIN 64821, accounted for the key difference between these compounds, supporting the absolute configuration of 1 as shown (Figure 1).
Figure 3.
Experimental ECD spectrum for 1 overlaid with ECD data reported for ditryptophenaline (5).
The extract of MSX19583 exhibited moderate cytotoxic activity (54% cell viability at 20 μg/mL) against MDA-MB-435 human melanoma cells using procedures described in detail previously.[15–17] As such, compounds 1–4 were tested against two cancer cell lines, MDA-MB-435 and HT-29 (human colon cancer). All compounds were found to be inactive, displaying IC50 values >20 μM. Although the initial activity observed with the extract could not be attributed to the major compounds, it is possible that minor components present in trace amounts could contribute to the moderate cytotoxic effects. Also, various other biological activities have been reported for the known compounds isolated during this study. For example, S-sydonol has been reported to have anti-diabetic and anti-inflammatory activities.[18] R-sydonol has been reported to show selective antibacterial activity against Staphylococcus albus and Micrococcus tetragenus, while R-sydonic acid exhibited broad spectrum activity against several species.[19] Cyclo-(L-phenylalaninyl-L-tryptophanyl) has been previously reported as a plant growth regulator.[4]
Experimental
General experimental procedures
Optical rotation data were acquired on a Rudolph Research Autopol III polarimeter (Rudolph Research Analytical, Flanders, NJ, USA). ECD data were collected using an Olis DSM 17 CD spectrophotometer (Olis, Bogard, GA, USA). UV data were obtained using a Varian Cary 100 Bio UV-vis spectrophotometer (Varian Medical Systems, Palo Alto, CA, USA). HRESIMS data were collected using an electrospray ionization (ESI) source coupled to a LTQ Orbitrap XL system (Thermo Fisher Scientific, San Jose, CA, USA) in positive and negative ionization modes via a liquid chromatography/autosampler system comprised of an Acquity UPLC system (Waters Corp., Milford, MA, USA). A CombiFlash Rf system using a 12 g RediSep Rf Si-gel Gold column (both from Teledyne-Isco, Lincoln, NE, USA) was employed for normal-phase flash column chromatography. High-performance liquid chromatography (HPLC) separations were performed utilizing Varian ProStar HPLC systems equipped with ProStar 210 pumps and a ProStar 335 photodiode array detector, using Galaxie Chromatography Workstation software (version 1.9.3.2, Varian Inc.). YMC ODS-A (Waters Corp.; 5μm; 250 × 10 mm for semi-preparative HPLC column) or Kinetex C18 (Phenomenex, Torrance, CA, USA; 5μm; columns of dimensions 250 × 21.2 mm for preparative HPLC and 250 × 4.6 mm for analytical HPLC) were used for HPLC. For UPLC analysis, a BEH C18 (Waters Corp.; 1.7 μm; 50 × 2.1mm column) was used with data collected and analyzed using Empower 3 software (Waters Corp.). The solvents were obtained from Fisher Scientific.
Fungal strain and fermentation
Mycosynthetix fungal strain MSX19583 (Aspergillus sydowii; See Supporting Information and Figure S5) was isolated from spruce litter collected in 1984 near Cumbres Pass, Colorado, USA at an elevation of 2985 m. A fresh culture of this isolate was grown on a malt extract slant, and a piece was transferred to a medium containing 2% soy peptone, 2% dextrose, and 1% yeast extract (YESD media). After incubation at 22 °C for 7 days (with agitation), the culture was used to inoculate 50 mL of rice medium [containing rice, vitamin solution and water (twice the volume of rice)] in a 250 mL Erlenmeyer flask. The culture was incubated at 22 °C until sufficient fungal growth (~ 14 d) was observed. The scaled-up culture was grown in a 2.8 L Fernbach flask containing 150 g of rice and 300 mL of H2O and inoculated using a seed culture grown in YESD medium followed by incubation at 22 °C for 14 d. Details for molecular identification and phylogenetic analysis of fungal strain MSX19583 can be found in the supporting information (Figure S5).
Extraction and isolation
To one Fernbach flask containing rice with fungal growth (MSX19583) was added 500 mL of 1:1 CH3OH/CHCl3. The culture was chopped with a spatula and shaken overnight (~16 h; rt) at ~100 rpm. After filtration, the remaining residues were washed with CH3OH. To the filtrate, 900 mL of CHCl3 and 1.5 L of H2O were added. The mixture was stirred for 30 min and then transferred to a separatory funnel. The lower layer was drawn off into round-bottom flasks and evaporated to dryness. This dried material was re-constituted in 200 mL of 1:1 CH3OH/CH3CN and 200 mL of hexanes and transferred to a separatory funnel. The biphasic solution was shaken vigorously. The CH3OH/CH3CN layer was evaporated to dryness under vacuum to obtain 795 mg of the organic extract.
A portion of the organic extract (770 mg) was adsorbed onto a minimal amount of Celite 545 (Acros Organics, Geel, Belgium). After drying, the adsorbed mixture was loaded into a cartridge and subjected to normal-phase silica gel flash column chromatography (RediSep Rf Gold Si-gel column; 12g) using a step gradient with hexanes, CHCl3, and CH3OH (30 mL/min flow rate and 61.0 column volumes over 31.4 min). The resulting fractions were pooled according to UV and ELSD data to afford four fractions. Fraction three (80 mg) was subjected to preparative RP-HPLC (gradient elution using CH3CN in H2O (w 0.1% HCOOH): 40–80% for 20 min and 80–100% CH3CN for 10 min; λ = 210 and 254 nm; flow rate = 21.2 mL/min) affording cyclo-(L-phenylalaninyl-L-tryptophanyl) [2; 1.2 mg, tR 4.0 min], 1 (3.0 mg, tR 7.5 min), S-sydonol (3; 2.4 mg, tR 9.7 min), and S-sydonic acid (4; 12.2 mg, tR 10.5 min). Compounds 1 and 2 were further subjected to semi-preparative HPLC [isocratic elution using 50% CH3CN in H2O for 30 min in the case of 2 and 60% CH3CN in H2O for 30 min in the case of 1; flow rate = 3 mL/min; YMC ODS-A (Waters Corp.; 5μm; 250 × 10 mm)] affording 1.1 mg (tR 7.5 min) and 0.51 mg (tR 6.0 min), respectively. Purity was determined by UPLC using a gradient elution of 20% CH3CN in H2O (w 0.1% HCOOH) to 100% CH3CN over 3 min. All of the known compounds (2–4) were identified by comparison of their 1H NMR, 13C NMR, and/or MS data with literature values.[4–6]
Compound 1: White powder; [α]23D −25 [c 0.04, 4:1 CH3OH: (CH3)2SO], [α]24D −343 [c 0.04, CH2Cl2]; UV/Vis (CH3OH) λmax (log ε) 303 (3.5), 245 (3.8), 218 (3.7) nm; ECD (50 μM, CH3OH) λmax (Δε) 303 (−18), 246 (−27), 226 (+12), 220 (+23) nm; 1H NMR (CDCl3, 400 MHz) and 13C NMR (CDCl3, 100 MHz) data, see table 1; Key NOESY data: H-5 ↔ H-6, H-17a, H-17b; H-8 ↔ H-9a, H-9b; H-9a ↔ H-2; H-19/H-23 ↔ H-6, H-17a, H-17b; HRESIMS obsd. m/z 665.2839 [M+H]+ (calcd. for C40H37N6O4, 665.2871).
Cyclo-(L-phenylalaninyl-L-tryptophanyl) [2]: White powder; [α]23D −110 (c 0.05, CH3OH); HRESIMS obsd. m/z 334.1548 [M+H]+ (calcd. for C20H20N3O2, 334.1550); 1H NMR data were fully consistent with those reported in literature.[4, 5]
S-Sydonic acid (3): Colorless oil; [α]23D +4 (c 1.22, CH3OH); HRESIMS obsd. m/z 249.1476 [M−H2O+H]+ (calcd. for C15H21O3, 249.1485); 1H NMR data were fully consistent with those reported in literature; structure was also confirmed by analysis of 2D NMR data.[6] S-Sydonol (4): Colorless oil; [α]23D +6 (c 0.24, CH3OH); HRESIMS obsd. m/z 235.1684 [M−H2O+H]+ (calcd. for C15H23O2, 235.1693); 1H NMR data were fully consistent with those reported in literature.[6]
NMR data
NMR spectra (1H, 13C, 1H-13C HSQC, and 1H-13C HMBC) were recorded at 25 °C in CDCl3 on a JEOL ECS-400 NMR spectrometer (399.78 MHz for 1H and 100.53 MHz for 13C; JEOL Ltd., Tokyo, Japan) equipped with an auto tune 5 mm field gradient tunable Royal probe (NM-03810RO5/UPG). The 1H and 13C chemical shifts were referenced to the residual solvent peak of CDCl3 at 7.24 ppm and 77.2 ppm, for proton and carbon, respectively. The 1H sweep width was set at 5997 Hz for all experiments with a 90° pulse for 1H of 6.4 μs and 13C sweep width 25131 Hz with a 90° pulse for 13C of 11.6 μs. The digital resolution of 1H NMR was 0.37 Hz and that of 13C NMR was 0.77 Hz. The edited-gradient 1H-13C HSQC was acquired with 13C sweep width of 16084 Hz and 256 t1 increments. Each increment was acquired with 16 transients. The one-bond coupling constant delay was set using 145 Hz and MPF8 decoupling was applied during acquisition. The gradient 1H-13C HMBC was acquired using 64 transients per increment with 256 t1 increments. A sweep width of 20105 Hz was used for the 13C dimension. One-bond coupling constant of 145 Hz and long-range coupling constant of 8 Hz were used to set the delays in the pulse sequence.
Supplementary Material
Acknowledgments
This research was supported by program project Grant P01 CA125066 from the National Cancer Institute/National Institutes of Health, Bethesda, MD, USA. Assistance from the staff of the NMR (Dr. Franklin J. Moy) and Mass Spectrometry (Dr. Brandie M. Ehrmann) Facilities at the University of North Carolina at Greensboro is gratefully acknowledged.
Footnotes
Supporting Information Additional supporting information may be found in the online version of this article at the publisher's website.
References
- [1].El-Elimat T, Figueroa M, Raja HA, Adcock AF, Kroll DJ, Swanson SM, Wani MC, Pearce CJ, Oberlies NH. Tetrahedron Lett. 2013;54:4300. doi: 10.1016/j.tetlet.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].El-Elimat T, Figueroa M, Raja HA, Graf TN, Adcock AF, Kroll DJ, Day CS, Wani MC, Pearce CJ, Oberlies NH. J. Nat. Prod. 2013;76:382. doi: 10.1021/np300749w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Figueroa M, Raja H, Falkinham JO, III, Adcock AF, Kroll DJ, Wani MC, Pearce CJ, Oberlies NH. J. Nat. Prod. 2013;76:1007. doi: 10.1021/np3008842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kimura Y, Tani K, Kojima A, Sotoma G, Okada K, Shimada A. Phytochemistry. 1996;41:665. [Google Scholar]
- [5].Tullberg M, Grøtli M, Luthman K. Tetrahedron. 2006;62:7484. [Google Scholar]
- [6].Kudo S, Murakami T, Miyanishi J, Tanaka K, Takada N, Hashimoto M. Biosci. Biotech. Bioch. 2009;73:203. doi: 10.1271/bbb.80535. [DOI] [PubMed] [Google Scholar]
- [7].Ayers S, Ehrmann BM, Adcock AF, Kroll DJ, Carcache de Blanco EJ, Shen Q, Swanson SM, Falkinham JO, Wani MC, Mitchell SM. J. Pept. Sci. 2012;18:500. doi: 10.1002/psc.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Dictionary of Natural Products, Online 23.1. Taylor & Francis Group; London: 2014. [Google Scholar]
- [9].Barrow CJ, Cai P, Snyder JK, Sedlock DM, Sun HH, Cooper R. J. Org. Chem. 1993;58:6016. [Google Scholar]
- [10].Varoglu M, Corbett TH, Valeriote FA, Crews P. J. Org. Chem. 1997;62:7078. doi: 10.1021/jo970568z. [DOI] [PubMed] [Google Scholar]
- [11].Ovenden SP, Sberna G, Tait RM, Wildman HG, Patel R, Li B, Steffy K, Nguyen N, Meurer-Grimes BM. J. Nat. Prod. 2004;67:2093. doi: 10.1021/np0497494. [DOI] [PubMed] [Google Scholar]
- [12].Pérez-Balado C, Rodríguez-Graña P, de Lera ÁR. Chem.-Eur. J. 2009;15:9928. doi: 10.1002/chem.200901056. [DOI] [PubMed] [Google Scholar]
- [13].Hiramoto M, Miyata H, Shibazaki M, Saita Y. Symposium on the Chemistry of Natural Products. 1994;557 [Google Scholar]
- [14].Springer JP, Bűchi G, Kobbe B, Demain AL, Clardy J. Tetrahedron Lett. 1977;18:2403. [Google Scholar]
- [15].Ayers S, Graf TN, Adcock AF, Kroll DJ, Matthew S, Carcache de Blanco EJ, Shen Q, Swanson SM, Wani MC, Pearce CJ. J. Nat. Prod. 2011;74:1126. doi: 10.1021/np200062x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].El-Elimat T, Raja HA, Day CS, Chen WL, Swanson SM, Oberlies NH. J. Nat. Prod. 2014;77:2088. doi: 10.1021/np500497r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Alali FQ, Amrine CSM, El-Elimat T, Alkofahi A, Tawaha K, Gharaibah M, Swanson SM, Falkinham JO, III, Cabeza M, Sánchez A. Phytochem. Lett. 2014;9:96. [Google Scholar]
- [18].Chung YM, Wei CK, Chuang DW, El-Shazly M, Hsieh CT, Asai T, Oshima Y, Hsieh TJ, Hwang TL, Wu YC. Bioorg. Med. Chem. 2013;21:3866. doi: 10.1016/j.bmc.2013.04.004. [DOI] [PubMed] [Google Scholar]
- [19].Li D, Xu Y, Shao CL, Yang RY, Zheng CJ, Chen YY, Fu XM, Qian PY, She ZG, de Voogd NJ. Mar. Drugs. 2012;10:234. doi: 10.3390/md10010234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.