Abstract
To facilitate the use of lignocellulosic biomass as an alternative bioenergy resource, during biological conversion processes, a pretreatment step is needed to open up the structure of the plant cell wall, increasing the accessibility of the cell wall carbohydrates. Lignin, a polyphenolic material present in many cell wall types, is known to be a significant hindrance to enzyme access. Reduction in lignin content to a level that does not interfere with the structural integrity and defense system of the plant might be a valuable step to reduce the costs of bioethanol production. In this study, we have genetically down-regulated one of the lignin biosynthesis-related genes, cinnamoyl-CoA reductase (ZmCCR1) via a double stranded RNA interference technique. The ZmCCR1_RNAi construct was integrated into the maize genome using the particle bombardment method. Transgenic maize plants grew normally as compared to the wild-type control plants without interfering with biomass growth or defense mechanisms, with the exception of displaying of brown-coloration in transgenic plants leaf mid-ribs, husks, and stems. The microscopic analyses, in conjunction with the histological assay, revealed that the leaf sclerenchyma fibers were thinned but the structure and size of other major vascular system components was not altered. The lignin content in the transgenic maize was reduced by 7-8.7%, the crystalline cellulose content was increased in response to lignin reduction, and hemicelluloses remained unchanged. The analyses may indicate that carbon flow might have been shifted from lignin biosynthesis to cellulose biosynthesis. This article delineates the procedures used to down-regulate the lignin content in maize via RNAi technology, and the cell wall compositional analyses used to verify the effect of the modifications on the cell wall structure.
Keywords: Bioengineering, Issue 89, Zea mays, cinnamoyl-CoA reductase (CCR), dsRNAi, Klason lignin measurement, cell wall carbohydrate analysis, gas chromatography (GC)
Introduction
The production of biofuels from lignocellulosic biomass is highly desirable due to its present abundance in the U.S.1, and in the case of the sustainable harvest of agricultural and forestry residues, the ability to not compete directly for cropland used for food and animal feed production. However, unlike maize grain, which is the main source of biofuel currently generated in the U.S., lignocellulosic materials are significantly more complex and difficult to break down. In addition to the long-chain carbohydrates, cellulose and hemicellulose, which are the main sources of sugars during fermentation of lignocellulosic materials, many types of plant cell walls also contain lignin, a phenylpropanoid polymer that provides strength, defense against pathogen attack, and hydrophobicity to cell walls. While necessary for plant growth and survival, lignin also presents a significant barrier to the successful enzymatic conversion of the cellulose and hemicellulose to soluble sugars. Materials with high lignin contents are generally less desirable materials for both the biofuel (through biological conversion pathways) and the pulp and paper industries due to the negative impacts on processing characteristics and product quality. Hence, genetic manipulation of plant materials for lignin reduction at a level that does not interfere with crop structural strength and defense systems might be important for the reduction of production costs for both the lignocellulosic biofuel and the pulp and paper industries.
In maize (Zea mays), lignin is covalently cross-linked to hemicellulose in the primary cell wall via ferulate and diferulate bridges2. The lignin-hemicellulose complex binds to cellulose microfibrils through hydrogen bonds, forming a complex matrix that confers integrity and strength to the secondary cell wall. The mechanical strength of plant cell walls is largely determined by the type of lignin subunits3-5. In previous studies, altering the proportions of lignin subunits has shown no clear trend on enzymatic digestibility6-11. However, reductions in lignin content generally show an improvement in conversions12,13 and may be a key to increasing the digestibility of plant material by hydrolytic enzymes including endocellulases, cellobiohydrolases, and β-glucosidases14.
Genetic engineering to regulate the expression level of transcripts has been extensively practiced to improve crop traits. Advanced techniques, including anti-sense15 and co-suppression16 technologies, enable effective down-regulation of target genes. Complete gene knock-out has also been achieved using gene constructs encoding intron-spliced RNA with a hairpin structure17. Furthermore, a double stranded RNA interference (dsRNAi) technique, i.e. a powerful and effective gene expression mediator that works by either targeting transcript degradation or translation repression, provides a potent means to induce a wide range of suppression effects on the target mRNA18. Gene silencing techniques show several limitations. These techniques do not precisely regulate the level of transcription and it could cause unexpected silencing effects on other homologous genes.
In this method, we employed particle bombardment to carry the dsRNAi constructs into the maize genome. To date, a vast array of plant species have been successfully transformed using particle bombardment, Agrobacterium mediated transformation, electroporation, and microinjection methods. In maize genetic transformation, the particle bombardment method is advantageous over all the other methods because it is the most efficient. Particle bombardment is not dependent on bacteria, so the method is free of biological constraints such as the size of the gene, species of gene origin, or the plant genotype. The physical transgene delivery system enables high molecular weight DNA and multiple genes to be introduced into plant genomes and in certain cases into chloroplasts at high transformation efficiency19. The lignin reduction in the vascular system of the leaf mid-rib can be visualized via scanning electron microscopy (SEM) which is beneficial for examining the topography and composition of samples.
In maize plants, two of the cinnamoyl-CoA reductase (ZmCCR1: X98083 and ZmCCR2: Y15069) genes were found in the maize genome20. Cinnamoyl-CoA reductase catalyzes the conversion of the hydroxycinnamoyl-CoA esters into cinnamyl aldehydes. We chose the ZmCCR1 gene to down-regulate this enzyme because the gene is expressed in all lignifying tissues. The 523 nucleotides at the 3’ terminus of the ZmCCR1 gene were chosen for a dsRNAi construct because the sequences appeared to be more diverse as compared to those of ZmCCR2. Thus, the dsRNAi construct would precisely bind only to ZmCCR1, avoiding off-target silencing21. A ZmCCR1_RNAi construct was engineered into the cytoplasmic expression system ImpactVector1.1-tag (IV 1.1) containing the green tissue specific promoter, ribulose-1, 5-bisphosphate carboxylase oxygenase (RuBisCO).
To study the effects of the dsRNAi construct on transgenic plants, the lignin content was quantified. The Klason (acid-insoluble) lignin measurement is known to be more accurate compared to the acid detergent lignin quantification methods which solubilize some of the lignin22. Therefore, the Klason lignin was measured in transgenic maize stalks. This procedure consists of a two-step acid hydrolysis that converts polymeric carbohydrates into soluble monosaccharides23. The hydrolyzed biomass was then fractionated into acid soluble and insoluble materials and the acid insoluble lignin was measured according to previous studies23,24. Ideally, lignin analysis should include extractions with water and ethanol prior to the hydrolysis step, in order to remove soluble materials that can interfere with the results, and a post-hydrolysis combustion of the lignin residue to account for any ash present in the residue. Without these steps, the lignin content of the sample could be artificially inflated. The full method is presented here, however for our experiments we were unable to perform both of these steps due to the small volume of material available for testing
Two other cell wall components, cellulose and hemicellulose were also analyzed in the lignin down-regulated transgenic maize lines. It has been reported that transgenic plants that have been down-regulated in either their phenylalanine ammonia-lyase (PAL)25, 4-coumarate:CoA ligase (4CL)26, or cinnamyl alcohol dehydrogenase (CAD)27 show an increase in other cell wall structural components. As a first step in our studies, crystalline cellulose was measured using the Updegraff method28. This method was originally devised for determination of cellulose in a large number of cellulolytic bacteria and fungi. Briefly, the milled maize stocks were treated with Updegraff reagent (acetic acid: nitric acid: water) to remove hemicellulose, lignin, and xylosans. The crystalline cellulose was completely hydrolyzed into glucose via Saeman hydrolysis by adding H2SO4. The crystalline cellulose was then assayed using the colorimetric anthrone method29. To verify if the hemicellulose contents were changed, the monosaccharide extracts from milled stalks were hydrolyzed using trifluoroacetic acid, derivatized using the alditol acetate method and then analyzed by gas chromatography (GC)30. The detailed procedures for crystalline cellulose content and matrix polysaccharides composition analyses are described in Foster et al. (2010)31.
Here, we describe the procedures used for lignin down-regulation in maize via a RNAi technology, particle bombardment transformation, and lignin analysis for accelerated deconstruction of maize lignocellulosic biomass into fermentable sugars for biofuels.
Protocol
1. Preparation of dsRNAi Constructs Used for the Down-regulation of ZmCCR1
Design gene specific primers including necessary restriction enzyme sites for making a dsRNAi construct to knock-out the ZmCCR1 gene. Two primer sets were designed to amplify two fragment segments of ZmCCR1 cDNA: a 523 bp fragment from nucleotide 748 to 1271, and a 285 bp fragment from nucleotide 986 to 1271. The ZmCCR1 cDNA was provided from the Arizona Genome Institute (AGI). More details are described in Figure 1.
Amplify the large fragment by polymerase chain reaction (PCR) from the ZmCCR1 cDNA template using the primers ZmCCR1_748F_BglII (5’-AGATCTACATCCTCAAGTACCTGGAC-3’) and ZmCCR1_1271R_NcoI (5’-CCATGGTTTACACAGCAGGGGAAGGT-3’). Amplify the smaller fragment (285 bp) using the primers ZmCCR1_986F_BglII (5’-AGATCTGGAAGCAGCCGTACAAGTTC-3’) and a ZmCCR1_1271R_SacI (5’-GAGCTCTTTACACAGCAGGGGAAGGT-3’).
Individually ligate the fragments into pGEM-T Easy following the manufacturer’s instructions.
Perform mini-prep plasmid DNA isolation from individual transformants, each containing the pGEM-T constructs using a commercial mini-prep plasmid kit.
Digest both the pGEM-T::ZmCCR1 (523 bp) and ImpactVector (IV)-1.1 (cytoplasm expression vector) with both BglII and NcoI.
Ligate the large digested gel purified ZmCCR1 fragment (523 bp) into the digested gel purified IV-1.1.
Digest the pGEM-T::ZmCCR1(285 bp ) and IV-1.1::ZmCCR1(523 bp) with both BglII and SacI in order to insert the small fragment into the IV-1.1::ZmCCR1(523 bp).
Ligate the small digested gel purified ZmCCR1 fragment (285 bp) into the digested gel purified IV-1.1::ZmCCR1(523 bp).
Clone both 523 bp and 285 bp fragments into IV-1.1 to make the ZmCCR1 RNAi construct, which has a 285 bp inverted repeat sequence with a 238 bp spacer in the middle of the inverted repeat fragments (see Figure 1).
Transfer this construct into Escherichia coli (E. coli), grow them and perform a midi-prep size plasmid DNA isolation to obtain enough plasmid DNA for maize genetic transformation.
2. Maize Genetic Transformation
- Preparation of Tungsten Particles
- Place 60 mg of tungsten beads (M10) in a 1.5 ml tube and wash with 1 ml of 70% ethanol by vortexing for 2 min. Incubate for 10 min at 23 °C then centrifuge at 18,894 x g for 2 min and discard the supernatant.
- Wash 3x with 1 ml of 100% ethanol, centrifuging for 2 min and discarding supernatant. Add 1 ml of sterile 50% glycerol to bring the microparticle concentration to 60 mg/ml.
- Preparation of DNA for Bombardment
- Place the 50 μl (3 mg) of tungsten beads prepared in 50% glycerol into a 1.5 ml tube. Add 5 μl (1 μg) of IV-1.1::ZmCCR1 RNAi plasmid DNA, 50 μl of 2.5 M CaCl2, and 20 μl of 0.1 M spermidine. Vortex briefly between each addition of the above reagents.
- Vortex the tungsten bead-DNA mixture briefly and centrifuge at 18,894 x g for 30 sec. Pour off the supernatant and resuspend the pellets in 140 μl of 70% ethanol. Remove the liquid and discard. Add 140 μl of 100% ethanol. Remove the liquid and discard.
- Add 48 μl of 100% ethanol. Use immediately or store on ice for up to 4 hr prior to bombardment.
- Bombardment
- Place a 3-5 cm diameter Hi-II embryogenic maize calli (provided from Maize transformation Center of Iowa State University) in the middle of 100 mm Petri dishes containing N6OSM media (as osmotium) at least 4 hr prior to the bombardment.
- Prepare the PSD-1000/He Particle Delivery device according to the manufacturer’s instructions32.
- Sterilize chamber wall with 70% ethanol. Load sterile 650 psi rupture disk into sterile retaining cap. Spread 5-6 μl of the M10-DNA solution onto the surface of a macrocarrier, dry briefly. Load macrocarrier and stopping screen into microcarrier launch assembly.
- Place microcarrier launch assembly and maize calli in chamber at a selected distance from the stopping screen (L2 = 6 cm) and close door. Accelerate in a vacuum of 27 psi against a wire mesh screen.
- Press the fire button until rupture disk bursts and helium pressure gauge drops to zero. Release the fire button.
- Incubate the bombarded calli in a Petri dish containing N6OSM (osmotic medium) for 16 hr in the dark at 27 °C. Break the calli into about ten pieces and transfer to N6E (callus induction medium) in Petri dishes and incubate for 5 days in the dark at 27 °C.
- Selection
- After 5 days on N6E, transfer the calli onto N6S medium (selection media). Subculture all calli on selection medium every 30 days for 8-12 weeks without disturbing the calli structure.
- After about 8-10 weeks, white fast-growing sectors will grow out of the non-proliferating and partially necrotic mother calli. Excise the white fast-growing tissues and subculture them to fresh selection medium (N6S) and continue to incubate as above.
- Regeneration
- Transfer the white and fast growing embryonic calli onto regeneration medium and incubate as above for 1 week. Switch the regenerating embryogenic calli to a period of 16 hr daylight and 8 hr dark at 25-27 °C
- Transfer the regenerating shoots onto the rooting medium in a glass test tube after 3-4 weeks, continue to incubate as above. After substantial root development appears, wash roots carefully under water tap, then transplant the plantlets to 4” pots with soil. Cover the pots with plastic bags to keep moist. After 2 days make small holes the plastic bags. After 5-6 days remove the plastic bags. Continue to incubate as above for another 5-6 days.
- Greenhouse
- Transfer the seedlings into 18” pots with soil and maintain in full summer sunlight or greenhouse light. The initial regenerated plants are called T0 while the first seeds belong to the T1 generation.
3. Histological Assay
- Fix the maize leaf mid-ribs in 5 ml of 10% neutral buffered formalin.
- Process and vacuum infiltrate with paraffin on a tissue processor using a tissue processor.
- Embed the tissues in paraffin using a HistoCentre III embedding station.
- Remove the excess paraffin from the edges once blocks are cooled.
- Section sample at 4-5 μm with a microtome using a microtome.
- Place sections on microscope slides and dry in a 56 °C incubator for 2-24 hr. Make sure sections are fully adhered to the slide.
- Deparaffinize sections in two changes of xylene for 5 min at 23 °C.
- Hydrate slides through two changes of 100% ethanol for 2 min and two changes of 95% ethanol for 2 min at 23 °C.
- Rinse the sections in running tap water for 2 min.
- Stain with 0.05% toluidine blue O for 1-2 min and rinse briefly with ddH2O.
- Place a coverslip on the samples with immersion oil and visualization with light microscopy.
- Scanning Electron Microscopy (SEM)
- Fix the cross-sectioned maize leaf mid-ribs in 4% glutaraldehyde and 0.1 M sodium phosphate buffer (pH 7.4) at 4 °C for 1-2 hr.
- Briefly rinse the samples in the buffer, dehydrated them in an ethanol series (25%, 50%, 75%, and 95%) for 10-15 min at each gradation and 100% ethanol for 10 min, 3x.
- Dry the dehydrated cross-sectioned maize leaf mid-ribs in a critical point dryer using liquid carbon dioxide as the transitional fluid.
- Mount the dried samples on aluminum stubs using high vacuum carbon tabs
- Coat the maize leaf mid-ribs mounted on aluminum stubs with gold (approximately 20 nm thickness) in a sputter coater purged with argon gas.
- Examine the coated samples in a JEOL JSM-6400V (lanthanum hexaboride electron emitter) scanning electron microscope.
- Digital images were pictured using an analySIS Pro software (version 3.2).
4. Klason Lignin Measurement
Mill the samples through a 2 mm screen.
Use a moisture analyzer to determine the moisture content of each sample and record the value.
Weigh out ~1.5 g of each sample and record the mass. Extract the samples using water for the first extraction, followed by ethanol for the second extraction using either an automated solvent extractor (3 cycles per extraction, ~14 min per cycle) or soxhlet apparatus (8 hr per extraction). (Note: This step removes extractives that can condense during acid hydrolysis and interfere with accurate lignin measurement, increasing the apparent Klason lignin content.)
Dry the extracted samples at 45 °C overnight, then allow them to cool in the desiccator, and weigh again.
Set an incubator to 30 °C. Measure 0.3 g of each dry, extracted sample into screw-top high pressure tubes (triplicates per sample is recommended) and record the weights to the nearest 0.1 mg. Add 3 ml of 72% H2SO4 to each pressure tube.
Mix the sample using a glass or Teflon stir rod. Leave the stir rod in the tube until water is added following incubation.
Place vials in an incubator set to 30 °C and 150 rpm for 60 min. After 1 hr add 84 ml of deionized water to dilute the acid concentration to 4% and mix with the stir rod. Be careful not to leave large amounts of sample on the sides of the vial above the water line.
Tightly seal the stoppers on all vials and place them into a metal rack or large beakers. Autoclave at 121 °C using a liquid sterilization cycle for 1 hr. Allow them to cool to room temperature before opening.
Pre-ash the filtering crucibles in a furnace at 575 °C for at least 4 hr. Allow crucibles to cool in a desiccator for at least an hour.
Vacuum filter the solution from each tube through a separate crucible, using a rubber adaptor to secure the crucible. Use deionized water to rinse any remaining particles from the tube.
Dry the lignin residue at 105 °C for a minimum of 4 hr. Record the weight of the dry crucible and residue.
If using a 575 °C, pre-fire the samples over a Bunsen burner until there is no smoke or ash and then place in the furnace for 24 hr, or if using a programmable furnace, do not pre-ash and use the following program:
Ramp from room temperature to 105 °C and hold for 12 min.
Ramp to 250 °C at 10 °C/min and hold for 30 min.
Ramp to 575 °C at 20 °C/min and hold for at least 180 min.
Remove the crucibles from the furnace and cool in a desiccator. Weigh the crucible and ash.
Calculate the acid insoluble residue using the following equation:
![]()
MPRE = Mass of pre-extracted biomass MPOST = Mass of post-extracted biomass MVIAL = Mass of extracted biomass added to the vial MRESIDUE = Mass of crucible and lignin residue MASH = Mass of crucible and ash MC = Moisture content of pre-extracted biomass, total weight basis
5. Carbohydrate Analysis
Perform cell wall carbohydrate analyses based on the Foster et al. (2010) protocol31. In brief, Prepare alcohol insoluble residue from freeze dried plant material. Then hydrolyze the material with trifluoroacetic acid and match the solubilized monosaccharide derivatives to their corresponding alditol acetates. Analyze these volatile derivatives by gas chromatography (GC) connected to a quadruple mass spectrometer.
Representative Results
We have demonstrated a reduction in the lignin content in maize plants via RNAi. The particle bombardment transformation method yielded around 30% trnasformation efficiency. The gene silencing of ZmCCR1 was consistently observed in T0-T2 generations. The lignin reduced transgenics grew similarly to wildtype maize plants except for displaying brown coloration in the leaf mid-rib, husk, and stem. The histological assay has shown that the mutant lines exhibit a significant reduction in the cell wall thickness of the sclerenchyma fibers in the maize leaf mid-rib18. Despite the reduction of cell wall thickness, the structure of other major vascular systems including the xylem vessel, phloem, and sheath cells did not reveal any differences compared to the wild-type control (Figure 2A)18. This implies that there are no detrimental effects for either water transport, nutrient transfer, or mechanical strength to the stems in the ZmCCR1_RNAi mutant lines.
The lignin content was quantified using a Klason lingin measurement. Figure 3 shows the amount of acid-insoluble Klason lignin (g/kg con stover) in wildtype maize and ZmCCR1_RNAi transgenic lines (T1). Three transgenic lines (1c-4, -5, and -6) showed a statistically significant lignin reduction (8.1%, 7.0%, and 8.7% respectively) compared to that of the wild-type maize plants18. To determine whether carbon flow was shifted from the lignin biosynthetic pathway to cell wall carbohydrate biosynthesis pathways, cellulose was analyzed via the Updegraff method. Figure 3A shows that two ZmCCR1_RNAi mutant lines (1b-6 and 1c-6) contained significantly increased levels of crystalline cellulose (1.5 and 1.8 fold respectively)18. The hemicellulose content was also analyzed. Figure 4B shows the amount of four main hemicellulose components (arabionose, xylose, galactose, and glucose). None of the four carbohydrate groups revealed any changes in the mutant lines18.
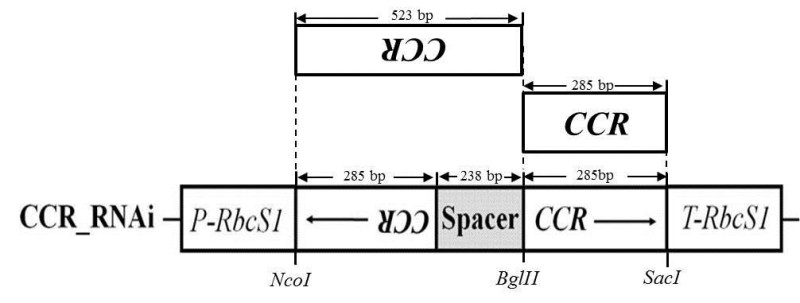 Figure 1. Cloning strategy for dsRNAi plasmid constructs for the down-regulation of the ZmCCR1. PRbcS1:Ribulose bisphosphate carboxylase promoter from Chrysanthemum morifolium Ramat. T-RbcS1: Ribulose bisphosphate carboxylase small unit terminator from Asteraceous chrysanthemum. This figure has been modified from Park et al (2012)18.
Figure 1. Cloning strategy for dsRNAi plasmid constructs for the down-regulation of the ZmCCR1. PRbcS1:Ribulose bisphosphate carboxylase promoter from Chrysanthemum morifolium Ramat. T-RbcS1: Ribulose bisphosphate carboxylase small unit terminator from Asteraceous chrysanthemum. This figure has been modified from Park et al (2012)18.
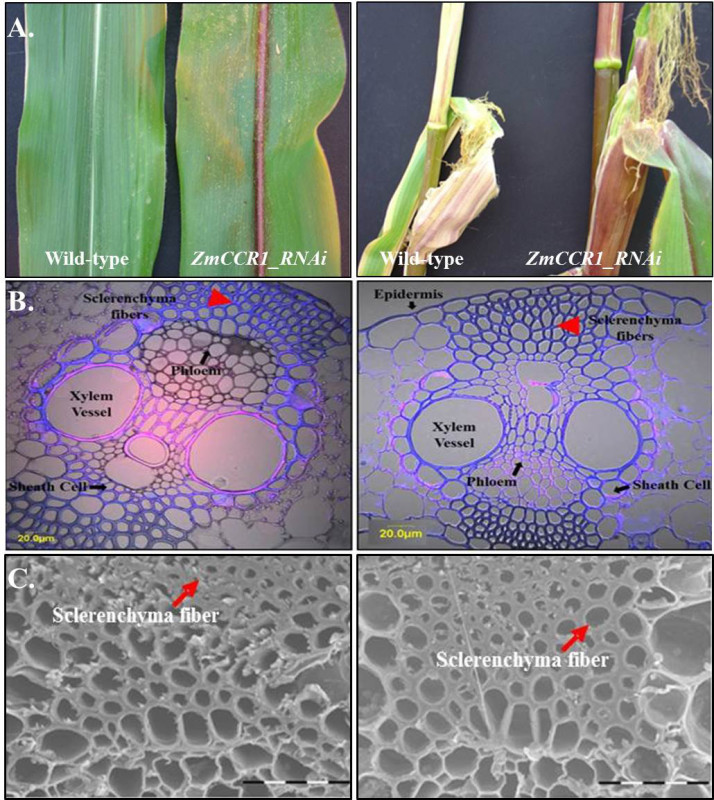 Figure 2. Phenotypic analyses of wild-type corn (Hi-II) and ZmCCR1_RNAi mutant leaf midrib. A) Brown coloration was seen in ZmCCR1 down-regulated corn leaves, stems and corn husks. B) The cross-sectioned maize leaf midribs of wild-type HI-II (left) and ZmCCR1_RNAi mutant line (right) were stained with 0.05 % toluidine blue O for 1 min to visualize secondary xylem tissues. The red arrowhead indicates the cell walls of sclerenchyma fibers of the leaf midrib. (C)Scanning electron microscopy (SEM) of ZmCCR1 down-regulated transgenic maize leaf midrib (right) as compared to that of wild-type non-transgenic control plant (left). The red arrow indicates sclerenchyma fibers. This figure has been modified from Park et al. (2012)18.
Figure 2. Phenotypic analyses of wild-type corn (Hi-II) and ZmCCR1_RNAi mutant leaf midrib. A) Brown coloration was seen in ZmCCR1 down-regulated corn leaves, stems and corn husks. B) The cross-sectioned maize leaf midribs of wild-type HI-II (left) and ZmCCR1_RNAi mutant line (right) were stained with 0.05 % toluidine blue O for 1 min to visualize secondary xylem tissues. The red arrowhead indicates the cell walls of sclerenchyma fibers of the leaf midrib. (C)Scanning electron microscopy (SEM) of ZmCCR1 down-regulated transgenic maize leaf midrib (right) as compared to that of wild-type non-transgenic control plant (left). The red arrow indicates sclerenchyma fibers. This figure has been modified from Park et al. (2012)18.
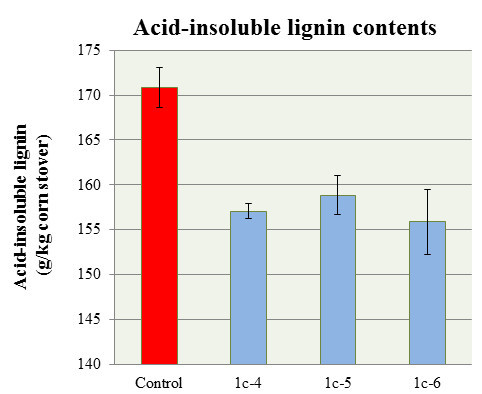 Figure 3. Klason lignin measurements (acid-insoluble lignin contents) of wild-type HI-II and ZmCCR1_RNAi mutant. The three mutant lines 1c, 1c-5, and 1c-6 had statistically lower lignin content, 8.5%, 7.5%, and 9.2% respectively, as percent of dry matter compared with the wild-type control plants. Mean ± standard deviation (P < 0.05, n = 3). This figure has been modified from Park et al. (2012)18.
Figure 3. Klason lignin measurements (acid-insoluble lignin contents) of wild-type HI-II and ZmCCR1_RNAi mutant. The three mutant lines 1c, 1c-5, and 1c-6 had statistically lower lignin content, 8.5%, 7.5%, and 9.2% respectively, as percent of dry matter compared with the wild-type control plants. Mean ± standard deviation (P < 0.05, n = 3). This figure has been modified from Park et al. (2012)18.
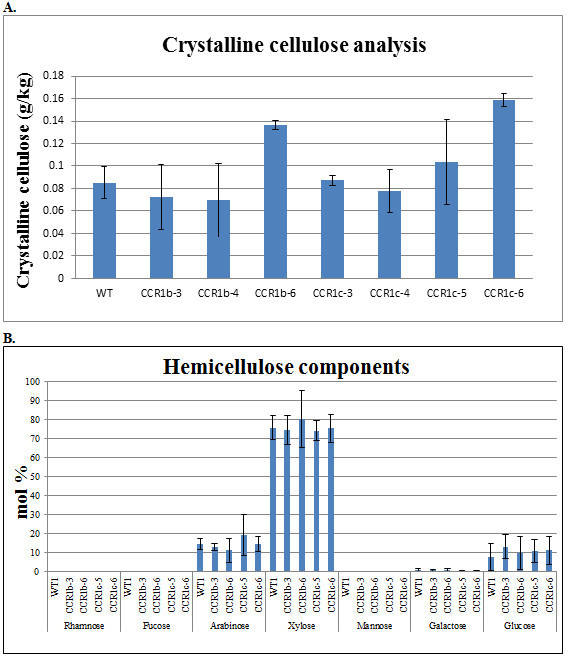 Figure 4. Cell wall compositional analyses. A) Crystalline cellulose analysis of ZmCCR1_RNAi lines (Tukey’s pairwise comparisons, * P < 0.05, n=3). B) Hemicellulose compositional analysis of wild-type maize and ZmCCR_RNAi transgenic maize lines (T1) via gas chromatography (GC). The main peaks from the chromatograms were integrated, identified based on retention times and fragment ion signatures, and expressed as mol percentage (P > 0.05, n = 3) (Tukey’s pairwise comparisons, P > 0.05; n = 3). This figure has been reused from Park et al. (2012)18.
Figure 4. Cell wall compositional analyses. A) Crystalline cellulose analysis of ZmCCR1_RNAi lines (Tukey’s pairwise comparisons, * P < 0.05, n=3). B) Hemicellulose compositional analysis of wild-type maize and ZmCCR_RNAi transgenic maize lines (T1) via gas chromatography (GC). The main peaks from the chromatograms were integrated, identified based on retention times and fragment ion signatures, and expressed as mol percentage (P > 0.05, n = 3) (Tukey’s pairwise comparisons, P > 0.05; n = 3). This figure has been reused from Park et al. (2012)18.
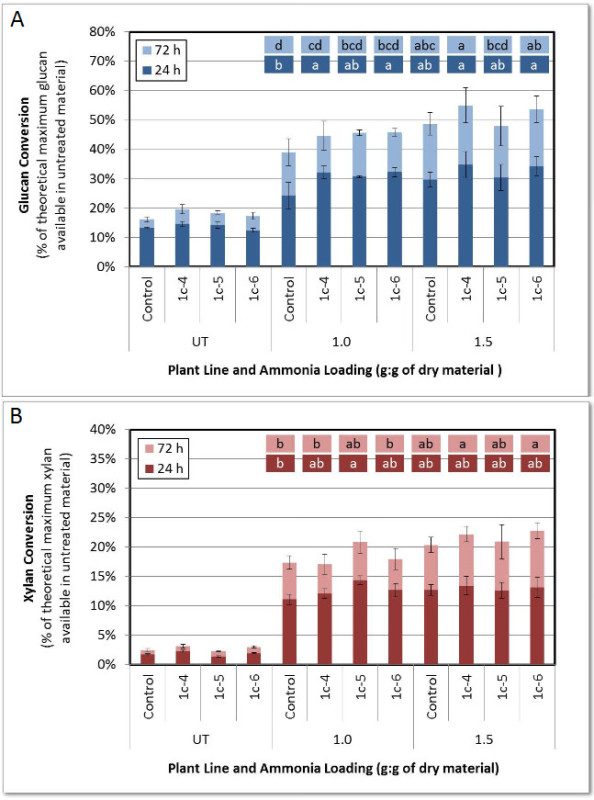 Figure 5. Percent sugar (glucan and xylan) conversions for untreated (UT) and AFEXTM-pretreated (90 °C, 5 min) maize stover at different concentrations of ammonia (1.0: 1.0 g NH3:g dry biomass 1.5: 1.5 g NH3:g dry biomass). Error bars represent the standard deviation of the mean and are based on two replicates for the untreated samples and four replicates (two pretreatment replicates with two hydrolysis replicates each) for the pretreated samples. Pretreated sugar conversions (24 hr or 72 hr) labeled with different letters are statistically different based on Tukey’s pairwise comparisons (P < 0.05). This figure has been reused from Park et al. (2012)18.
Figure 5. Percent sugar (glucan and xylan) conversions for untreated (UT) and AFEXTM-pretreated (90 °C, 5 min) maize stover at different concentrations of ammonia (1.0: 1.0 g NH3:g dry biomass 1.5: 1.5 g NH3:g dry biomass). Error bars represent the standard deviation of the mean and are based on two replicates for the untreated samples and four replicates (two pretreatment replicates with two hydrolysis replicates each) for the pretreated samples. Pretreated sugar conversions (24 hr or 72 hr) labeled with different letters are statistically different based on Tukey’s pairwise comparisons (P < 0.05). This figure has been reused from Park et al. (2012)18.
| Media | Chemical compositions |
| N6OSM | 4 g/L N6 salts |
| (Osmotic medium) | 1 ml/L N6 vitamin stock |
| 2 mg/L 2,4-D | |
| 100 mg/L myo-inositol | |
| 0.69 g/L proline | |
| 30 g/L sucrose | |
| 100 mg/L casein hydrolysate | |
| 36.4 g/L sorbitol | |
| 36.4 g/L mannitol | |
| 2.5 g/L gelrite, pH 5.8 | |
| Add filter sterilized silver nitrate (25 μM) after autoclaving | |
| N6E | 4 g/L N6 salts |
| (Callus induction) | 1 ml/L (1,000x) N6 vitamin stock |
| 2 mg/L 2,4-D | |
| 100 mg/L myo-inositol | |
| 2.76 g/L proline | |
| 30 g/L sucrose | |
| 100 mg/L casein hydrolysate | |
| 2.5 g/L gelrite, pH 5.8 | |
| Add filter sterilized silver nitrate (25 μM) after autoclaving | |
| N6S media | 4 g/L N6 salts |
| (Selection media) | 1 ml/L N6 vitamin stock |
| 2 mg/L 2,4-D | |
| 100 mg/L myo-inositol | |
| 0.69 g/L proline | |
| 30 g/L sucrose | |
| 100 mg/L casein hydrolysate | |
| 36.4 g/L sorbitol | |
| 36.4 g/L mannitol | |
| 2.5 g/L gelrite, pH 5.8 | |
| Add filter sterilized silver nitrate (25 μM) after autoclaving | |
| Regeneration medium | 4.3 g/L MS salts |
| 1 ml/L (1000x) MS vitamin stock | |
| 100 mg/L myo-inositol | |
| 60 g/L sucrose | |
| 3 g/L gelrite, pH 5.8 (100 x 25 mm Petri plates) | |
| Add filter sterilized bialaphos (3 mg/L) added after autoclaving | |
| Rooting medium | 4.3 g/L MS salts |
| 1 ml/L MS vitamin stock | |
| 100 mg/L myo-inositol | |
| 30 g/L sucrose | |
| 3 g/L gelrite, pH 5.8 (100 x 25 mm Petri plates) | |
| 10% Neutral buffered formalin | 100 ml of formalin |
| (1 liter) | 900 ml of ddH2O |
| 4.0 g of Sodium dihydrogen phosphate, monohydrate (NaH2PO4·H2O) |
Table 1. Table of specific reagents.
Discussion
The accessibility of microbial cellulases to plant cell wall polysaccharides is largely dependent on the degree to which they are associated with phenolic polymers23. The conversion rate from lignocellulosic biomass to fermentable sugar is negatively correlated with lignin content deposited in plant secondadry cell walls. This correlation is ascribed to the physical properties of lignin such as hydrophobicity24, chemical heterogeneity, and the absence of regular hydrolysable intermonomeric linkages25.
In this study, a dsRNAi technique induced various levels of gene down-regulation on genetic targets. Lignin down-regulation, mediated by a ZmCCR1_RNAi construct, has resulted in the brown-coloration in T1 transgneic lines. Brown-midrib (bm) coloration is a naturally occurring phenomenon that is caused by reduced lignin content and altered lignin composition. Unlike other naturally occurred bm mutants, which show the brown coloration only in leaf mid-ribs, the ZmCCR1_RNAi mutant lines revealed the phenotype in other parts of the plant, including the stems and husks. The histological assay also indicated that the sclerenchyma cell wall thickness of ZmCCR1 down-regulated leaves was much less than those of the wild-type control plants (Figure 2A). However, the sturucture and cell wall thickness of the main vascular systems inclduing xylem vessels, phloem, or sheath cells was not changed. This could explain the normal growth of the ZmCCR1_RNAi transgenic lines which grew normally in terms of plant height and stem diameter.
A reduction of more than 20% in the lignin content has generally caused a loss of biomass and made the plants more vulnerable against microbial pathogens and pests27,28. However, the mutant lines produced in this research, expressing less than a 10% lignin reduction, did not compromise the plant biomass and defensive mechanism against abiotic and biotic stresses.
Previous studies have shown that transgenic tobacco lines with significantly reduced CCR expression also showed an increase of other cell wall constituents such as glucose, xylose, and wall-bound phenolic compounds (e.g., sinapic and ferulic acids). In this study, the mild lignin reduction increased the level of crystalline cellulose in some of the ZmCCR1 down-regulated maize plants. Conversely, a cellulose compensation mechanism was also observed in Arabidopsis mutants which exhibited ectopic lignification when cellulose synthesis genes were defective34. The quantitative or qualitative changes of one cell wall carbohydrate component induces the alternation of other components35. Such compensation mechansims are important to maintan the homeostasis of plant vascular systems. However, in this study, hemicelluloses showed no statistically significant changes in ZmCCR1 down-regulated mutant lines. This result may be because the observed lignin reduction was not sufficient to trigger additional hemicellulose synthesis.
The decreased level of lignin and the increased crystalline cellulose level would be doubly beneficial for biofuel production. The lower lignin contents would require fewer inputs (e.g., H2SO4, cellulases, etc.) during processing and facilitate the biomass conversion process. The extra cellulose may increase the yield of fermentable sugars. The genetic manipulation of ZmCCR1 detailed in this study can be implemented to help make lignocelluosic bimoass derived bioethanol more commercially competitive.
Disclosures
No conflicts of interest declared.
Acknowledgments
The microscopic imaging was conducted via the services of the Michigan State University Center for Advanced Microscopy. Maize callus was purchased from the Maize Transformation Center of Iowa State University. The authors would like to thank Jeffrey R. Weatherhead of the MSU Plant Research Laboratory for his technical assistance on the carbohydrate analysis. This research was generously funded by the Corn Marketing Program of Michigan (CMPM) and the Consortium for Plant Biotechnology Research (CPBR).
References
- Perlack RD, et al. USDA-DOE. Biomass as feedstock for a bioenergy and bioproducts industry: The technical feasibility of a billion-ton annual supply. 2005. Available from: http://feedstockreview.ornl.gov/pdf/billion_ton_vision.pdf.
- Ralph J, Grabber JH, Hatfield RD. Lignin-ferulate cross-links in grasses - Active incorporation of ferulate polysaccharide esters into ryegrass lignins. Carbohydrate research. 1995. pp. 275pp. 167–178.
- Park S-H. Expediting cellulosic biofuels agenda: Production of high value-low volume co-products and lignin down-regulation of bioenergy crops [Ph.D. thesis] Michigan State University; 2011. [Google Scholar]
- Boerjan W, Ralph J, Baucher M. Lignin biosynthesis. Annual review of plant biology. 2003;54:519–546. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- Gibson LJ. The hierarchical structure and mechanics of plant materials. Journal of the Royal Society, Interface / the Royal Society. 2012;9:2749–2766. doi: 10.1098/rsif.2012.0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dien BS, et al. Enhancing alfalfa conversion efficiencies for sugar recovery and ethanol production by altering lignin composition. Bioresource technology. 2011. pp. 102pp. 6479–6486. [DOI] [PubMed]
- Fu CX, et al. Downregulation of Cinnamyl Alcohol Dehydrogenase (CAD) Leads to Improved Saccharification Efficiency in Switchgrass. Bioenerg Res. 2011;4:153–164. [Google Scholar]
- Grabber JH, Ralph J, Hatfield RD, Quideau S. p-hydroxyphenyl, guaiacyl, and syringyl lignins have similar inhibitory effects on wall degradability. Journal of agricultural and food chemistry. 1997;45:2530–2532. [Google Scholar]
- Li X, et al. Lignin monomer composition affects Arabidopsis cell-wall degradability after liquid hot water pretreatment. Biotechnology for biofuels. 2010;3 doi: 10.1186/1754-6834-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield SD, Kang KY, Chapple C. Designed for deconstruction--poplar trees altered in cell wall lignification improve the efficacy of bioethanol production. The New phytologist. 2012;194:91–101. doi: 10.1111/j.1469-8137.2011.04031.x. [DOI] [PubMed] [Google Scholar]
- Studer MH, et al. Lignin content in natural Populus variants affects sugar release. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:6300–6305. doi: 10.1073/pnas.1009252108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Dixon RA. Lignin modification improves fermentable sugar yields for biofuel production. Nature. 2007;25:759–761. doi: 10.1038/nbt1316. [DOI] [PubMed] [Google Scholar]
- Ziebell A, et al. Increase in 4-coumaryl alcohol units during lignification in alfalfa (Medicago sativa) alters the extractability and molecular weight of lignin. The Journal of biological chemistry. 2010;285:38961–38968. doi: 10.1074/jbc.M110.137315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S-H, et al. The quest for alternatives to microbial cellulase mix production: corn stover-produced heterologous multi-cellulases readily deconstruct lignocellulosic biomass into fermentable sugars. Journal of Chemical Technolog., and Biotechnology. 2011;86:633–641. [Google Scholar]
- Mol JN, et al. Regulation of plant gene expression by antisense RNA. FEBS letters. 1990;268:427–430. doi: 10.1016/0014-5793(90)81298-3. [DOI] [PubMed] [Google Scholar]
- Adamo A, et al. Transgene-mediated cosuppression and RNA interference enhance germ-line apoptosis in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3440–3445. doi: 10.1073/pnas.1107390109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith NA, et al. Total silencing by intron-spliced hairpin RNAs. Nature. 2000;407:319–320. doi: 10.1038/35030305. [DOI] [PubMed] [Google Scholar]
- Park S-H, et al. Downregulation of Maize Cinnamoyl-Coenzyme A Reductase via RNA Interference Technology Causes Brown Midrib and Improves Ammonia Fiber Expansion-Pretreated Conversion into Fermentable Sugars for Biofuels. Crop Sci. 2012;52:2687–2701. [Google Scholar]
- Altpeter F, et al. Particle bombardment and the genetic enhancement of crops: myths and realities. Mol Breeding. 2005;15:305–327. [Google Scholar]
- Pichon M, Courbou I, Beckert M, Boudet AM, Grima-Pettenati J. Cloning and characterization of two maize cDNAs encoding cinnamoyl-CoA reductase (CCR) and differential expression of the corresponding genes. Plant molecular biology. 1998;38:671–676. doi: 10.1023/a:1006060101866. [DOI] [PubMed] [Google Scholar]
- Mansoor S, Amin I, Hussain M, Zafar Y, Briddon RW. Engineering novel traits in plants through RNA interference. Trends in plant science. 2006;11:559–565. doi: 10.1016/j.tplants.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Hatfield RD, Jung H-JG, Ralph J, Buxton DR, Weimer PJ. A comparison of the insoluble residues produced by the Klason lignin and acid detergent lignin procedures. J Sci Food Agr. 1994;65:51–58. [Google Scholar]
- Sluiter JB, Ruiz RO, Scarlata CJ, Sluiter AD, Templeton DW. Compositional analysis of lignocellulosic feedstocks. 1. Review and description of methods. Journal of agricultural and food chemistry. 2010;58:9043–9053. doi: 10.1021/jf1008023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluiter A, Hames B, Ruiz R, Scarlata C, Sluiter J, Templeton D, Crocker D. Determination of structural carbohydrates and lignin in biomass. Laboratory Analytic Procedure. 2008.
- Bate NJ, et al. Quantitative Relationship between Phenylalanine Ammonia-Lyase Levels and Phenylpropanoid Accumulation in Transgenic Tobacco Identifies a Rate-Determining Step in Natural Product Synthesis. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:7608–7612. doi: 10.1073/pnas.91.16.7608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu WJ, et al. Repression of lignin biosynthesis promotes cellulose accumulation and growth in transgenic trees. Nature. 1999;17:808–812. doi: 10.1038/11758. [DOI] [PubMed] [Google Scholar]
- Lapierre C, et al. Signatures of cinnamyl alcohol dehydrogenase deficiency in poplar lignins. Phytochemistry. 2004;65:313–321. doi: 10.1016/j.phytochem.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Updegraff DM. Semimicro determination of cellulose in biological materials. Anal Biochem. 1969;32:420–424. doi: 10.1016/s0003-2697(69)80009-6. [DOI] [PubMed] [Google Scholar]
- Yemm EW, Willis AJ. The estimation of carbohydrates in plant extracts by anthrone. The Biochemical journal. 1954;57:508–514. doi: 10.1042/bj0570508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filomena AP, Cherie W, Geoffrey BF, Antony B. Determining the polysaccharide composition of plant cell walls. Nature. 2012;7:1590–1607. doi: 10.1038/nprot.2012.081. [DOI] [PubMed] [Google Scholar]
- Foster CE, Martin TM, Pauly M. Comprehensive Compositional Analysis of Plant Cell Walls (Lignocellulosic biomass) Part II: Carbohydrates. J. Vis. Exp. 2010. [DOI] [PMC free article] [PubMed]
- Department of Agronomy, Iowa State University. Particle bombardment of Hi II immature zygotic embryos and recovery of transgenic maize plants. 2005.
- Bio-Rad. Biolistic PDS-1000/He Particle Delivery System. 1997. Available from: http://www.bio-rad.com/webroot/web/pdf/lsr/literature/Bulletin_9075.pdf.
- Cano-Delgado A, Penfield S, Smith C, Catley M, Bevan M. Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. The Plant journal : for cell and molecular biology. 2003;34:351–362. doi: 10.1046/j.1365-313x.2003.01729.x. [DOI] [PubMed] [Google Scholar]
- Boudet AM, Kajita S, Grima-Pettenati J, Goffner D. Lignins and lignocellulosics: a better control of synthesis for new and improved uses. Trends in plant science. 2003;8:576–581. doi: 10.1016/j.tplants.2003.10.001. [DOI] [PubMed] [Google Scholar]


