Abstract
The major genetic cause of frontotemporal dementia and amyotrophic lateral sclerosis is a G4C2 repeat expansion in C9ORF72. Efforts to combat neurodegeneration associated with “c9FTD/ALS” are hindered by a lack of animal models recapitulating disease features. We developed a mouse model to mimic both neuropathological and clinical c9FTD/ALS phenotypes. We expressed (G4C2)66 throughout the murine central nervous system by means of somatic brain transgenesis mediated by adeno-associated virus. Brains of 6-month-old mice contained nuclear RNA foci, inclusions of poly(Gly-Pro), poly(Gly-Ala), and poly(Gly-Arg) dipeptide repeat proteins, as well as TDP-43 pathology. These mouse brains also exhibited cortical neuron and cerebellar Purkinje cell loss, astrogliosis, and decreased weight. (G4C2)66 mice also developed behavioral abnormalities similar to clinical symptoms of c9FTD/ALS patients, including hyperactivity, anxiety, antisocial behavior, and motor deficits.
An intronic G4C2 repeat expansion in the chromosome 9 open reading frame 72 (C9ORF72) gene is the major genetic cause of frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS) (1, 2). Although FTD and ALS are characterized, respectively, by cognitive and behavioral symptoms and by motor symptoms, there is clinical and neuropathological overlap between the two diseases. The precise mechanisms by which the C9ORF72 mutation contributes to “c9FTD/ALS” remain elusive, but toxicity mediated by RNA bidirectionally transcribed from the expansion [r(G4C2)exp and r(G2C4)exp] is thought to play an important role (3–12). Repeat-containing transcripts form intranuclear RNA foci believed to sequester various RNA-binding proteins (8, 9, 11, 13–17), and they are also susceptible to repeat-associated non-ATG (RAN) translation resulting in the synthesis of “c9RAN proteins” of repeating dipeptides (18–21). Despite advances made toward elucidating c9FTD/ALS pathogenesis, many questions remain because of the lack of mouse models recapitulating key disease features.
To investigate the neurotoxic effects linked to the expanded G4C2 repeat and to create a model for testing new therapies in vivo, we sought to generate mice that develop clinical and pathological features of c9FTD/ALS. We used an adeno-associated viral vector to mediate robust expression of either 2 or 66 G4C2 repeats, lacking an ATG start codon, in the central nervous system (CNS) of mice. Six months after intracerebroventricular (ICV) administration of AAV2/9-(G4C2)66 (n = 11) or AAV2/9-(G4C2)2 (n = 12) to postnatal day 0 mice, which results in predominantly neuronal transduction (22), a thorough characterization of the mice was undertaken.
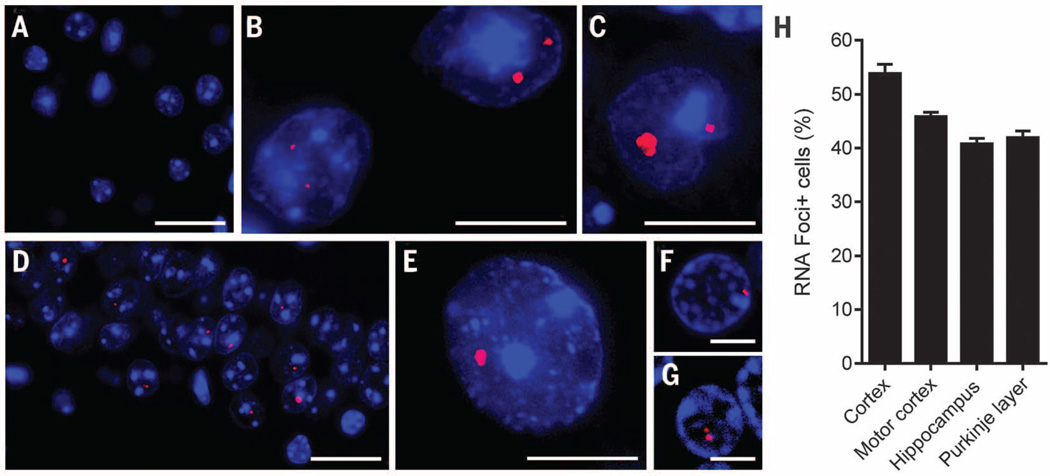
To assess whether RNA foci are formed in (G4C2)66 mice, we performed RNA fluorescence in situ hybridization using a probe against r(G4C2). As anticipated, no foci were detected in control (G4C2)2 mice (Fig. 1A), but nuclear foci were detected throughout the CNS of (G4C2)66 mice (Fig. 1, B to E), reminiscent of those observed in c9FTD/ALS patients (Fig. 1, F and G). Foci were present across all layers of the cortex (Fig. 1B), in Purkinje cells of the cerebellum (Fig. 1C), in the CA1 to CA3 fields of the hippocampus (Fig. 1D), and in the thalamus (table S1). Foci were also observed, albeit to a lesser extent, in the ventral horn of the spinal cord (Fig. 1E), as well as in the hippocampal dentate gyrus, cerebellar granular and molecular layers, and the amygdala (table S1). The number of foci-positive cells ranged from 40 to 54% in the cortex, motor cortex, hippocampus, and cerebellar Purkinje layer (Fig. 1H).
Fig. 1. Intranuclear RNA foci were detected in the CNS of (G4C2)66 mice.
The presence of r(G4C2) RNA foci in brain and spinal cord of 6-month-old (G4C2)2 and (G4C2)66 mice was evaluated by RNA fluorescence in situ hybridization. (A) Foci were not detected in (G4C2)2 mice; shown is a representative image of the cortex. (B to G) In (G4C2)66 mice, RNA foci were detected in various regions—including the cortex (B), cerebellum (C), hippocampus (D), and spinal cord (E)—and were similar to foci observed in the cortex (F) and cerebellum (G) of c9FTD/ALS patients. (H) The average percentage of cells containing nuclear RNA foci in the indicated brain regions of (G4C2)66 mice (n = 11) was calculated. The total number of cells counted per region was 500 for cortex, motor cortex, and hippocampus (CA1 to CA3) and 100 for Purkinje cells. Scale bars: (A), (D), (F), and (G), 25 µm; (B), (C), and (E), 10 µm.
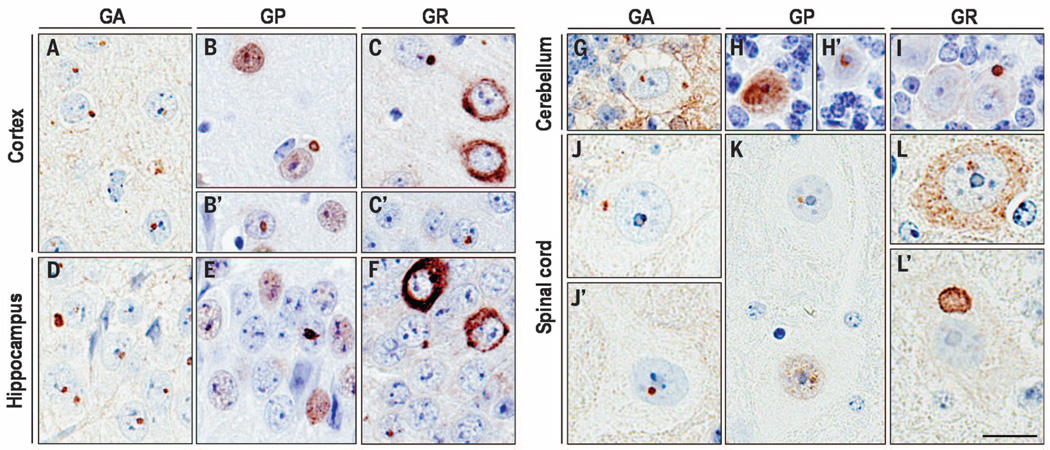
To investigate RAN translation in (G4C2)66 mice, we first used a poly(Gly-Pro) [poly(GP)] immunoassay and observed robust poly(GP) expression in brain homogenates of (G4C2)66 mice but not (G4C2)2 mice (fig. S1). In addition, c9RAN protein inclusions were specifically expressed in the CNS of (G4C2)66 mice (Fig. 2 and fig. S2), as seen in c9FTD/ALS patients (fig. S3). Indeed, r(G4C2)66 was RAN translated in all frames. In the cortex and hippocampus of (G4C2)66 mice, and less frequently in the cerebellum and spinal cord, globular poly(Gly-Ala) [poly(GA)], poly(GP), or poly(Gly-Arg) [poly(GR)] inclusions were detected most frequently in the nucleus, but cytoplasmic inclusions were also present. We also observed cells with diffuse nuclear poly(GP) staining and diffuse cytoplasmic poly(GR) staining. On the basis of semiquantitative analysis, cells immunopositive for poly(GA) or poly(GP) were more frequent than those immunopositive for poly(GR) (table S1), consistent with c9FTD/ALS c9RAN protein pathology (23). The majority of c9RAN protein inclusions in (G4C2)66 mice were ubiquitin-positive (fig. S4), as in c9FTD/ALS (24), and localized to microtubule-associated protein 2–positive neurons but rarely to glial fibrillary acidic protein (GFAP)–positive cells (fig. S5). About 70% of cortical cells immunopositive for poly(GP) inclusions also contained at least one RNA focus (fig. S6).
Fig. 2. c9RAN protein pathology was detected throughout the CNS of (G4C2)66 mice.
Intranuclear and cytoplasmic inclusions immunopositive for poly(GA), poly(GP), or poly(GR) were observed in various regions of the CNS of 6-month-old (G4C2)66 mice, including the cortex (A to C′), hippocampus (D to F), cerebellum (G to I), and spinal cord (J to L′). Note also the diffuse nuclear poly(GP) staining [(B), (B′), (E), (H), and (K)] and diffuse cytoplasmic poly(GR) staining [(C), (F), and (L)]. Semiquantitative analysis of c9RAN protein staining in different neuroanatomical regions is provided in table S1. No c9RAN proteins were detected in 6-month-old control (G4C2)2 mice (fig. S3). Scale bar, 20 µm.
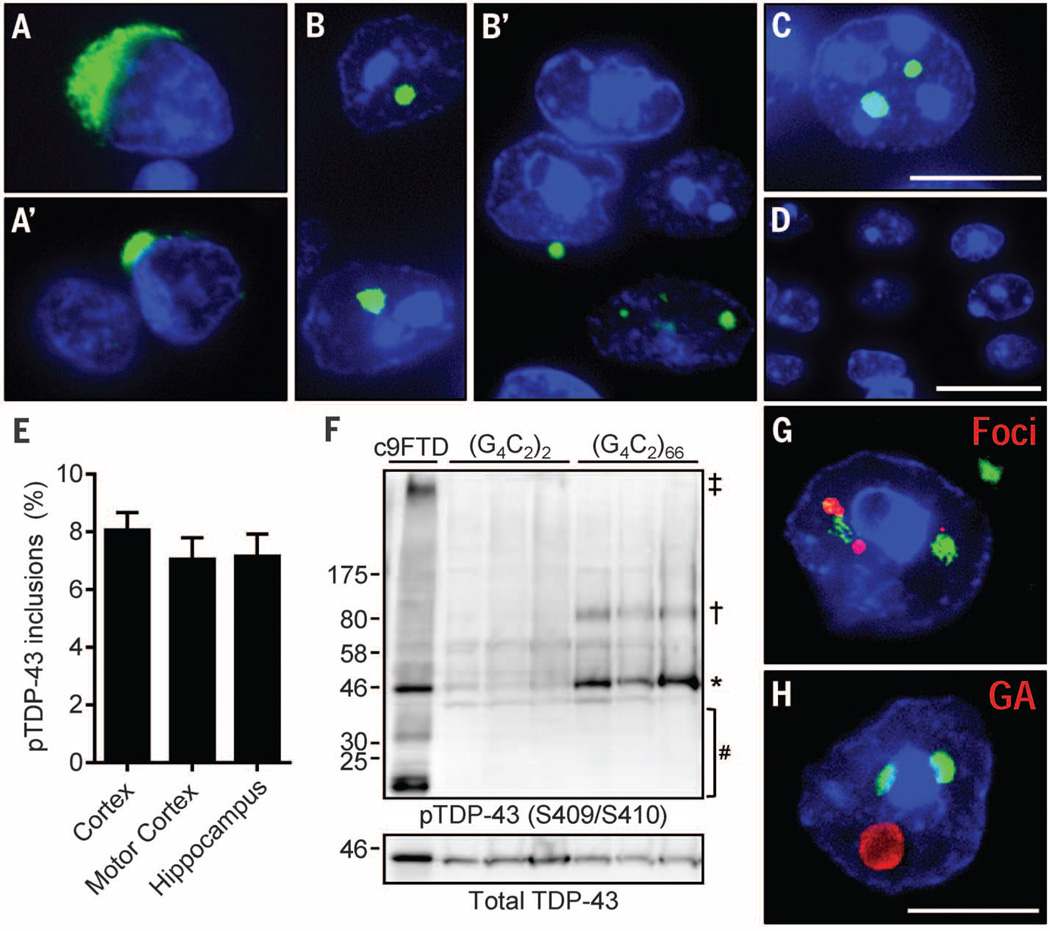
Inclusions of phosphorylated TDP-43 (pTDP-43) are yet another neuropathological feature of c9FTD/ALS (Fig. 3, A and A′) (23). Remarkably, nuclear, and occasionally cytoplasmic, inclusions of endogenous pTDP-43 were observed in the cortex (Fig. 3, B and B′) and hippocampus (Fig. 3C) of (G4C2)66 mice that were present in about 7 to 8% of cells (Fig. 3E) but were not observed in (G4C2)2 mice (Fig. 3D). Immunoblot analysis of hemibrain urea fractions confirmed the presence of insoluble pTDP-43 in (G4C2)66 mice (Fig. 3F). Insoluble pTDP-43 was predominantly monomeric, but oligomers of ~80 kD were also observed. The high-molecular-weight or truncated pTDP-43 species seen in c9FTD were not detected in (G4C2)66 mice under these conditions. Of the 250 cortical cells with pTDP-43 pathology examined among five (G4C2)66 mice, all harbored at least one nuclear RNA focus (Fig. 3G). Similarly, ~75% of cells with pTDP-43 pathology also contained poly(GA) inclusions. Despite the coexistence of both proteins in a given cell, they did not localize to the same inclusions (Fig. 3H).
Fig. 3. (G4C2)66 mice developed inclusions composed of pTDP-43.
(A and A′) Representative pTDP-43 inclusions in postmortem c9FTD/ALS frontal cortex sections. (B to D) Similarly, multiple cells with nuclear, and occasionally cytoplasmic pTDP-43, inclusions were observed in the cortex (B and B′) and hippocampus (C) of 6-month-old (G4C2)66 mice. No pTDP-43 immunoreactivity was detected in control (G4C2)2 mice; shown is a representative image of the cortex (D). (E) The average percentage of cells with pTDP-43 inclusions in the indicated regions was calculated on the basis of evaluations of 500 cells per mouse (n = 6 mice). (F) pTDP-43 was detected in brain urea fractions from (G4C2)66 mice and a c9FTD patient but not from (G4C2)2 mice. Symbols: ‡high molecular weight species; †oligomers; *full-length protein; #cleavage products. (G) Of 250 cells containing pTDP-43 inclusions examined among five (G4C2)66 mice, all were found to contain nuclear foci. (H) Of 250 cells containing pTDP-43 inclusions examined among five (G4C2)66 mice, ~75% were found to be positive for poly(GA) inclusions. Nuclei are stained blue in (A) to (D), (G), and (H). Scale bars: (A) to (C), (G), and (H), 10 µm; (D), 20 µm.
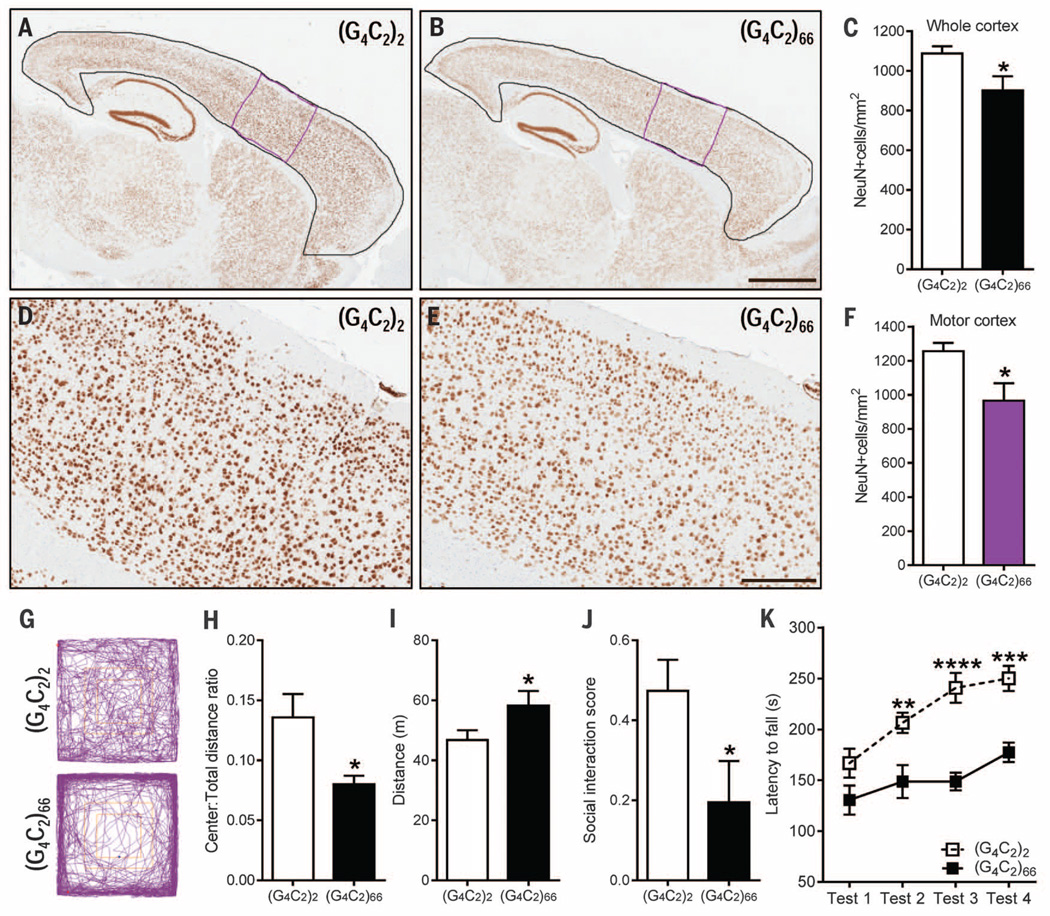
Next, we evaluated whether expression of the expanded repeat caused neurodegeneration and gliosis. In (G4C2)66 mice, the number of NeuN-positive neurons in the whole cortex and motor cortex was significantly reduced by 17% and 23%, respectively (Fig. 4, A to F), and 11% fewer Purkinje cells were present in the cerebellum (fig. S7, A to C). No such cell loss was observed in the hippocampus, thalamus, or spinal cord. Immunoreactivity for GFAP, a marker of reactive gliosis, was increased in the cortex of (G4C2)66 mice (fig. S7, D to F).
Fig. 4. (G4C2)66 mice developed cortical neuron loss and exhibited behavior and motor deficits at 6 months of age.
(A and B) Representative images of NeuN-labeled cells in the cortex of (G4C2)2 mice (n = 6) and (G4C2)66 mice (n = 6). (C) Quantification of NeuN-positive cells in the whole cortex [area delineated in black in (A) and (B)]. (D and E) Enhanced magnification of the motor cortex [area delineated in purple in (A) and (B)]. (F) Quantification of NeuN-positive cells in the motor cortex. (G to I) In the open-field test, (G4C2)66 mice (n = 11) displayed signs of increased anxiety-like behavior compared with (G4C2)2 mice (n = 12), as evidenced by representative traces (G) and a significant decrease in distance traveled in the center area normalized to total distance traveled (H). (G4C2)66 mice traveled a greater distance overall (I), which is indicative of hyperactivity. (J) In the social interaction test, (G4C2)66 mice had a decreased interaction score, which is indicative of social deficits. (K) Motor deficits, manifesting as a decreased latency to fall from a rotating rod, were observed in (G4C2)66 mice on test day 2 to 4 of the Rota-Rod test. Scale bars: (A) and (B), 1 mm; (D) and (E), 300 µm. Data are presented as means T SEM; *P < 0.05, unpaired, two-tailed Student’s t-test. ****P < 0.0001, ***P < 0.001, **P < 0.01, two-way analysis of variance followed by Sidak’s multiple comparisons test. Abbreviated units: m, meters; s, seconds.
To determine whether the combination of pathological features described above affected behavior, 6-month-old (G4C2)2 and (G4C2)66 mice were subjected to a battery of behavioral tasks. At this time point, no significant difference in body weight was detected between male mice expressing 2 or 66 repeats, but female (G4C2)66 mice exhibited an 11% decrease in body weight compared with female (G4C2)2 mice (fig. S8A). A modest, but statistically significant, decrease in brain weight in (G4C2)66 mice was evident (fig. S8B), which again suggested that the expanded repeat caused brain atrophy. The open-field assay can be used to evaluate general locomotor activity, exploration, and anxiety-like behavior in a novel environment. (G4C2)66 mice showed a decreased tendency to explore the center of the open field (Fig. 4, G and H), which is suggestive of anxiety-like behavior. (G4C2)66 mice also traveled a longer distance in comparison with (G4C2)2-expressing mice (Fig. 4I) and at a greater speed (fig. S8C), indicative of hyperactivity. Such hyperactivity has been linked to behavioral disinhibition in a mouse model of Alzheimer’s disease (25).
To examine whether the behavioral phenotype of (G4C2)66 mice includes a social component, we used a three-chamber social interaction test. This test assesses the active interaction time of the test mouse, with a novel probe mouse housed in a cylinder within one of the chambers and an empty cylinder in the opposite chamber. Compared with (G4C2)2 mice, (G4C2)66 mice spent more time in the chamber with the empty cylinder than in the chamber containing the probe mouse (Fig. 4J), which suggested social abnormalities.
Finally, motor coordination and balance of mice were evaluated using the Rota-Rod test in which mice have to keep their balance on a rotating rod. Although no significant difference in time spent on the rod before falling off was seen between (G4C2)66 and (G4C2)2 mice on the first day of testing, from the second day onward (G4C2)66 mice fell significantly faster, which is indicative of motor impairments (Fig. 4K).
Here, we found that mice expressing (G4C2)66 throughout the CNS developed RNA foci, ubiquitin-positive inclusions of c9RANproteins, and pTDP-43 inclusions, as well as cortical neuron and cerebellar Purkinje cell loss. These abnormalities likely contributed to the behavioral phenotype of (G4C2)66 mice that suggested the disinhibition, anxiety, impaired social cognition, and motor skill deficits observed in c9FTD/ALS patients (26). These data demonstrate that the expression of expanded sense G4C2 repeats is sufficient to cause neuropathological changes and neurodegeneration; however, additional studies are required to determine the contribution of antisense repeats to disease.
(G4C2)66 mice should prove valuable in deciphering pathomechanisms associated with the C9ORF72 repeat expansion. Indeed, the observation of pTDP-43 inclusions in (G4C2)66 mice suggests that the repeat expansion is an initiator of TDP-43 pathology. Because all examined cells with TDP-43 pathology were found to contain foci, repeat-containing RNA or the foci themselves may be responsible for instigating TDP-43 abnormalities. As (G4C2)66 mice recapitulate neuropathological and clinical phenotypes of c9FTD/ALS, they also offer an attractive model for testing potential therapeutics targeting r(G4C2)exp, such as antisense oligonucleotides (8, 11, 12) and small molecules (27). In fact, our findings suggest that such approaches may not only mitigate foci formation and RAN translation, they may also alleviate TDP-43–mediated toxicity.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institute on Aging, NIH [P50AG016574 (L.P.)]; National Institute of Neurological Disorders and Stroke, NIH [R21NS089979 (T.F.G. and K.B.B.), R21NS084528 (L.P.), R21NS079807 (Y.-J.Z.), R01NS088689 (L.P.), R01NS063964 (L.P.), R01NS077402 (L.P.), and P01NS084974 (L.P., D.W.D., R.R., and K.B.B.)]; National Institute of Environmental Health Services, NIH [R01ES20395 (L.P.)]; U.S. Department of Defense [ALS Research Program AL130125 (L.P.)]; Mayo Clinic Foundation (L.P.); Mayo Clinic Center for Regenerative Medicine (P.O.B.); Mayo Graduate School (J.C. and K.F.B.); ALS Association (T.F.G., Y.-J.Z., P.O.B., K.B.B., and L.P.); Robert Packard Center for ALS Research at Johns Hopkins (L.P.); Target ALS (L.P.); Alzheimer’s Association [NIRP-14-304425 (Y.-J.Z.) and NIRP-12-259289 (J.D.F.)]; and European Research Council under European Union’s Seventh Framework Programme [FP7/2014-2019 no. 617198 [DPR-MODELS] (D.E.)]. L.P. and T.F.G. have filed a patent (no. 14/162,570) in the United States regarding the use of poly(GP) immunoassays as a diagnostic for C9ORF72 repeat expansion–related diseases. The data are tabulated in the manuscript and the supplementary materials.
Footnotes
www.sciencemag.org/content/348/6239/1151/suppl/DC1
Materials and Methods
Figs. S1 to S8
Tables S1
References (28–35)
REFERENCES AND NOTES
- 1.DeJesus-Hernandez M, et al. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Renton AE, et al. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizielinska S, et al. Science. 2014;345:1192–1194. doi: 10.1126/science.1256800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang YJ, et al. Acta Neuropathol. 2014;128:505–524. doi: 10.1007/s00401-014-1336-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.May S, et al. Acta Neuropathol. 2014;128:485–503. doi: 10.1007/s00401-014-1329-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwon I, et al. Science. 2014;345:1139–1145. doi: 10.1126/science.1254917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wen X, et al. Neuron. 2014;84:1213–1225. doi: 10.1016/j.neuron.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donnelly CJ, et al. Neuron. 2013;80:415–428. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Almeida S, et al. Acta Neuropathol. 2013;126:385–399. doi: 10.1007/s00401-013-1149-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee YB, et al. Cell Reports. 2013;5:1178–1186. doi: 10.1016/j.celrep.2013.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sareen D, et al. Sci. Transl. Med. 2013;5:208ra149. doi: 10.1126/scitranslmed.3007529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lagier-Tourenne C, et al. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E4530–E4539. doi: 10.1073/pnas.1318835110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mori K, et al. Acta Neuropathol. 2013;125:413–423. doi: 10.1007/s00401-013-1088-7. [DOI] [PubMed] [Google Scholar]
- 14.Reddy K, Zamiri B, Stanley SY, Macgregor RB, Jr, Pearson CE. J. Biol. Chem. 2013;288:9860–9866. doi: 10.1074/jbc.C113.452532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Z, et al. Proc. Natl. Acad. Sci. U.S.A. 2013;110:7778–7783. doi: 10.1073/pnas.1219643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooper-Knock J, et al. Brain. 2014;137:2040–2051. doi: 10.1093/brain/awu120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haeusler AR, et al. Nature. 2014;507:195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ash PE, et al. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mori K, et al. Acta Neuropathol. 2013;126:881–893. doi: 10.1007/s00401-013-1189-3. [DOI] [PubMed] [Google Scholar]
- 20.Mori K, et al. Science. 2013;339:1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- 21.Gendron TF, et al. Acta Neuropathol. 2013;126:829–844. doi: 10.1007/s00401-013-1192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chakrabarty P, et al. PLOS ONE. 2013;8:e67680. doi: 10.1371/journal.pone.0067680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mackenzie IR, Frick P, Neumann M. Acta Neuropathol. 2014;127:347–357. doi: 10.1007/s00401-013-1232-4. [DOI] [PubMed] [Google Scholar]
- 24.Mann DM, et al. Acta Neuropathol. Commun. 2013;1:68. doi: 10.1186/2051-5960-1-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gil-Bea FJ, Aisa B, Schliebs R, Ramírez MJ. Behav. Neurosci. 2007;121:340–344. doi: 10.1037/0735-7044.121.2.340. [DOI] [PubMed] [Google Scholar]
- 26.Boeve BF, Graff-Radford NR. Alzheimers Res. Ther. 2012;4:29. doi: 10.1186/alzrt132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su Z, et al. Neuron. 2014;83:1043–1050. doi: 10.1016/j.neuron.2014.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.