Abstract
As immunization programs for human papillomavirus (HPV) are implemented more widely around the world, interest is increasing in measuring their impact. One early measurable impact of HPV vaccine is on the prevalence of specific HPV types in a population. In low-resource settings, a potentially attractive strategy would be to monitor HPV prevalence using clinical cervical cancer screening test results to triage specimens for HPV typing. We assessed this approach in a nationally representative population of U.S. females aged 14–59 years. Using self-collected cervico-vaginal swab specimens from 4,150 women participating in the National Health and Nutrition Examination Survey during 2003–2006, we evaluated type-specific HPV prevalence detected by the Roche linear array (LA) research test on all specimens, compared with type-specific HPV prevalence detected by LA conducted only on specimens positive by the digene hybrid capture 2 (HC-2) clinical test. We calculated weighted prevalence estimates and their 95% confidence intervals (CIs), and examined relative type-specific HPV prevalence according to the two testing approaches. The population prevalence of oncogenic HPV vaccine types 16/18 was 6.2% (CI:5.4–7.1) by LA if all specimens were tested, and 2.4% (CI:1.9–3.0) if restricted to positive HC-2. Relative prevalence of individual HPV types was similar for both approaches. Compared with typing all specimens, a triage approach would require testing fewer specimens, but a greater reduction in HPV prevalence or a larger group of specimens would be needed to detect vaccine impact. Further investigation is warranted to inform type-specific HPV monitoring approaches around the world.
Keywords: papillomavirus infections, population surveillance, papillomavirus vaccines, DNA probes, HPV, laboratories, epidemiology
Despite slow initial adoption of vaccines against human papillomaviruses (HPV), global implementation of HPV immunization programs is likely to accelerate as vaccine prices decrease.1,2 One of the earliest measurable effects of these vaccines is expected to be a reduced prevalence of HPV vaccine types (HPV types 6, 11, 16 and/or 18) detected in cervico-vaginal specimens.3 While a variety of different study designs are being considered, monitoring type-specific HPV prevalence is an early way to assess the impact of HPV vaccine in a population, since reductions in cervical cancer or other HPV-related diseases may not be detectable for years or even decades following vaccine introduction. Although impact monitoring is not considered a precondition for HPV vaccine introduction, demonstrating vaccine impact may help justify the use of funds for vaccination programs, particularly in low-resource settings such as low or middle income countries or other settings where resources are limited.4 However, the optimal approaches to monitoring the type-specific prevalence of HPV in a population after HPV vaccine implementation have not been established.
Various testing methods exist for detecting HPV infections, and molecular tests for HPV are available for either research or clinical use. It is important to note that the purposes of the tests used in each of these two contexts differ substantially.5 Generally, the goals of research using HPV molecular tests are to understand natural history, risk factors and distribution of HPV infection at the population level.6 For this reason, HPV molecular tests used for research have high analytic sensitivity and can be used to identify specific HPV types present in a specimen. Commercially available research tests include Roche linear array (LA) assay (Roche, Pleasanton, CA) and Innogenetics line probe assays (Innogenetics, Gent, Belgium) for HPV genotyping, and others. In comparison, the goal of a clinical HPV molecular test is to identify persons likely to have HPV-associated disease. For HPV testing, clinical assays are approved by the U.S. Food and Drug Administration (FDA) based on their ability to detect infections with oncogenic HPV types that have good correlation with clinical cervical disease but lower sensitivity than the research assays; such tests are designed for use in clinical laboratories and currently include digene hybrid capture 2 (HC-2) assay (Quiagen, Valencia, CA), Cervista HPV (Hologic, Bedford, MA) and cobas HPV (Roche, Pleasanton, CA) tests.7–10 The U.S. Preventive Services Task Force recently recommended the use of clinical HPV testing in combination with cervical cytology (Papanicolau, or Pap smear) in women aged 30–65 years.11
In low-resource settings, a potentially attractive strategy for monitoring HPV vaccine impact would involve using clinical HPV test results from cervical cancer screening specimens. Clinical HPV tests such as HC-2, or careHPV, which was modeled on HC-2 and developed specifically for use in low-resource settings, do not provide type-specific results, and so positive specimens require additional testing to determine HPV type.12,13 Working with an existing program doing cervical cancer screening could have the advantages of supporting cervical cancer screening, eliminating the added costs of collecting the initial specimen for HPV testing, and reducing overall testing costs, since only a subset of samples would require additional HPV testing.
In the USA, HPV prevalence has been measured using both clinical and epidemiologic research tests since 2003 as part of the National Health and Nutrition Examination Survey (NHANES).14 NHANES examines a nationally representative sample of U.S. civilians. As part of the survey, both the LA research test and the HC-2 clinical test are performed on cervico-vaginal specimens. We used this convenient dataset to analyze whether clinical HPV test results might be able to play a useful role in monitoring HPV vaccine impact.
To do so, we evaluated the type-specific HPV prevalence detected by the LA research test on all specimens, compared with the type-specific HPV prevalence detected by LA only on specimens that were positive by the HC-2 clinical test. We also examined relative type-specific HPV prevalences according to these two testing approaches.
Methods
Study population and data collection
NHANES is an ongoing series of cross-sectional surveys that use a complex, multistage, probability sample design with unequal probabilities of selection to obtain a nationally representative sample of the non-institutionalized civilian population in the USA. NHANES methodology has been described in detail elsewhere.14 Our study included females aged 14–59 years who participated in NHANES during 2003–2006.
Informed consent was obtained from all participants, and parental permission was received for persons <18 years old. Human subjects approval was given by the National Center for Health Statistics (NCHS) research ethics review board.
Participants had a household interview to ascertain demographic information including age, marital status, race/ethnicity, income and education. In a mobile examination center, participants also answered a sexual behavior questionnaire administered in a private room using an audio computer-assisted self-interview in English or Spanish. Those who reported ever having sex (vaginal, anal or oral) were asked additional sexual history questions, including numbers of sex partners and age at first sex. Cervico-vaginal swabs were self-collected and submitted for laboratory testing.
Laboratory methods
In order to isolate HPV DNA from cervico-vaginal swab specimens, DNA extractions were performed within 1 month of specimen collection using modified QIAmp Mini Kit (Qiagen, Valencia, CA), as previously described.15,16 Extract (100 μL total volume) was tested immediately or stored at −20° C. With every batch of specimens, one water blank was processed through all steps of extraction to detect potential contamination. The same extract was used for both the clinical and research HPV assays.
The clinical HC-2 assay is designed to detect any of the 14 HPV types known to be oncogenic (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66 and 68), without indicating the presence of any specific HPV type/s in the specimen.17,18 Of note, the HC-2 test includes probes for only 13 types yet is considered cross-reactive with HPV type 66.19 The FDA-approved format was modified to accommodate DNA extracts from self-collected cervico-vaginal swabs. A 50 μL aliquot of extracted DNA was added to 200 μL physiological saline and 125 μL denaturing solution from the HC-2 kit. Of this mixture, 75 μL was subjected to the assay following the manufacturer’s protocol.
The research use only LA genotyping assay uses PGMY09/11 L1 consensus PCR and reverse line blot hybridization to detect any of 37 HPV types (6, 11, 16, 18, 26, 31, 33, 35, 39, 40, 42, 45, 51, 52, 53, 54, 55, 56, 58, 59, 61, 62, 64, 66, 67, 68, 69, 70, 71, 72, 73, 81, 82, 83, 84, 89 and IS39); results indicate whether each specific HPV type can be detected within a specimen. Laboratory methods for the use of this test in NHANES have been described previously.20
Additional positive and negative control specimens were processed each time an assay was performed. Women whose results for either LA or HC-2 tests were unavailable or inadequate were excluded from this analysis.
Statistical methods
We used data from NHANES to estimate the overall prevalence of oncogenic HPV types detected by a positive HC-2 test according to selected demographic and sexual behavior correlates. Weighted percentages and 95% confidence intervals (CIs) were calculated. We compared the estimated HPV prevalence of oncogenic vaccine types (HPV 16 and 18), vaccine types (HPV 6, 11, 16 and 18) and oncogenic types detectable by HC-2 (HPV 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66 and 68) based on LA results from all specimens, or LA on the subset of HC-2 positive specimens.
Prevalence estimates were weighted using 4-year weights constructed from the examination weights provided by NCHS to account for the unequal probabilities of selection and adjustment for non-response. Variance of the estimates was calculated using a Taylor series linearization that incorporated the complex survey design. We used F-statistics transformed from Wald χ2 statistics to test bivariate associations between HC-2 test results and selected characteristics. Any characteristic with a p-value <0.05 was considered statistically significant. No adjustments were made for multiple comparisons. Estimates with relative standard error ≥30% were considered unstable.
To compare the prevalence of oncogenic HPV detected by using LA on all specimens versus only on the HC-2-positive subset, we plotted the relative prevalence for 14 oncogenic HPV types, according to testing approach. The first testing approach was LA on all specimens. The second testing approach was LA only on specimens that were positive by HC-2, so that clinical test results were used as an initial triage for type-specific testing. To estimate the reduction in the true population prevalence of HPV that would be needed to detect a statistically significant difference by using each of these two testing approaches, with a power of 0.8 and a Type I error rate of 0.05, we assumed that all participants would be recruited using a convenience sampling strategy, and based our calculations on a sample size of 4,150 subjects. Statistical analyses were conducted using SAS (version 9.1; SAS Institute, Cary, NC, 2002) and SAS-callable SUDAAN (RTI, Research Triangle Park, NC).
Results
During 2003–2006, 5,178 females aged 14–59 years were interviewed for NHANES; 4,990 (96.4%) then received an examination in the mobile examination center and 4,233 (81.7%) also submitted a self-collected cervico-vaginal swab. Eighty-three swabs gave inadequate results in HPV DNA testing. Thus, we analyzed results from 4,150 women.
The HPV prevalence as detected by the HC-2 clinical test was 10.1% (CI: 8.8–11.6) for any of 14 oncogenic HPV types. Demographic and behavioral characteristics of women with a positive HC-2 test are reported in Table 1. Factors associated with HC-2 positivity, including younger age, black or Hispanic race and higher numbers of recent or lifetime sex partners, are consistent with those previously reported with LA.15,16 As reported elsewhere, the overall population prevalence of any of 37 HPV types detected by the LA research test was 42.5% (CI: 40.3–44.7).15
Table 1.
Demographic and behavioral characteristics of women aged 14–59 years with a positive HC-2 test for HPV—USA, 2003–2006
| Characteristic | Positive HC-2 test for HPV | |
|---|---|---|
| Sample size |
Prevalence % (95% CI) |
|
| Total | 4150 | 10.1 (8.8–11.6) |
| Age (years) | ||
| 14–29 | 2198 | 15.2 (13.3–17.3) |
| 30–59 | 1952 | 7.6 (6.2–9.4) |
| Race/ethnicity | ||
| Non-Hispanic black | 1134 | 14.2 (11.7–17.1) |
| Hispanic | 991 | 10.5 (7.8–14.1) |
| Other | 320 | 10.2 (7.1–14.5) |
| Non-Hispanic white | 1705 | 9.3 (7.7–11.1) |
| Number of sex partners in past year | ||
| 3+ | 246 | 23.1 (17.4–30.1) |
| 2 | 247 | 16.0 (12.3–20.5) |
| 1 | 1983 | 8.4 (6.9–10.2) |
| 0 | 434 | 8.5 (5.4–13.1) |
| Lifetime number of sex partners | ||
| ≥6 | 1141 | 12.7 (11.1–14.6) |
| 3–5 | 902 | 11.4 (9.0–14.2) |
| 2 | 385 | 9.5 (6.4–13.9) |
| 1 | 709 | 5.8 (4.1–8.2) |
| 0 | 664 | 3.7 (2.0–6.7) |
Population prevalences of specific HPV types by testing approach are presented in Table 2. Based on LA testing of specimens, the prevalence of HPV oncogenic vaccine types (HPV 16 and 18) was 6.2% (CI: 5.4–7.1), and for HPV vaccine types (HPV 6, 11, 16 and 18) it was 8.8% (CI: 7.8–10.0).16 The total prevalence of any oncogenic HC-2 HPV types (HPV 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66 and 68) was 23.7% (CI: 21.6–26.0). When LA typing was restricted to specimens with HC-2-positive results, the observed prevalence of HPV oncogenic vaccine types was 2.4% (CI: 1.9–3.0), and for HPV vaccine types it was 3.0% (CI: 2.5–3.6). The detected prevalence of any oncogenic HC-2 HPV types was 7.3% (CI: 6.4–8.2).
Table 2.
Population prevalence of HPV, as detected by LA conducted without or with triage by a positive HC-2 test among women aged 14–59 years—USA, 2003–2006
| HPV types | Population prevalence detected by LA conducted on all specimens (without triage) % (95% CI) |
Population prevalence detected by LA conducted on the subset of specimens positive by HC-2 (with triage) % (95% CI) |
|---|---|---|
| Oncogenic HPV vaccine types (HPV 16, 18) | 6.2 (5.4–7.1) | 2.4 (1.9–3.0) |
| HPV vaccine types (HPV 6, 11, 16, 18) | 8.8 (7.8–10.0) | 3.0 (2.5–3.6) |
| Oncogenic HPV types detected by HC-2 (HPV 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, 68) |
23.7 (21.6–26.0) | 7.3 (6.4–8.2) |
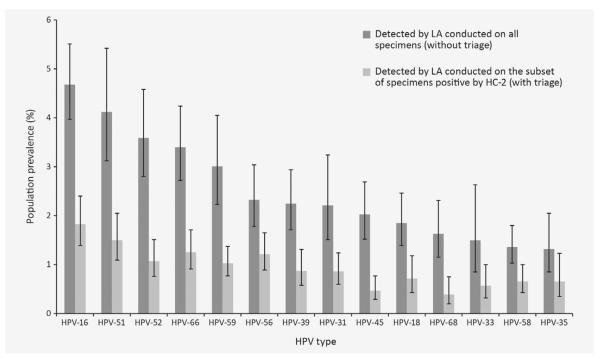
Relative type-specific HPV prevalences detected by LA based on all specimens on the HC-2-positive subset of specimens are shown in Figure 1. In both testing approaches, oncogenic HPV 16 was detected most commonly, followed by HPV 51. Although the percentage of HPV infections detected was lower with the testing approach involving HC-2, the relative type-specific prevalence (i.e., rank order of HPV types detected by LA) was similar for both testing approaches, whether or not HC-2 result was used initially.
Figure 1.
Population prevalence of 14 oncogenic HPV types, as detected by LA conducted without or with triage by a positive HC-2 test among women aged 14–59 years—USA, 2003–2006.
Discussion
Most studies investigating type-specific HPV prevalence have used PCR-based testing on all specimens. We investigated using type-specific PCR testing only on specimens initially positive by a clinical HPV test. Such a triage testing approach might be more compatible with cervical cancer screening programs and could reduce costs by greatly reducing the number of specimens requiring type-specific testing; however, it would detect fewer HPV infections. In certain low-resource or other settings, this tradeoff in sensitivity may be acceptable given that the relative HPV type-specific prevalences detected are comparable by both approaches.
Compared with typing all specimens, a testing approach using a clinical test as an initial triage would require typing fewer specimens, but would require a greater reduction in the true population prevalence of HPV or else a larger group of specimens would be needed to detect HPV vaccine impact after implementation. For example, we estimate that, in order to have an 80% chance of detecting a significant reduction by using LA testing in two samples of similar size, the true population prevalence of HPV 16/18 would need to decrease by at least 24% (from 6.2 to 4.7% or lower). However, to detect a significant reduction by using the triage testing approach, the true population prevalence of HPV 16/18 would need to decrease by at least 38% (from 2.4 to 1.5% or lower) or else it would be necessary to triage a much larger group of specimens. In this analysis, if LA testing had been restricted to specimens with an initially positive HC-2 test, nearly 90% of the specimens in this sample would not have been tested by LA, with a corresponding reduction in reagent costs.
In some populations where HPV studies might be conducted (e.g., younger women; visitors to sexually transmitted disease clinics), baseline HPV prevalence may be higher than in this representative sample of the U.S. population, so decreases in HPV prevalence due to HPV vaccination might be detectable with smaller sample sizes or a smaller decrease in prevalence. Compared to testing all samples by LA, though, the triage approach would still require a higher percentage reduction in HPV prevalence or larger sample sizes. Furthermore, cervical cancer screening may not be recommended routinely in these populations.
This report is also the first to use NHANES data to estimate that, nationally, one in 10 U.S. women between the ages of 14–59 years is infected with at least one of 14 oncogenic HPV types detectable by HC-2 on a self-collected cervico-vaginal swab. This prevalence is lower than that detected by LA for the same HPV types in the same study population, as expected based on the lower sensitivity of the HC-2 test compared to the LA test.7,21
Our analysis is subject to several limitations. First, U.S. NHANES data may not be generalizable to low-resource settings in other countries. Where available, local or regional data may be helpful, as HPV type distribution may vary by country and location. In addition, studies with different sampling approaches, data collection strategies, or other design differences, might produce different results. Second, biological specimens tested in NHANES are self-collected cervico-vaginal swabs, rather than cervical specimens described in the package insert instructions for clinical use of the HC-2 test, although other evaluations have shown no significant differences in performance of self-collected versus clinician-collected cervico-vaginal specimens with HPV assays including LA and HC-2.22,23 Third, HC-2 is the only clinical test that is part of NHANES testing; other clinical tests for HPV might result in different findings in our analysis. Of note, correlation with cervical cytopathology or HPV-associated disease outcomes could not be measured as NHANES is a cross-sectional survey that does not conduct Pap smear screening, collect cervical specimens, or follow participants over time. However, results from other studies conducted in primary care settings in the USA suggested that most women with detectable HPV infections did not have current cervical disease indicated by any abnormal cytology findings on a Pap screening test.24
These results indicate limitations in using clinical HPV tests to monitor population prevalence of HPV infections following HPV vaccine implementation. A triage testing approach might be more useful in settings where baseline HPV prevalence is high. Further investigation is warranted to inform and improve type-specific HPV monitoring strategies around the world.
What’s new?
The impact of human papillomavirus (HPV) immunization on cervical cancer incidence in populations of vaccinated women may not be apparent for decades, complicating evaluation of the early effectiveness of HPV vaccination programs. Evidence presented here indicates that it may be possible to do this by performing HPV-type analysis on cervical specimens, since one of the earliest measurable effects of HPV vaccine in a population should be a reduction in specific HPV types. The triage of cervical specimens for HPV typing based on results of a clinical HPV test such as the digene hybrid capture 2 (HC-2) could complement existing cervical cancer screening programs and reduce overall testing costs, and therefore may be appealing for use in low-resource settings.
Acknowledgments
EM, EU, LM and SH designed the study and participated in all aspects of the work. CL performed the statistical analyses. MS and SP performed the laboratory testing. All authors collaborated in writing the manuscript, had full access to the data, and take responsibility for the integrity of that data and the accuracy of the data analysis. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Grant sponsor: Centers for Disease Control and Prevention
Abbreviations
- CI
confidence interval
- FDA
Food and Drug Administration
- HPV
human papillomavirus
- HC-2
Hybrid Capture
- LA
Linear Array
- NCHS
National Center for Health Statistics
- NHANES
National Health and Nutrition Examination Survey
References
- 1.Agosti JM, Goldie SJ. Introducing HPV vaccine in developing countries—Key challenges and issues. N Engl J Med. 2007;356:1908–10. doi: 10.1056/NEJMp078053. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention Progress toward implementation of human papillomavirus vaccination—The Americas, 2006–2010. Morb Mortal Wkly Rep. 2011;60:1382–4. [PubMed] [Google Scholar]
- 3.Muñoz N, Kjaer SK, Sigurdsson K, et al. Impact of human papillomavirus (HPV)–6/11/16/18 vaccine on all HPV-associated genital diseases in young women. J Natl Cancer Inst. 2010;102:325–39. doi: 10.1093/jnci/djp534. [DOI] [PubMed] [Google Scholar]
- 4.World Health Organization Monitoring the coverage and impact of human papillomavirus vaccine—report of WHO meeting, November 2009. Wkly Epidemiol Rec. 2010;85:237–43. [PubMed] [Google Scholar]
- 5.Hubbard RA. Human papillomavirus testing methods. Arch Pathol Lab Med. 2003;127:940–5. doi: 10.5858/2003-127-940-HPTM. [DOI] [PubMed] [Google Scholar]
- 6.Hariri S, Unger ER, Dunne EF, Swan DC, Patel SS, Sternberg M, Markowitz LE. Prevalence of human papillomavirus (HPV) types detected by digene high-risk and low-risk HPV DNA tests among a nationally representative sample of females in the United States. International Society for Sexually Transmitted Disease Research, 17th Annual Meeting; Seattle, WA, USA. July 2007. [Google Scholar]
- 7.Ermel A, Qadadri B, Morishita A, Miyagawa I, Yamazaki G, Weaver B, Tu W, Tong Y, Randolph M, Cramer H, Brown D. Human papillomavirus detection and typing in thin prep cervical cytologic specimens comparing the digene hybrid capture II assay, the Roche linear array HPV genotyping assay, and the Kurabo GeneSquare microarray assay. J Virol Methods. 2010;169:154–61. doi: 10.1016/j.jviromet.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 8.Stevens MP, Garland SM, Rudland E, Tan J, Quinn MA, Tabrizi SN. Comparison of the digene hybrid capture 2 assay and Roche amplicor and linear array human papillomavirus (HPV) tests in detecting high-risk HPV genotypes in specimens from women with previous abnormal Pap smear results. J Clin Microbiol. 2007;45:2130–7. doi: 10.1128/JCM.02438-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gravitt PE, Schiffman M, Solomon D, Wheeler CM, Castle PE. A comparison of linear array and hybrid capture 2 for detection of carcinogenic human papillomavirus and cervical precancer in the ASCUS-LSIL triage study. Cancer Epidemiol Biomarkers Prev. 2008;17:1248–54. doi: 10.1158/1055-9965.EPI-07-2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wright TC, Stoler MH, Behrens CM, Apple R, Derion T, Wright TL. The ATHENA human papillomavirus study: design, methods, and baseline results. Am J Obstet Gynecol. 2012;206:46, e1–11. doi: 10.1016/j.ajog.2011.07.024. [DOI] [PubMed] [Google Scholar]
- 11.Moyer VA. Screening for cervical cancer: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med. 2012;156:880–91. doi: 10.7326/0003-4819-156-12-201206190-00424. [DOI] [PubMed] [Google Scholar]
- 12.Qiao YL, Sellors JW, Eder PS, et al. A new HPV-DNA test for cervical-cancer screening in developing regions: a cross-sectional study of clinical accuracy in rural China. Lancet Oncol. 2008;9:929–36. doi: 10.1016/S1470-2045(08)70210-9. [DOI] [PubMed] [Google Scholar]
- 13.Gage JC, Ajenifuja KO, Wentzensen N, et al. Effectiveness of a simple rapid human papillomavirus DNA test in rural Nigeria. Int J Cancer. 2012;131:2903–9. doi: 10.1002/ijc.27563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Markowitz LE, Sternberg M, Dunne EF, McQuillan G, Unger ER. Seroprevalence of human papillomavirus types 6, 11, 16, and 18 in the United States: National Health and Nutrition Examination Survey 2003–2004. J Infect Dis. 2009;200:1059–67. doi: 10.1086/604729. [DOI] [PubMed] [Google Scholar]
- 15.Hariri S, Unger ER, Sternberg M, Dunne EF, Swan D, Patel S, Markowitz LE. Prevalence of genital human papillomavirus among females in the United States, the National Health And Nutrition Examination Survey, 2003–2006. J Infect Dis. 2011;204:566–73. doi: 10.1093/infdis/jir341. [DOI] [PubMed] [Google Scholar]
- 16.Dunne EF, Sternberg M, Markowitz LE, McQuillan G, Swan D, Patel S, Unger ER. Human papillomavirus 6, 11, 16, and 18 prevalence among females in the United States—National Health And Nutrition Examination Survey, 2003–2006: opportunity to measure HPV vaccine impact? J Infect Dis. 2011;204:562–5. doi: 10.1093/infdis/jir342. [DOI] [PubMed] [Google Scholar]
- 17.Poljak M, Marin IJ, Seme K, Vince A. Hybrid capture II HPV test detects at least 15 human papillomavirus genotypes not included in its current high-risk probe cocktail. J Clin Virol. 2002;25(Suppl 3):S89–97. doi: 10.1016/s1386-6532(02)00187-7. [DOI] [PubMed] [Google Scholar]
- 18.Bouvard V, Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F, Benbrahim-Tallaa L, Guha N, Freeman C, Galichet L, Cogliano V. A review of human carcinogens—Part B: biological agents. Lancet Oncol. 2009;10:321–2. doi: 10.1016/s1470-2045(09)70096-8. [DOI] [PubMed] [Google Scholar]
- 19.Peyton CL, Schiffman M, Lorincz AT, et al. Comparison of PCR- and hybrid capture-based human papillomavirus detection systems using multiple cervical specimen collection strategies. J Clin Microbiol. 1998;36:3248–54. doi: 10.1128/jcm.36.11.3248-3254.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Onyekwuluje JM, Steinau M, Swan DC, Unger ER. A real-time PCR assay for HPV52 detection and viral load quantification. Clin Lab. 2012;58:61–6. [PubMed] [Google Scholar]
- 21.Plummer M, Vaccarella S, Franceschi S. Multiple human papillomavirus infections: the exception or the rule? J Infect Dis. 2011;203:891–3. doi: 10.1093/infdis/jiq146. [DOI] [PubMed] [Google Scholar]
- 22.Gage JC, Partridge EE, Rausa A, Gravitt PE, Wacholder S, Schiffman M, Scarinci I, Castle PE. Comparative performance of human papillomavirus DNA testing using novel sample collection methods. J Clin Microbiol. 2011;49:4185–9. doi: 10.1128/JCM.01254-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gravitt PE, Lacey JV, Jr, Brinton LA, et al. Evaluation of self-collected cervicovaginal cell samples for human papillomavirus testing by polymerase chain reaction. Cancer Epidemiol Biomarkers Prev. 2001;10:95–100. [PubMed] [Google Scholar]
- 24.Datta SD, Koutsky LA, Ratelle S, Unger ER, Shlay J, McClain T, Weaver B, Kerndt P, Zenilman J, Hagensee M, Suhr CJ, Weinstock H. Human papillomavirus infection and cervical cytology in women screened for cervical cancer in the United States, 2003–2005. Ann Intern Med. 2008;148:493–500. doi: 10.7326/0003-4819-148-7-200804010-00004. [DOI] [PubMed] [Google Scholar]