Abstract
The majority of structural efforts addressing RNA’s catalytic function have focused on natural ribozymes, which catalyze phosphodiester transfer reactions. By contrast, little is known about how RNA catalyzes other types of chemical reactions. We report here the crystal structures of a ribozyme that catalyzes enantioselective carbon-carbon bond formation by the Diels-Alder reaction in the unbound state and in complex with a reaction product. The RNA adopts a λ-shaped nested pseudoknot architecture whose preformed hydrophobic pocket is precisely complementary in shape to the reaction product. RNA folding and product binding are dictated by extensive stacking and hydrogen bonding, whereas stereoselection is governed by the shape of the catalytic pocket. Catalysis is apparently achieved by a combination of proximity, complementarity and electronic effects. We observe structural parallels in the independently evolved catalytic pocket architectures for ribozyme- and antibody-catalyzed Diels-Alder carbon-carbon bond-forming reactions.
The discovery of the catalytic activity of RNA1,2 and the hypothesis of a prebiotic ‘RNA world’3 have expanded the scope of enzymology to include other biopolymers than proteins. The currently known natural ribozymes catalyze only a narrow range of chemical reactions, namely the hydrolysis and transesterification of internucleotide bonds4,5, and probably peptide bond formation6. However, in vitro selection and evolution have demonstrated that ribozymes are capable of accelerating a much broader reaction spectrum7. This finding and recent discoveries of metabolite-controlled RNA switches and ribozymes8,9 suggest that RNA might have performed an even broader range of activities in the preprotein realm, and that in vitro–selected ribozymes could be analogs of the missing links in the transition from an RNA world to modern protein-dominated life10. Whereas high-resolution structures and biochemical investigations of several natural ribozymes provide a basic understanding of how RNA carries out phosphodiester chemistry5,11, little is known about how RNA catalyzes other reactions. To obtain a comprehensive picture of the catalytic abilities and limitations of ribozymes, it is thus important to expand structural and mechanistic investigations to in vitro–selected ribozymes12–14. Such structural information can be especially valuable in the determination of the minimal RNA folds required for catalysis and, therefore, could be helpful in the investigation of the origin and evolution of natural ribozymes15.
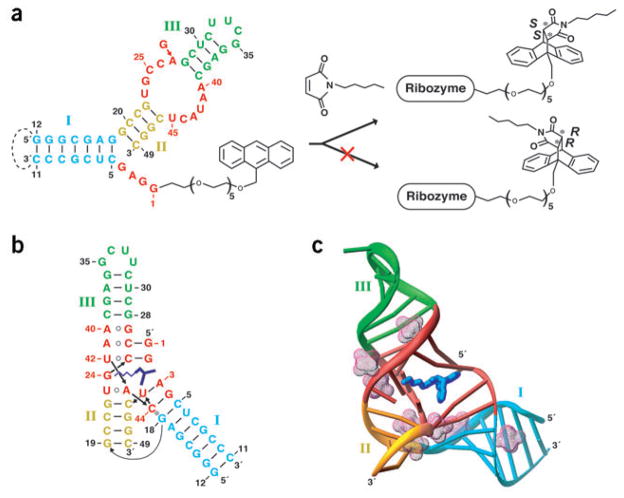
Two examples describe the in vitro selection of ribozymes that accelerate the formation of carbon-carbon bonds by the Diels-Alder reaction16,17, a [4 + 2] cycloaddition reaction between a tethered diene and a biotinylated dienophile. This reaction type, creating two carbon-carbon bonds and up to four new stereo centers in one step, has broad applications in organic chemistry for the stereospecific synthesis of complex molecules18. In one case, a minimal 49-nucleotide ribozyme was rationally deduced from the sequences of the active RNA molecules16, and this ribozyme was later found to accelerate the cycloaddition reaction free in solution (in trans) in a truly bimolecular fashion19. The minimum ribozyme adopted a secondary fold composed of three helical segments, a central element defined by an asymmetric bubble containing opposing invariant UGCCA and AAUACU fragments, and a 5′-terminal GGAG sequence (red, Fig. 1a). The minimal ribozyme could furthermore be engineered to give catalytically active bipartite (Fig. 1a) and tripartite systems16,20. Catalysis proceeds with multiple turnover and high enantioselectivity, making this a unique ribozyme system19.
Figure 1.
Secondary and tertiary folds of the Diels-Alder ribozyme. (a) Ribozyme secondary structure (left) with stems I–III in cyan, gold and green, respectively, and residues found invariant in the selection procedure in red. The same colors are used throughout this figure. The ribozyme consists of a 11-mer (G1–C11) HEG-linked with anthracene and a 38-mer (G12–C49). The dashed line indicates a loop present in the selected ribozyme and deleted in the bipartite system. The ribozyme-catalyzed Diels-Alder reaction between anthracene and N-pentyl maleimide results in formation of a single enantiomer of the complex (right). (b,c) Tertiary fold (b) and three-dimensional topology (c) in the crystal structure of the ribozyme–product complex. The RNA bases and bound product (blue) are cylinders in c. The backbone is depicted with a ribbon, whereas the hydrated Mg2+ cations are in a mesh representation.
A first insight into the principles of substrate recognition and the molecular determinants of stereoselectivity has been obtained by a study of the interactions of the ribozyme with a panel of 44 systematically varied substrate analogs21. The results suggest a major role for hydrophobic and van der Waals interactions, whereas hydrogen bonding and metal ion coordination seem less important for catalysis. Mutation analysis and probing identified interactions between nucleotides of the 5′ end and the internal asymmetric bubble, suggesting a pseudoknot topology20. In contrast to most known RNA aptamers22, the probing data indicate a preformed tertiary structure that shows no major changes upon substrate or product binding. These results lead to the proposal that the opposite sides of the asymmetric internal loop are most likely clamped by the four 5′-terminal nucleotides20.
To obtain insights into the mechanism by which the Diels-Alder ribozyme catalyzes formation of carbon-carbon bonds, we determined the crystal structures of the ribozyme both in the unbound state and bound to the reaction product. These structures define underlying principles by which an RNA molecule carries out chemical reactions with small organic substrates and highlight the mechanism used to achieve stereo-selectivity in catalysis.
RESULTS
Structure determination
Our structural efforts focused on the Diels-Alder ribozyme containing 38-mer and 11-mer RNA strands, with the shorter strand either unmodified or containing hexaethyleneglycol (HEG)-linked cyclo-addition product (Fig. 1a). The product in complex with fully active A27G-substituted RNA gave the best crystals, and therefore initial efforts focused on determination of this structure. Because suitable heavy atom derivatives could not be identified using either soaking or halogen substitution approaches, we decided to rely entirely on the selenium (Se)-modification approach23,24, which has been successfully applied for structure solution of a DNA duplex25. Owing to the rapid radiation-induced decay of the crystals, six 2′-Se-methyl uridine (USe) and cytidine (CSe) substitutions24 were required for determination of the structure at a resolution of 3.0 Å by SAD (see Methods and Table 1). Next we determined the isomorphous crystal structure of the ribozyme–product complex lacking Se modifications and carrying adenine at position 27. These nearly identical structures provide details of the catalytic pocket, including bound product, surrounding RNA and coordinated divalent cations.
Table 1.
Data collection and refinement statistics
| Product complex (Se and A27G) | Product complex | Free ribozyme | |
|---|---|---|---|
| Data collection | |||
| Space group | P21 | P21 | P21 |
| Cell dimensions | |||
| a, b, c (Å) | 77.87, 43.27, 79.93 | 78.28, 44.04, 80.00 | 77.69, 44.24, 79.63 |
| β (°) | 106.19 | 106.87 | 107.12 |
| Resolution (Å) | 30.0–3.0 (3.11–3.00) | 20.0–3.3 (3.42–3.30) | 20.0–3.5 (3.62–3.5) |
| Rmerge | 8.4 (52.0) | 9.2 (16.2) | 15.4 (72.9) |
| I / σI | 11.2 (2.6) | 14.0 (8.0) | 8.1 (1.8) |
| Completeness (%) | 99.3 (98.2) | 97.7 (88.9) | 97.7 (98.2) |
| Redundancy | 3.6 (3.6) | 4.9 (4.4) | 3.8 (3.8) |
| Refinement | |||
| Resolution (Å) | 20.0–3.0 | 20.0–3.3 | 20.0–3.5 |
| No. reflections | 10,481 | 7,909 | 6,563 |
| Rwork / Rfree | 0.209 / 0.234 | 0.228 / 0.262 | 0.286 / 0.324 |
| No. atoms | |||
| RNA | 2,106 | 2,092 | 2,078 |
| Cycloaddition product | 56 | 56 | – |
| Cations (Mg2+ and Cd2+) | 13 | 7 | 12 |
| Water | 83 | – | – |
| B-factorsa | |||
| RNA | 25.0 | 23.9 | – |
| Cycloaddition product | 26.0 | 23.8 | – |
| Cations (Mg2+ and Cd2+) | 51.3 | 10.6 | – |
| Water | 48.0 | – | – |
| B-Patterson / B-Wilsonb | 42.4 / 37.9 | 46.3 / 25.8 | 70.4 / 91.2 |
| R.m.s deviations | |||
| Bond lengths (Å) | 0.017 | 0.016 | 0.015 |
| Bond angles (°) | 1.539 | 1.480 | 1.612 |
| Estimated coordinate errorc | 0.34 | 0.38 | 0.73 |
| Real-space R-factord | 0.138 | 0.122 | 0.104 |
Values in parentheses are for the highest-resolution shell.
Owing to low resolution, individual B-factors were assigned to 40 Å2 for all atoms during refinement.
Overall B-factor, estimated from Patterson origin peak, and Wilson B-factors were calculated using SFCHECK49.
Estimated overall coordinate error based on maximum likelihood (ESU) was calculated with REFMAC50.
The mean real-space R-factor for composite simulated-annealing omit maps was calculated using CNS51.
Overall topology and structural features
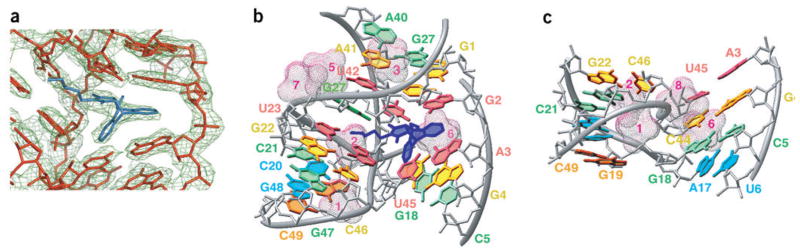
The molecule adopts a λ-shaped topology (Fig. 1b,c), with colinear stacking of stems II and III bridged by a zippered-up asymmetric bubble, whereas stem I is extended by two base pairs. Segments of the two arms of the tilted λ-shaped scaffold are positioned in close proximity, such that the sugar-phosphate backbone of one of the strands of stem I is positioned within the major groove of stem II. The electron density centered about the catalytic pocket, built by the G1–G4 tetranucleotide and asymmetric bubble segments, is shown in Figure 2a, and establishes that only the S,S stereo-isomer of the Diels-Alder product can be readily traced into the map. In the structure of the product complex, we have found eight hydrated Mg2+ cations, which participate in stabilization of the RNA architecture and in crystal packing (Fig. 1c).
Figure 2.
Details of the structure of the Diels-Alder ribozyme–product complex. (a) Final 2Fo – Fc electron density map (green), contoured at 1 σ, for the cycloaddition product and surrounding RNA. The product (blue) and RNA (red), superimposed with the density, are from the refined model. (b) RNA tertiary structure surrounding the ribozyme catalytic pocket containing bound product. The product is blue, adjacent A3-U45 and U23•A43 base pairs and U42•(G2-C25) base triple are light red, G24 is green, with additional layers moving farther out from the pocket in gold (A41•(C26-G2), G22-C46 and C44-G4), light green (A40•G27, C21-G47 and G18-C5), cyan (C20-G48) and orange (G19-C49). The hydrated Mg2+ cations are in magenta-colored mesh. (c) Tertiary structure in the region of stems I and II. The RNA is slightly tilted compared with b to highlight the relative alignment of the stems. Individual base pairs and hydrated Mg2+ cations are shown as in b.
The 5′-G1-G2-A3-G4 segment plays a critical role in shaping both the RNA scaffold and the catalytic pocket (Figs. 1b and 2). All four residues align through Watson-Crick base pairing with residues within the asymmetric bubble segment. The G1 and G2 residues form pairs with C26 and C25 respectively of one strand of the bubble, whereas the A3 and G4 residues form pairs with U45 and C44 respectively, of the opposite strand of the bubble. Notably, the 5′-GGAG segment bridges opposite strands of the internal bubble, thereby generating a complex nested pseudoknot topology.
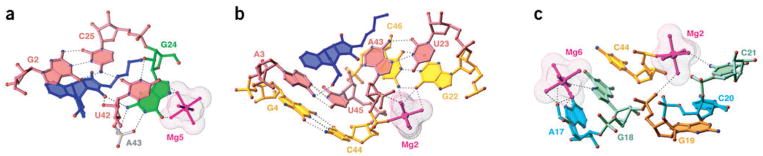
The asymmetric bubble (residues 23–27 and 40–45) is zippered up through noncanonical pair formation between residues across its opposing strands. These involve a G/A27•A40 pair with purines aligned through their Watson-Crick edges (Fig. 2b and Supplementary Fig. 1 online), and a reversed Hoogsteen U23•A43 pair thereby extending both stems III and II, respectively, by one step. The A41 residue aligns with the G1-C26 pair to form a three-base platform (Fig. 2b and Supplementary Fig. 1 online), with potential for the formation of a C-H…O hydrogen bond. U42 participates in a U42•(G2-C25) base triple (Fig. 3a), where the alignment of U42 is dictated by formation of base-base and base-sugar hydrogen bonds. The unpaired G24 residue is sandwiched between U23 and U42 (Fig. 3a and Supplementary Fig. 1 online), and forms three hydrogen bonds along its Watson-Crick edge with the phosphate oxygens of A43 and the carbonyl group of the maleimide ring of the bound product, while its 2′ OH is hydrogen-bonded with U42.
Figure 3.
Structural elements of the Diels-Alder ribozyme–product complex. The view is rotated 180° along the vertical axis relative to the schematic in Figure 1b. The color code is the same as in Figure 2, unless specified otherwise. Oxygen, nitrogen and phosphorus atoms are, respectively, red, blue and yellow balls. Hydrogen bonds are dotted lines. Coordination bonds to Mg atom are magenta sticks. (a) RNA segment aligned above the product. The A43 backbone is silver. (b) RNA segment aligned below the product. (c) Structural motif highlighting splaying out of G18 and G19.
All bases of the 5′-GGAG segment and all residues within the asymmetric internal bubble are involved in stacking interactions. The majority of the stacking is within individual strands but cross-strand stacking is found between U42 and G24 in the vicinity of the RNA catalytic pocket (Supplementary Fig. 1 online). Extensive stacking is observed among the adjacent G27•A40 pair, A41•(G1-C26) three-base platform and U42•(G2-C25) base triple (Fig. 2b), between adjacent Watson-Crick A3-U45 and Watson-Crick G4-C44 pairs (Fig. 3b), and between reversed Hoogsteen A43•U23 and Watson-Crick G22-C46 pairs (Fig. 3b), associated with the three stems that converge on the junction. Notably, the A43-C44-U45-C46 segment forms a sharp turn stabilized by two coordinated magnesium cations (Fig. 3b and Supplementary Fig. 1 online), with stacking of the first (A43) and last (C46) residues. The C44 residue, involved in turn formation, stacks on G18 (Fig. 3c). The G18 and G19 bases are splayed apart (Figs. 2c and 3c), thereby preventing stems I and II from adopting a colinear alignment, a feature recently observed in the structure of a group I intron26. A disruption in stacking is also observed at the G24-C25 step (Fig. 3a and Supplementary Fig. 1 online), as G24 intercalates into the extended U42-A43 step. Finally, portions of stems I and II pack against each other, such that the backbone at the C44-U45 step of stem I is directed toward the major groove edge of stem II, with interactions mediated through bridging hydrated magnesium ions Mg1 and Mg2 (Supplementary Fig. 1 online).
Divalent cation-binding sites
Among the eight Mg2+ cations found in the structure, six participate in the stabilization of the RNA architecture (Mg1–Mg6) and two (Mg7 and Mg8) mediate contacts between RNA molecules in the crystal lattice. Mg2 seems to make the most important contribution to the structural scaffold because it makes multiple direct and water-mediated contacts that bridge bases and sugar-phosphate backbones of six nucleotides situated beneath the product (Fig. 3b), and, together with Mg1, mediates the packing of helices I and II against each other (Supplementary Fig. 1 online). Mg3 and Mg5 stabilize nonpaired G24 and the three-base platform and triple above the bound product (Fig. 3a), whereas Mg4 and Mg6 are located in the groove of helix I. There are no Mg2+ cations positioned in the immediate vicinity of the binding pocket that could potentially participate in the catalytic process. The divalent cation–dependence of the imino proton NMR spectra of the ribozyme–product complex has established that a minimum of two Mg2+ cations are required for RNA structure folding (Supplementary Fig. 2 online). Although the direct evidence is missing, good candidates for such cations are Mg1 and Mg2 found in all Diels-Alder ribozyme structures reported in this article.
RNA-product interactions
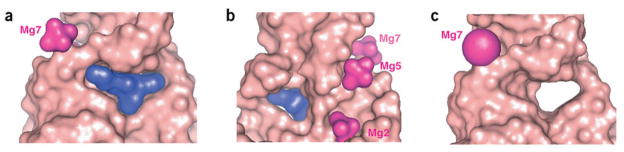
The catalytic pocket is wedge-shaped (~31° angle) and bracketed by the Watson-Crick A3-U45 pair, the noncanonical A43•U23 pair, and the U42•(G2-C25) triple (Figs. 2b and 3a,b). Base G24 is positioned opposite to the opened edge of the pocket, and shares the space with the side chain projecting from the five-membered maleimide ring (Figs. 2b and 3a). The cycloaddition product, which binds to the ribozyme with a dissociation constant in the 10 μM range21, is positioned within the catalytic pocket with one of its aromatic rings sandwiched between purines G2 and A3, and the other wedged between the base and sugar components of A43 and U45 (Figs. 2a and 3a,b). The HEG-linked bridgehead position is directed inward into the RNA scaffold, whereas the unmodified bridgehead position points outward toward solvent. The maleimide ring is stacked over C25, and oriented through a pair of hydrogen bonds made by one of its two carbonyl oxygens with the NH2 group of G24 and the 2′ OH of U42 (Fig. 3a). The maleimide side chain runs inside a hydrophobic canyon created by bases C25 (top), A43 (bottom), and G24 (back) (Fig. 3a,b). The catalytic pocket, which is primarily formed by base edges with minimal contribution of the sugar-phosphate backbone, is accessible to the product from the front but not from the back (Fig. 4a,b). The 9′ O atom (the one next to the bridgehead carbon) forms hydrogen bonds to the NH2 of A3 (Fig. 3b). The hydrophobic pocket has room along the long edges of the cycloaddition product, but there is a snug fit along the short edges. The catalytic pocket contains not only a cavity to accommodate the cycloaddition product but also a direction-specific surface canyon to hold the maleimide side chain, thus allowing binding of only one product enantiomer.
Figure 4.
Surface representation of the catalytic pocket. (a,b) Front (a) and back (b) views of the catalytic pocket of the ribozyme–product complex. (c) Front view of the free ribozyme. Mg2+ ions are shown as hydrated cations in a and b for the 3.0-Å structure, and as balls in c for the 3.5-Å structure.
Comparison of ribozyme structures
We have also determined the structure of the free Diels-Alder ribozyme at 3.5 Å, which is virtually identical to the structure of the product complex, indicative of a preformed architecture of the catalytic pocket (Figs. 4c and 5). This conclusion is strongly supported by the imino proton NMR spectral patterns of the free ribozyme, its anthracene conjugate and product complex in Mg2+-containing solution, all of which exhibit resonance patterns associated with pairing of the G1-G2-A3-G4 segment (Supplementary Fig. 2 online). This pairing probably generates the key interactions defining the geometry of the catalytic pocket. The imino proton NMR spectra of the samples with 38-mer plus 11-mer and the 38-mer plus 11-mer HEG-linked anthracene are very similar, whereas the sample of 38-mer plus 11-mer HEG-linked product shows a better-resolved spectrum with a total of 20 imino protons. The unusually well-resolved spectrum for the product complex probably reflects both its compact scaffold and the slow exchange of imino protons lining the pocket, as a consequence of the positioning of the Diels-Alder reaction product within the catalytic cavity.
Figure 5.
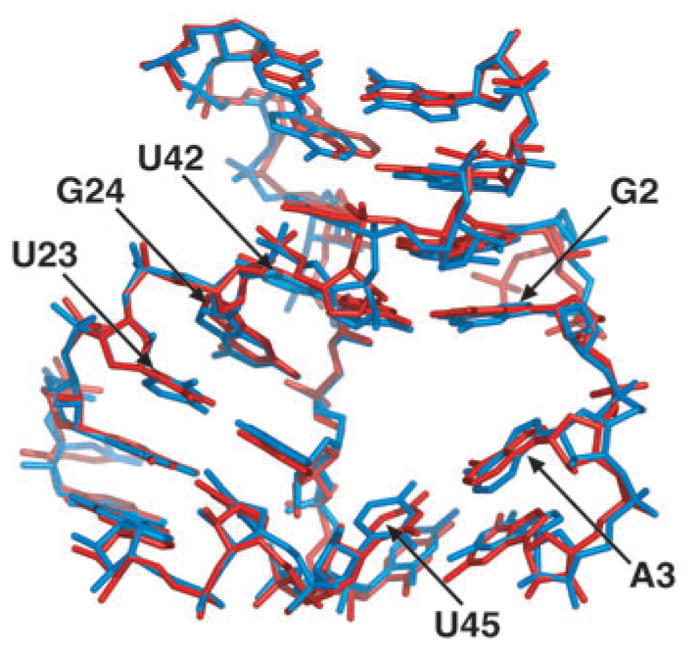
Superposition of the Diels-Alder ribozyme structures around the catalytic pocket. The RNA structures in stick representation are from the ribozyme–product complex (red) and the free ribozyme (blue).
DISCUSSION
Architecture of the Diels-Alder ribozyme
This is the first report on the three-dimensional architecture of a ribozyme catalyzing a synthetic reaction between small organic molecules. The catalytic pocket is centered about a λ-shaped three-helical junction that is distinct from the reported topologies of natural ribozymes. In the hammerhead ribozyme27, three helices converge toward a conserved sequence junction in a Y-shaped folding topology including a four-nucleotide turn element. The Varkud satellite ribozyme builds an active site using two three-way junctions28. In the Diels-Alder ribozyme the three stems abut directly against the catalytic pocket, and are in part involved in the formation of an unusual nested pseudoknot. A double-pseudoknot architecture has been reported previously for the hepatitis delta helper virus (HDV) ribozyme29, and both systems share common structural features, despite differences in pseudoknot topologies (Supplementary Fig. 3 online) and catalytic activities. Both ribozymes have preorganized active sites located in crevices, and both show a marked intertwining of secondary structure elements, leading to a densely packed, stable three-dimensional architecture. It is a notable coincidence that the HDV and Diels-Alder ribozymes, two of the fastest known ribozymes, both use nested pseudoknot architectures. The same pseudoknot topology is common to both the Diels-Alderase ribozyme and the putative fold in the regulatory region of the Escherichia coli α operon (Supplementary Fig. 3 online), which is specifically recognized and bound by ribosomal protein S4, leading to translational repression of the operon30. Thus, the Diels-Alder pseudoknot topology may have relevance in a different biological context.
Correlation with biochemical data
The crystal structure is in excellent agreement with the observed structure-activity relations reported for ribozyme mutants20. The bridging role of the G1-G2-A3-G4 segment correlates with the observation that essentially no single substitutions are tolerated within this sequence20, whereas pairwise-complementary double substitutions lead to fully active mutants, at least for the A3-U45 and the G4-C44 base pairs. For G1 and G2, no compensatory double mutants were found, consistent with their involvement in the A41•(G1-C26) three-base platform and the U42•(G2-C25) base triple. The unpaired G24 residue is anchored in place by three specific hydrogen bonds involving its Watson-Crick edge; therefore, it is not surprising that its replacement by any other nucleotide results in loss of >90% of the catalytic activity20.
Chemical probing has established that the N7 position of A43 is protected from modification, whereas the N1 position of this nucleotide becomes more exposed upon tertiary structure formation20. This is consistent with the observation of the important reversed Hoogsteen U23•A43 pair in the crystal structure, resulting in pairing of the N7 position along the Hoogsteen edge and exposure to solvent of the N1 position along the Watson-Crick edge. Related modification studies also established that the N3 positions of C25, C26 and C44 become increasingly protected from methylation upon tertiary structure formation, in agreement with formation of Watson-Crick G1-C26, G2-C25 and G4-C44 pairs.
Preformed catalytic pocket
The Diels-Alderase ribozyme forms a wedge-shaped catalytic pocket lined predominantly by bases and base pairs that are involved in extensive stacking interactions with neighboring residues. There are no backbone phosphates in the vicinity of the pocket, nor were divalent cations identified that could participate in the catalytic process. Sugar rings participate in defining the dimensions of the pocket, but not in hydrogen bonding with the product. The formation of a well-defined hydrophobic pocket, and its use to bind the reactants, is generally thought to be very difficult for RNA to achieve31.
The catalytic pocket architectures for the ribozyme in the free state and in the product complex are very similar. Independent chemical probing experiments support this fact, indicating that overall RNA folding is not changed upon addition of the anthracene substrate or the cycloaddition product20. The observation of a preformed catalytic pocket for the Diels-Alder ribozyme contrasts with known examples of adaptive recognition found in ligand–RNA complexes22,32,33.
The catalytic pocket must be able to accommodate both the diene and maleimide components of the reactants and the product. Previous studies have shown that substitutions along the longer edges of the diene ring are tolerated whereas substitutions along the shorter edges result in loss of catalytic activity21, consistent with the dimensions of the catalytic pocket in the crystal structure. The maleimide component is positioned within the walls of a canyon that seems to be wide enough to accommodate an unbranched alkyl or an aryl chain. Substitutions at the maleimide double bond positions result in total loss in activity21, as they would clash against the pocket-forming face of the tertiary G2-C25 base pair. Side chain reduction from pentyl down to ethyl results in decreased acceptance of the substrates, probably owing to reduced hydrophobic interactions with the walls of the canyon. By contrast, the open-ended nature of the canyon predicts that it should readily accommodate longer side chains, in agreement with experimental findings16,21.
A proposed reaction mechanism
The Diels-Alder ribozyme structures, coupled with extensive chemical and biochemical experiments16,19–21, suggest that the ribozyme should bind the substrates in trans in a precisely defined orientation within the catalytic pocket, thereby facilitating the reaction by reducing translational and rotational degrees of freedom. The anthracene substrate could be bound by stacking interactions between G2 and the A3-U45 pair. The reaction requires that the maleimide substrate be stacked on top of the anthracene, and parallel to it, at a distance of ∼3.5 Å. In this position, maleimide can form hydrogen bonds from its carbonyl oxygen to the exocyclic amino group of G24 and to the 2′ OH of U42. The N-alkyl side chain can be placed within the hydrophobic canyon (Supplementary Fig. 4 online), which would allow the approach of the maleimide to the bound anthracene from only one direction, thereby providing the stereoselectivity of the reaction.
In the transition state of the reaction, computed using MOPAC (http://www.cachesoftware.com/mopac/index.shtml) and published geometries of similar systems34,35, two new single bonds are partly formed (bond lengths ∼2.2 Å), and the anthracene ring system is bent out of planarity to yield an angle of ∼154° (Supplementary Fig. 4 online), thereby further reducing the minimal unoccupied space underneath the bridgehead carbons seen in the structure of the ribozyme–product complex (Fig. 4a). Notably, the transition state has near-perfect shape complementarity with the catalytic pocket, a feature of prime importance in antibody catalysis of the Diels-Alder reactions36,37. In addition to steric factors, both stacking and hydrogen bonding may contribute the energetics of the reaction. Stacking of the anthracene with nucleotides A3 and U45 could accelerate the reaction by increasing the diene’s electron density, whereas hydrogen bonding of the maleimide would make it more electron-deficient, thereby increasing catalytic reactivity38. Thus, it seems that ribozyme-based catalysis of Diels-Alder reactions reflects a combination of proximity, shape complementarity and energetic contributions to the catalytic process.
Comparison with protein-catalyzed reactions
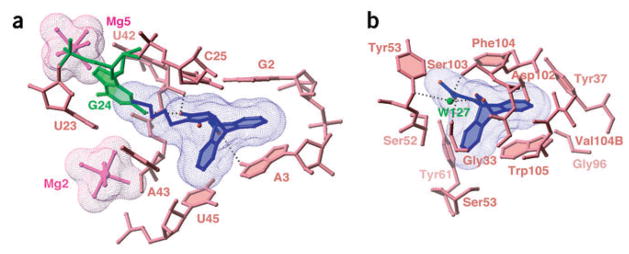
The structural and mechanistic principles outlined above can be compared with the strategies used by antibodies catalyzing Diels-Alder and retro-Diels-Alder reactions. Several such antibodies have been crystallized in complex with either products or transition state analogs36,39–41. Common features observed in these structures are the formation of hydrophobic cores lined predominantly by the side chains of tyrosine, phenylalanine, valine and tryptophan residues, and the stacking of one reactant over an aromatic residue, preferably tryptophan. Notably, the retro-Diels-Alderase catalytic antibody 10F11, the only antibody using anthracene as the diene41, forms a hydrophobic catalytic pocket with an overall shape similar to the Diels-Alder ribozyme pocket (Fig. 6). The anthracene ring is stacked over a tryptophan, similar to the position of A3 in the ribozyme, with a few direct and water-mediated hydrogen bonds observed between heteroatoms on the bicyclic adduct and the amide backbone41.
Figure 6.
Comparison of the RNA and protein catalytic sites for the Diels-Alder reaction. The surface of the bound ligand is in mesh representation. (a) Catalytic pocket from the Diels-Alder ribozyme–product complex. (b) Catalytic pocket of the antibody catalyzing the retro-Diels-Alder reaction41 (PDB entry 1LO2). Anchoring of the ligand is achieved through direct and water-mediated (W127) hydrogen bonds.
The natural Diels-Alder enzyme also has a mainly hydrophobic catalytic pocket and interacts with the substrates by hydrogen bonding. Major differences, however, lie in the use of a bound Mg2+ cation, which directly coordinates to the carbonyl oxygens of the dienophile and participates in the catalytic mechanism, and in the generally lower importance of stacking in enzyme-substrate interactions42,43.
Another Diels-Alder ribozyme requires both a covalent modification of the RNA and the presence of Cu2+ ions for catalytic activity, and apparently uses a different catalytic strategy17,44. The structural prerequisites, however, are yet unknown.
The obvious similarities between antibody and RNA catalytic systems lead us to conclude that similar structural principles and catalytic mechanisms underlie macromolecule-mediated catalysis of Diels-Alder reactions. Notably, although RNA is equipped with a much less varied arsenal of functional groups than proteins, it has independently evolved matching criteria for generating catalytic pockets capable of facilitating carbon-carbon bond-forming reactions with comparable catalytic efficiency and enantioselectivity.
METHODS
RNA preparation
RNA (38-mer) was ordered from Dharmacon and CSS. Anthracene-HEG-linked 11-mer RNA was synthesized as described16. Synthesis of the stereochemically correct 11-mer with the HEG-linked Diels-Alder product was achieved by reaction between N-pentyl maleimide and anthracene-linked 11-mer annealed with 38-mer RNA16, and subsequent HPLC purification. RNAs containing Se modifications were chemically synthesized based on the 2′-O-TOM methodology, implementing USe and CSe phosphoramidites according to published procedures24. The 5′-anthracene-HEG-linked 11-mer with USe at position 6 and CSe at position 10 was converted into the product as described above. The 38-mer was prepared with CSe at positions 15 and 20 and USe at positions 30 and 33. Details of preparation and purification of the RNAs are summarized in Supplementary Methods online.
Complex formation and crystallization
For annealing, 11-mer and 38-mer RNAs (200 μM) were heated in 5 mM Tris-HCl, pH 8.0, 30 mM NaCl, 5 mM MgCl2, at 95 °C for 2 min and cooled on ice. The RNA sample was combined with the same volume of the reservoir solution, and crystals were grown by hanging-drop vapor diffusion. Reservoir solutions were: (i) Se-modified ribozyme–product complex, 30% (v/v) PEG400, 10% (v/v) 2-methyl-2,4-pentadiol, 70 mM Na-HEPES, pH 7.5, 200 mM MgCl2, 200 mM NaCl, 50 mM RbCl; (ii) unmodified ribozyme–product complex, 30% (v/v) PEG400, 100 mM Na-HEPES, pH 7.5, 200 MgCl2, 0.5% (v/v) ethylacetate; (iii) free ribozyme, 30% (v/v) PEG400, 50 mM Na-cacodylate, pH 6.5, 80 mM Mg-acetate, 3% (v/v) 1,6-hexandiol, 50 mM NaCl, 15 mM CdCl2. Crystals of the RNA–product complex grew to a maximal size of 0.4 × 0.1 × 0.02 mm in ∼20 d at 4°C. Free ribozyme crystals were grown at 20 °C.
Structure determination
X-ray data on the modified Diels-Alder ribozyme–product complex (containing six Se sites and A27G substitution) were collected at beamline X25 at the Brookhaven National Synchrotron Light Source (NSLS) from a single crystal cooled at 100 K. Data were processed with HKL2000 (HKL Research)45. Owing to substantial radiation damage inflicted on the crystal, only the peak-wavelength data were used for phasing and all subsequent calculations.
Selenium sites were identified with SHELXD46 run against all Bijvoet differences up to a resolution of 3.5 Å. The SAD phasing was carried out with SHARP47, including the solvent flattening procedure with SOLOMON48,49, assuming an optimized solvent content of 63% (Supplementary Fig. 5 online). Anomalous phasing power was 1.08, and figure of merit after solvent flattening was 0.87. Data collection and refinement statistics are listed in Table 1. The initial RNA model was built using the SAD electron density map and TURBO-FRODO (http://afmb.cnrs-mrs.fr/rubrique113.html), and then refined with REFMAC50 for the resolution range 20.0–3.0 Å. The noncrystallographic symmetry restraints were applied between two molecules in the asymmetric unit. The Diels-Alder product and hydrated Mg2+ cations were added to the model based on analysis of the 2Fo – Fc and Fo – Fc electron density maps (Supplementary Fig. 5 online). The asymmetric unit contains two ribozymes, each bound to its Diels-Alder product, and 13 hydrated Mg2+ cations. The ribozymes are composed of 38-mer and 11-mer RNA molecules. The HEG linker is disordered.
The isomorphous crystal structures of the unmodified ribozyme–product complex and the free ribozyme were refined at a resolution of 3.3 and 3.5 Å, respectively, using the data collected at beamlines 14-BM of Argonne Advanced Photon Source and X25 of NSLS. Owing to limited resolution, no B-factor refinement was carried out for the free ribozyme structure.
NMR experiments
Imino proton NMR spectra were recorded using Varian and Bruker NMR spectrometers at 25 °C, using jump-and-return water suppression for detection (Supplementary Fig. 2 online).
Graphics
The figures were prepared with Ribbons (http://sgce.cbse.uab.edu/ribbons/) and PyMOL (DeLano Scientific; http://pymol.sourceforge.net/).
Coordinates
Coordinates for the Se-modified A27G-substituted Diels-Alder ribozyme–product complex at a resolution of 3.0 Å have been deposited in the Protein Data Bank (accession code 1YLS). The structures of the free ribozyme (3.5 Å) and unmodified ribozyme–product complex (3.3 Å) have also been deposited (accession codes 1YKQ and 1YKV, respectively).
Supplementary Material
Supplementary Figure 1 Pairing alignments, turns and extensions within the structure of the ribozyme-product complex. (a) The alignment of the G27•A40 non-canonical pair stacked over the A41•(G1-C26) three-base platform. (b) The A41•(G1-C26) platform stacked over the U42•(G2-C25) triple. (c) Intercalation of G24 (in green) between the extended U42-A43 step, together with hydrogen bond alignments involving the Watson-Crick edge and sugar 2′-OH of G24. (d) The A43-C44-U45-C46 turn, stabilized by Mg1 and Mg2, with stacking of A43 and C46. (e) The U23-G24-C25-C26 segment, with splaying out of adjacent G24 and C25 bases. (f) Surface representation of the packing of the C44-U45 backbone segment of stem I (blue) positioned against the major groove of stem II (gold), mediated by bridging Mg1 and Mg2 cations.
Supplementary Figure 2 Folding of the Diels-Alder ribozyme monitored by imino proton NMR spectra (9 to 15 ppm). (a) Mg2+-dependence of the spectra of the 38-mer plus 11-mer-HEG-linked-product. Titration data are summarized in the insert, which shows that the majority of the RNA is folded at RNA:Mg2+ ratio of 1:2. Peaks used for quantification of Mg-dependent RNA folding are indicated by stars. Note that many more imino protons of the Diels-Alder product are observed on addition of Mg2+ to a sample lacking divalent cations. Also note that the exchange between Mg-free and Mg-bound states is slow, resulting in doubling of spectra. NMR spectra are recorded in 30 mM NaCl, 5 mM Tris buffer at pH 7.2. The RNA concentration was 0.05 mM. (b) NMR spectra of the ribozyme components. The 7-mer lacks the G1-G2-A3-G4 segment. The 11-mer is either free or HEG-linked with anthracene or product. The pairing of G1-G2-A3-G4 segment is reflected by appearance of additional imino protons. NMR spectra are recorded in 5 mM MgCl2, 5 mM NaCl, 5 mM Tris buffer, pH 7.2. All NMR spectra were recorded at 25 °C, using jump-and-return water suppression for detection (Plateau & Gueron, (1982), J. Am. Chem. Soc. 104, 7310–7311; Phan et al., (2001), Methods Enzymology, 338, 341–371).
Supplementary Figure 3 Schematic drawings of RNA pseudoknot topologies. (a) The Diels-Alder ribozyme, which adopts a 1-2-3-1′-2′-3′ topology. For comparison, the same coloring scheme as in Figure 1 was used. Note that the blue and green connectors are partly double-stranded, but not involved in pseudoknot interactions. (b) The HDV ribozyme, which adopts a 1-2-3-4-3′-1′-4′-2′ topology29. (c) The regulatory region of E. coli α operon, which adopts a 1-2-3-1′-2′-3′ topology30.
Supplementary Figure 4 Proposed model for the catalytic mechanism of the Diels-Alder ribozyme. Enzyme-substrate (ES) complex and transition state models have been obtained by manually docking the molecules into the pocket. The transition state was computed using MOPAC. The product-RNA complex is derived from the crystallographic structure. The surfaces of the A3-U45 base pair, amino group of G24 and 2′-OH of U42 are orange, green and yellow, respectively. Angles showing the buckling of the anthracene ring system and key distances during formation of the carbon-carbon bonds are indicated.
Supplementary Figure 5 Electron density maps of the catalytic pocket of the Diels-Alder ribozyme. (a) Solvent-flattened SAD electron density map (green) for the cycloaddition product (blue) and surrounding RNA (red), contoured at 1σ level. (b) Fo-Fc map, contoured at 2σ (green) and 1σ (red) levels, calculated for the product region after the RNA refinement and prior to the addition of the product to the model. The product and RNA, superimposed with the density, are from the refined model.
Acknowledgments
We gratefully acknowledge support by the US National Institutes of Health, the DeWitt Wallace Foundation and the Abby Rockefeller Mauze Trust (D.J.P.), the Bundesministerium für Bildung und Forschung (BioFuture program), the Deutsche Forschungsgemeinschaft, HFSP and the Fonds der Chemischen Industrie (A.J.), and the Austrian Science Fund FWF (R.M.). We thank V. Kuryavyi for extensive discussions on graphic programs, A. Teplov for help with data collection, M. Becker and the staff of the X25 and X12c beamlines at National Synchrotron Light Source for assistance with data collection, and the personnel of beamlines 14-BM and 19-BM at the Advanced Photon Source for data collection support.
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
References
- 1.Kruger K, et al. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence in Tetrahymena. Cell. 1982;31:145–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 2.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 3.Gilbert W. The RNA world. Nature. 1986;319:618–620. [Google Scholar]
- 4.Cech TR. Ribozymes, the first 20 years. Biochem Soc Trans. 2002;30:1162–1166. doi: 10.1042/bst0301162. [DOI] [PubMed] [Google Scholar]
- 5.Doudna JA, Cech TR. The chemical repertoire of natural ribozymes. Nature. 2002;418:222–228. doi: 10.1038/418222a. [DOI] [PubMed] [Google Scholar]
- 6.Steitz TA, Moore PB. RNA, the first macromolecular catalyst: the ribosome is a ribozyme. Trends Biochem Sci. 2003;28:411–418. doi: 10.1016/S0968-0004(03)00169-5. [DOI] [PubMed] [Google Scholar]
- 7.Wilson DS, Szostak JW. In vitro selection of functional nucleic acids. Annu Rev Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 8.Mandal M, Breaker RR. Gene regulation by riboswitches. Nature Rev Mol Cell Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- 9.Nudler E, Mironov AS. The riboswitch control of bacterial metabolism. Trends Biochem Sci. 2004;29:11–17. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 10.Murray JM, Doudna JA. Creative catalysis: pieces of the RNA world jigsaw. Trends Biochem Sci. 2001;26:699–701. doi: 10.1016/s0968-0004(01)02019-9. [DOI] [PubMed] [Google Scholar]
- 11.Lilley DM. Analysis of global conformational transitions in ribozymes. Methods Mol Biol. 2004;252:77–108. doi: 10.1385/1-59259-746-7:077. [DOI] [PubMed] [Google Scholar]
- 12.Chapman KB, Szostak JW. In vitro selection of catalytic RNAs. Curr Opin Struct Biol. 1994;4:618–622. doi: 10.1016/s0959-440x(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 13.Famulok M, Mayer G, Blind M. Nucleic acid aptamers—from selection in vitro to applications in vivo. Acc Chem Res. 2000;33:591–599. doi: 10.1021/ar960167q. [DOI] [PubMed] [Google Scholar]
- 14.Jäschke A. Artificial ribozymes and deoxyribozymes. Curr Opin Struct Biol. 2001;11:321–326. doi: 10.1016/s0959-440x(00)00208-6. [DOI] [PubMed] [Google Scholar]
- 15.Joyce GF, Orgel LE. Prospects for understanding the origin of the RNA world. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. 2. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1998. pp. 49–77. [Google Scholar]
- 16.Seelig B, Jäschke A. A small catalytic RNA motif with Diels-Alderase activity. Chem Biol. 1999;6:167–176. doi: 10.1016/S1074-5521(99)89008-5. [DOI] [PubMed] [Google Scholar]
- 17.Tarasow TM, Tarasow SL, Eaton BE. RNA-catalyzed carbon-carbon bond formation. Nature. 1997;389:54–57. doi: 10.1038/37950. [DOI] [PubMed] [Google Scholar]
- 18.Nicolaou KC, Snyder SA, Montagnon T, Vassilikogiannakis GE. The Diels-Alder reaction in total synthesis. Angew Chem Int Ed. 2002;41:1668–1698. doi: 10.1002/1521-3773(20020517)41:10<1668::aid-anie1668>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 19.Seelig B, Keiper S, Stuhlmann F, Jäschke A. Enantioselective ribozyme catalysis of a bimolecular cycloaddition reaction. Angew Chem Int Ed. 2000;39:4576–4579. [PubMed] [Google Scholar]
- 20.Keiper S, Bebenroth D, Seelig B, Westhof E, Jäschke A. An architecture of a Diels-Alderase ribozyme with a preformed catalytic pocket. Chem Biol. 2004;11:1217–1227. doi: 10.1016/j.chembiol.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 21.Stuhlmann F, Jäschke A. Characterization of an RNA active site: interactions between a Diels-Alderase ribozyme and its substrates and products. J Am Chem Soc. 2002;124:328–344. doi: 10.1021/ja0167405. [DOI] [PubMed] [Google Scholar]
- 22.Hermann T, Patel DJ. Adaptive recognition by nucleic acid aptamers. Science. 2000;287:820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- 23.Du Q, et al. Internal derivatization of oligonucleotides with selenium for X-ray crystallography with MAD. J Am Chem Soc. 2002;124:2425. doi: 10.1021/ja0171097. [DOI] [PubMed] [Google Scholar]
- 24.Höbartner C, Micura R. Chemical synthesis of selenium-modified oligoribonucleotides and their enzymatic ligation leading to an U6 snRNA stem-loop segment. J Am Chem Soc. 2004;126:1141–1149. doi: 10.1021/ja038481k. [DOI] [PubMed] [Google Scholar]
- 25.Teplova M, et al. Covalent incorporation of selenium into oligonucleotides for X-ray crystal structure determination via MAD: proof of principle. Biochimie. 2002;84:849–858. doi: 10.1016/s0300-9084(02)01440-2. [DOI] [PubMed] [Google Scholar]
- 26.Adams PL, Stahley MR, Kosek AB, Wang J, Strobel SA. Crystal structure of a self-splicing group I intron with both exons. Nature. 2004;430:45–50. doi: 10.1038/nature02642. [DOI] [PubMed] [Google Scholar]
- 27.Pley H, Flaherty KM, McKay DB. Three-dimensional structure of a hammerhead ribozyme. Nature. 1994;372:68–74. doi: 10.1038/372068a0. [DOI] [PubMed] [Google Scholar]
- 28.Lilley DM. The Varkud satellite ribozyme. RNA. 2004;10:151–158. doi: 10.1261/rna.5217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferre-D’Amare AR, Zhou K, Doudna JA. Crystal structure of a hepatitis delta virus ribozyme. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- 30.Schlax PJ, Xavier KA, Gluick TC, Draper DE. Translational repression of the Escherichia coli α operon mRNA: importance of an mRNA conformational switch and a ternary entrapment complex. J Biol Chem. 2001;276:38494–38501. doi: 10.1074/jbc.M106934200. [DOI] [PubMed] [Google Scholar]
- 31.Majerfeld I, Yarus M. An RNA pocket for an aliphatic hydrophobe. Nat Struct Biol. 1994;1:287–292. doi: 10.1038/nsb0594-287. [DOI] [PubMed] [Google Scholar]
- 32.Williamson JR. Induced fit in RNA-protein recognition. Nat Struct Biol. 2000;7:834–837. doi: 10.1038/79575. [DOI] [PubMed] [Google Scholar]
- 33.Leulliot N, Varani G. Current topics in RNA-protein recognition: control of specificity and biological function through induced fit and conformational capture. Biochemistry. 2001;40:7947–7956. doi: 10.1021/bi010680y. [DOI] [PubMed] [Google Scholar]
- 34.Manoharan M, De Proft F, Geerlings P. A computational study of aromaticity-controlled Diels-Alder reactions. J Chem Soc Perkin Trans. 2000;2:1767–1773. [Google Scholar]
- 35.Wise KE, Wheeler RA. Donor-acceptor-assisted Diels-Alder reaction of anthracene and tetracyanoethylene. J Phys Chem. 1999;103:8279–8287. [Google Scholar]
- 36.Xu J, et al. Evolution of shape complementarity and catalytic efficiency from a primordial antibody template. Science. 1999;286:23455–2348. doi: 10.1126/science.286.5448.2345. [DOI] [PubMed] [Google Scholar]
- 37.Chen J, Deng Q, Wang R, Houk K, Hilvert D. Shape complementarity, binding-site dynamics, and transition state stabilization: a theoretical study of Diels-Alder catalysis by antibody 1E9. Chembiochem. 2000;1:255–261. doi: 10.1002/1439-7633(20001117)1:4<255::AID-CBIC255>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 38.Fleming I. Frontier Orbitals in Organic Chemical Reactions. Wiley; New York: 1976. [Google Scholar]
- 39.Heine A, et al. An antibody exo Diels-Alderase inhibitor complex at 1.95 Å resolution. Science. 1998;279:1934–1940. doi: 10.1126/science.279.5358.1934. [DOI] [PubMed] [Google Scholar]
- 40.Romesberg FE, Spiller B, Schultz PG, Stevens RC. Immunological origins of binding and catalysis in a Diels-Alderase antibody. Science. 1998;279:1929–1933. doi: 10.1126/science.279.5358.1929. [DOI] [PubMed] [Google Scholar]
- 41.Hugot M, et al. A structural basis for the activity of retro-Diels-Alder catalytic antibodies: Evidence for a catalytic aromatic residue. Proc Natl Acad Sci USA. 2002;99:9674–9678. doi: 10.1073/pnas.142286599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ose T, et al. Insights into a natural Diels-Alder reaction from the structure of macro-phomate synthase. Nature. 2003;422:185–189. doi: 10.1038/nature01454. [DOI] [PubMed] [Google Scholar]
- 43.Ose T, et al. Structure of macrophomate synthase. Acta Crystallogr D. 2004;60:1187–1197. doi: 10.1107/S0907444904008881. [DOI] [PubMed] [Google Scholar]
- 44.Tarasow TM, et al. The effect of mutation on RNA Diels-Alderases. J Am Chem Soc. 2004;126:11843–11851. doi: 10.1021/ja0494149. [DOI] [PubMed] [Google Scholar]
- 45.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 46.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 47.De La Fortelle E, Bricogne G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 48.Abrahams JP, Leslie AG. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr D. 1996;52:30–42. doi: 10.1107/S0907444995008754. [DOI] [PubMed] [Google Scholar]
- 49.Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 50.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–245. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 51.Brunger AT, et al. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 Pairing alignments, turns and extensions within the structure of the ribozyme-product complex. (a) The alignment of the G27•A40 non-canonical pair stacked over the A41•(G1-C26) three-base platform. (b) The A41•(G1-C26) platform stacked over the U42•(G2-C25) triple. (c) Intercalation of G24 (in green) between the extended U42-A43 step, together with hydrogen bond alignments involving the Watson-Crick edge and sugar 2′-OH of G24. (d) The A43-C44-U45-C46 turn, stabilized by Mg1 and Mg2, with stacking of A43 and C46. (e) The U23-G24-C25-C26 segment, with splaying out of adjacent G24 and C25 bases. (f) Surface representation of the packing of the C44-U45 backbone segment of stem I (blue) positioned against the major groove of stem II (gold), mediated by bridging Mg1 and Mg2 cations.
Supplementary Figure 2 Folding of the Diels-Alder ribozyme monitored by imino proton NMR spectra (9 to 15 ppm). (a) Mg2+-dependence of the spectra of the 38-mer plus 11-mer-HEG-linked-product. Titration data are summarized in the insert, which shows that the majority of the RNA is folded at RNA:Mg2+ ratio of 1:2. Peaks used for quantification of Mg-dependent RNA folding are indicated by stars. Note that many more imino protons of the Diels-Alder product are observed on addition of Mg2+ to a sample lacking divalent cations. Also note that the exchange between Mg-free and Mg-bound states is slow, resulting in doubling of spectra. NMR spectra are recorded in 30 mM NaCl, 5 mM Tris buffer at pH 7.2. The RNA concentration was 0.05 mM. (b) NMR spectra of the ribozyme components. The 7-mer lacks the G1-G2-A3-G4 segment. The 11-mer is either free or HEG-linked with anthracene or product. The pairing of G1-G2-A3-G4 segment is reflected by appearance of additional imino protons. NMR spectra are recorded in 5 mM MgCl2, 5 mM NaCl, 5 mM Tris buffer, pH 7.2. All NMR spectra were recorded at 25 °C, using jump-and-return water suppression for detection (Plateau & Gueron, (1982), J. Am. Chem. Soc. 104, 7310–7311; Phan et al., (2001), Methods Enzymology, 338, 341–371).
Supplementary Figure 3 Schematic drawings of RNA pseudoknot topologies. (a) The Diels-Alder ribozyme, which adopts a 1-2-3-1′-2′-3′ topology. For comparison, the same coloring scheme as in Figure 1 was used. Note that the blue and green connectors are partly double-stranded, but not involved in pseudoknot interactions. (b) The HDV ribozyme, which adopts a 1-2-3-4-3′-1′-4′-2′ topology29. (c) The regulatory region of E. coli α operon, which adopts a 1-2-3-1′-2′-3′ topology30.
Supplementary Figure 4 Proposed model for the catalytic mechanism of the Diels-Alder ribozyme. Enzyme-substrate (ES) complex and transition state models have been obtained by manually docking the molecules into the pocket. The transition state was computed using MOPAC. The product-RNA complex is derived from the crystallographic structure. The surfaces of the A3-U45 base pair, amino group of G24 and 2′-OH of U42 are orange, green and yellow, respectively. Angles showing the buckling of the anthracene ring system and key distances during formation of the carbon-carbon bonds are indicated.
Supplementary Figure 5 Electron density maps of the catalytic pocket of the Diels-Alder ribozyme. (a) Solvent-flattened SAD electron density map (green) for the cycloaddition product (blue) and surrounding RNA (red), contoured at 1σ level. (b) Fo-Fc map, contoured at 2σ (green) and 1σ (red) levels, calculated for the product region after the RNA refinement and prior to the addition of the product to the model. The product and RNA, superimposed with the density, are from the refined model.