Summary
Metabolite-sensing mRNAs, or “riboswitches,” specifically interact with small ligands and direct expression of the genes involved in their metabolism. Ribo-switches contain sensing “aptamer” modules, capable of ligand-induced structural changes, and downstream regions, harboring expression-controlling elements. We report the crystal structures of the add A-riboswitch and xpt G-riboswitch aptamer modules that distinguish between bound adenine and guanine with exquisite specificity and modulate expression of two different sets of genes. The riboswitches form tuning fork-like architectures, in which the prongs are held in parallel through hairpin loop interactions, and the internal bubble zippers up to form the purine binding pocket. The bound purines are held by hydrogen bonding interactions involving conserved nucleotides along their entire periphery. Recognition specificity is associated with Watson-Crick pairing of the encapsulated adenine and guanine ligands with uri-dine and cytosine, respectively.
Introduction
RNA-mediated regulation [1–3] and catalysis [4, 5] are critically dependent on both nucleic acid architecture [6] and recognition [7, 8]. RNA aptamer-based systems serve as exceptional modules for ligand recognition and catalysis and exhibit tunable specificities and en-antiomeric selectivities [9, 10]. Aptamer and catalytic RNA modular domains can be coupled through RNA linker elements to generate allosteric ribozymes [11]. The functional principle of such designed molecular switches reflects propagation of ligand-associated adaptive transitions in the aptamer domain to the ribozyme domain, which in turn can influence ribozyme-folding patterns. Such in vitro engineered tripartite RNA switches [12] have been successfully constructed to generate designed molecular sensors [13] and new families of genetic control elements [14].
The natural counterparts of in vitro engineered allosteric switches are recently discovered metabolite-sensing mRNAs that could potentially modulate the expression of many genes. A structure-function perspective of this new and unexpected role for mRNA could provide critical information for defining the molecular basis of allosteric mRNA transitions associated with the modulation of gene expression levels and metabolic homeostasis. Indeed, principles related to recognition of the aptamer scaffolds of metabolite-sensing mRNAs could underlie new approaches to drug design and to development of molecular sensors [15].
Recent reports describe the identification of RNA genetic control elements that bind coenzyme B12 [16], flavin mononucleotide [17, 18], thiamine pyrophosphate [17, 19], S-adenosylmethionine [20–22], guanine [23], adenine [24], lysine [25, 26], and glycine [27]. Interaction of these small organic molecules with mRNA domains, or riboswitches, modulates expression of the genes involved in metabolism of these compounds, accounting for approximately 2% of the bacterial genes [23]. The field of metabolite-sensing mRNAs continues to flourish as outlined in recent reviews [28, 29], with a recent entry being a glucosamine-6-phosphate-sensing mRNA, which introduces a new paradigm, namely that the riboswitch is also a ribozyme [30]. More generally, it is conceivable that yet-to-be-identified riboswitches could also control other RNA-associated processes, such as processing, transport, and degradation [28, 31]. Genetic control by metabolite-sensing mRNAs has also been recently extended to eukaryotes [32].
Bacterial riboswitches are typically positioned within the 5′-untranslated region of the mRNA under control. Riboswitches are composed of a ligand binding ap-tamer domain and an expression platform that interfaces with RNA elements involved in gene expression. The natural aptamers, which are highly conserved within organisms and among domains of life, form independently folded modular units, whereas the expression platforms vary in sequence, topology, and mechanism. The riboswitches can adopt two distinct conformations: the metabolite bound and metabolite-free folds, involving alternative base-pairing of the regulatory RNA region. If the specific ligand is present above the threshold concentration in the cell, it interacts with the aptamer domain and stabilizes the metabolite bound fold of the riboswitch, thereby preventing formation of the alternative conformation. This results in most cases in either stabilization or disruption of the regulatory hairpin within the expression platform, thereby influencing gene expression. The 70–170-nucleotide natural aptamer domains [29] are larger than their in vitro selected counterparts [9, 10], and the increased complexity and information content of the former may reflect their need to function as high-affinity and -selectivity RNA receptors. Indeed, the natural aptamers exhibit binding affinities in the nM to low μM range, with the affinities decreasing from 10- to 100-fold on proceeding from the aptamer domain alone to the intact riboswitch. These numbers reflect the need for riboswitches to dynamically and rapidly regulate gene expression in both a temporal and a spatial manner, while retaining the selectivity of the response to a specific subset of the organism’s genes.
A search was undertaken to identify purine-sensing mRNAs by examining the regulatory mechanisms of purine metabolism. These efforts focused on the xpt-pbuX Bacillus subtilis operon (hereafter termed xpt) that encodes genes involved in guanine metabolism and is regulated by certain purines, such as guanine, hypo-xanthine, and xanthine [23]. A database search for evolutionary conservation within the 5′-UTR of xpt identified five transcriptional units with closely corresponding sequences and predicted secondary folds, termed the G-box. This domain is composed of three stems (P1, P2, and P3) connected by the junction, with significant conservation within the junction, hairpin loops (L2 and L3), and junctional P1 residues (Figure 1A; shown in red; left panel). In addition, the optimal lengths of stems P2 and P3 are 7 and 6 bp, respectively, and the matching sequences of the hairpin loops of L2 and L3 could allow potential pseudoknot formation [23]. An “in-line” probing assay, based on the patterns of spontaneous RNA cleavage, was used to establish that a 93-mer G-box xpt mRNA construct underwent a pronounced conformational transition on addition of guanine [23]. Guanine bound this RNA with approximately 5 nM affinity and reduced spontaneous cleavage throughout the junctional segment. Hypoxanthine and xanthine bound the xpt mRNA with 10-fold reduced affinity, whereas adenine binding was reduced by six orders of magnitude. Further, substantial loss of binding affinity was associated after alteration of every functional group on the guanine ring, suggesting that the guanine is completely encapsulated within the RNA fold in the complex [23].
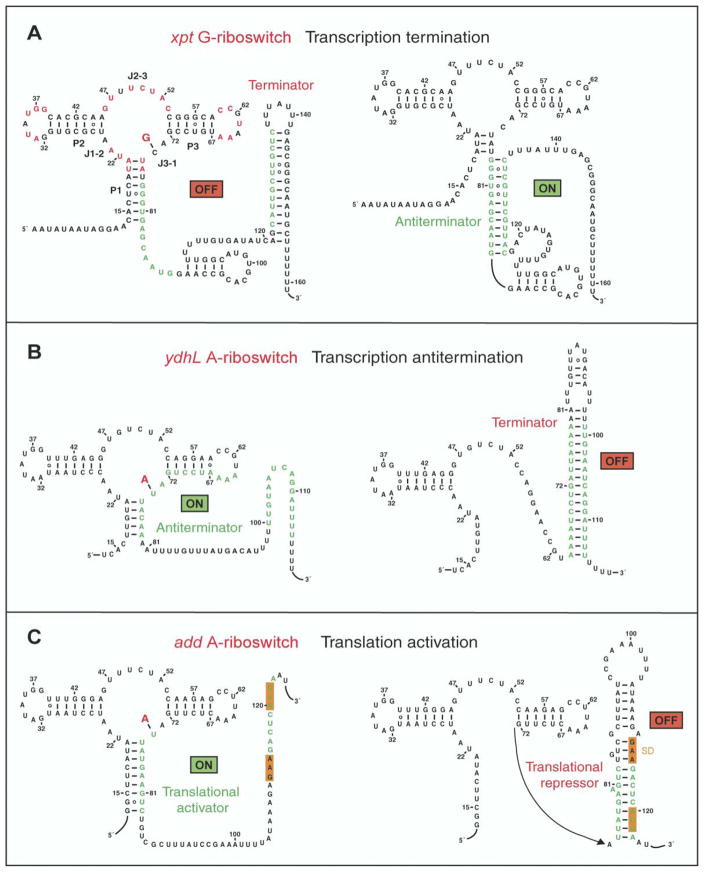
Figure 1. Mechanisms for Regulation of Gene Expression by Purine Riboswitches.
(A) Schematic showing control of gene expression by the xpt G-riboswitch through transcriptional termination [23]. Highly conserved residues in the G-box of the aptamer domain are marked in red.
(B) Gene expression regulation by the ydhL A-riboswitch through disruption of the terminator hairpin [24].
(C) Control of gene expression by the add A-riboswitch most likely through translational activation. The Shine-Dalgarno GAA sequence and the initiation codon are both shaded in orange.
It has been recently demonstrated that the ydhL gene, encoding for the putative purine efflux pump of B. subtilis, and the add gene, encoding for adenine deaminase from Clostridium perfringens and Vibrio vulnificus, harbor mRNA elements that sense adenine [24]. The secondary structure of the aptamer domain of the ydhL adenine-sensing mRNA (Figure 1B) is very similar to its xpt guanine-sensing mRNA counterpart. The ydhL mRNA binds most tightly to 2,6-diaminopurine (10 nM affinity) and less tightly (300 nM affinity) to adenine and 2-aminopurine. It also discriminates against guanine (>10,000 nM affinity) and purine (30,000 nM affinity). The guanine-sensing xpt and adenine-sensing ydhL mRNAs differ within the junction-connecting regions of their aptamer folds at three positions [24]. Two of these occur at positions that are known to be variable and are unlikely to influence ligand recognition. The third difference occurs at position 74, which is C in the guanine-sensing xpt mRNA and U in the adenine-sensing ydhL mRNA. Replacing this C with U in the xpt RNA alters its specificity from guanine to adenine; replacing the corresponding U with a C in the ydhL RNA alters its specificity from adenine to guanine [24]. Thus, remarkably, single-nucleotide substitutions can be used to interchange the guanine/adenine specificities within the aptamer domains.
Riboswitches appear to control gene expression by metabolite-modulated allosteric interconversions between alternate base-paired structures. The adaptive conformational transitions associated with metabolite binding to the aptamer domains are then harnessed through the expression platform. The xpt G-riboswitch has been shown to control gene expression through transcriptional termination [23]. Thus, the mRNA forms an antiterminator in the absence of guanine, thereby allowing RNA transcription elongation to proceed to completion (Figure 1A, right panel). The presence of guanine results in stabilization of the aptamer domain, thereby facilitating terminator formation and shutting down transcription (Figure 1A, left panel). In contrast, the ydhL A-riboswitch has been shown to control gene expression through transcriptional activation [24]. This mRNA forms a terminator in the absence of adenine (Figure 1B, right panel) but an antiterminator in the presence of adenine (Figure 1B, left panel). The add A-riboswitch does not have a stretch of uridines characteristic of transcriptional terminators but rather contains nonpaired Shine-Dalgarno GAA and initiation codon sequences, immediately downstream of the aptamer domain. Such sequences are likely to control gene expression through translational activation, whereby the Shine-Dalgarno and initiation codon sequences (both shaded in orange) are sequestered through pairing interactions in the absence of adenine (Figure 1C, right panel). The presence of adenine results in stabilization of the aptamer fold, thereby releasing these segments (Figure 1C, left panel) for interaction with ribosomal RNA and tRNA, resulting in initiation of translation. These results establish that the same mRNA aptamer fold can facilitate transcriptional termination and activation on the one hand and translational activation on the other, depending on the composition of the expression platform.
We now report the crystal structures of adenine bound to the aptamer domain of the adenine-sensing add mRNA and guanine bound to the aptamer domain of the guanine-sensing xpt mRNA. Our successful structure determination highlights the molecular principles by which a single nucleotide is capable of altering binding specificity [23, 24] and thereby switching gene expression patterns. Our structures complement the results in a just-published paper by Robert Batey and coworkers on the crystal structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine [33].
Results
Adenine- and Guanine-Riboswitch Complexes
Our structural studies were initially undertaken on the adenine-sensing B. subtilis ydhL and V. vulnificus add mRNA aptamer domains and the guanine-sensing B. subtilis xpt mRNA aptamer domain. The well-established B. subtilis xpt G-riboswitch and the ydhL A-riboswitch differ in over 20 nucleotide positions spanning the aptamer domain and are significantly different in the expression platform sequences, leading to either repression or activation of transcription, respectively (Figures 1A and 1B). Even though there are only a few differences between the adenine-sensing aptamer domains of the V. vulnificus add and B. subtilis ydhL A-riboswitches, their expression platforms are quite distinct, such that the former likely activates translation rather than transcription (Figure 1C).
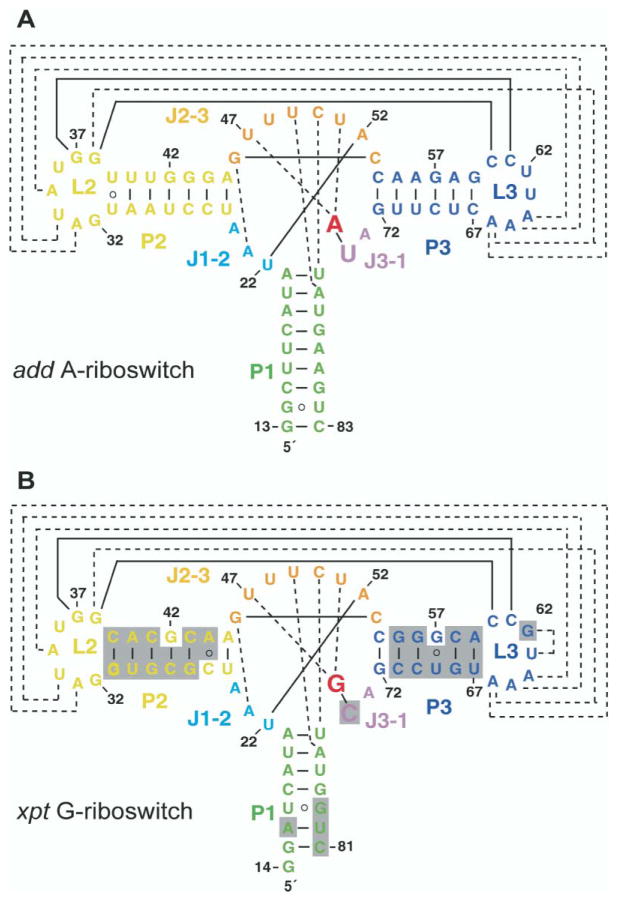
We were only successful in growing diffraction quality crystals of adenine bound to the 71-mer adenine-sensing add mRNA aptamer domain (hereafter designated A-riboswitch; Figure 2A) and guanine bound to the 68-mer guanine-sensing xpt mRNA aptamer domain (designated G-riboswitch; Figure 2B). We have adopted a simple nomenclature and color identification scheme for the aptamer domains, as outlined in the legend to Figure 2. Because the 5′ end of the add A-riboswitch was not experimentally determined, we have kept the xpt G-riboswitch numbering for both riboswitches.
Figure 2. Sequence and Stem Secondary Structures of Aptamer Domains of Purine-Sensing mRNAs.
(A) V. vulnificus 71-mer add A-riboswitch; (B) B. subtilis 68-mer xpt G-riboswitch. The color-coding scheme is as follows: Stems P1, P2, and P3 are green, yellow, and blue, respectively. Loops L2 and L3 are yellow and blue, respectively. Junction-connecting segments J1-2, J2-3, and J3-1 are cyan, orange, and violet, respectively. The specificity-determining pyrimidine residue at position 74 is highlighted with a larger lettering size. The bound adenine and guanine ligands are indicated in red lettering. Tertiary pairing alignments involving Watson-Crick and noncanonical base pairs are shown by full and dashed lines, respectively. The shaded regions in (B) highlight differences between the A- and G-riboswitches.
Complex Formation Monitored by NMR
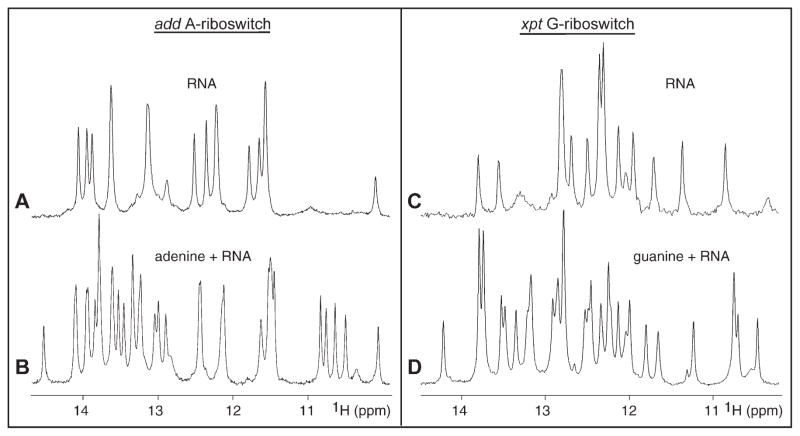
We have prepared the riboswitch RNAs by in vitro transcription and have incorporated a hammerhead ribozyme at the 3′ end to facilitate preparation of homogeneous transcripts. Binding of the ligand adenine and guanine to A- and G-riboswitches, respectively, has been monitored by nuclear magnetic resonance (NMR) spectroscopy. The imino proton NMR spectra (10.5–14.5 ppm) of the riboswitches in the absence and presence of one equivalent of bound ligand in 50 mM potassium acetate buffer (pH 6.8) are plotted in Figure 3. We observe exceptionally well-resolved imino proton (guanine N1H and uridine N3H) NMR spectra for both complexes (Figures 3B and 3D), consistent with formation of a single folded species in solution. Note the presence of new imino proton resonances between 13 and 14 ppm and between 10 and 11 ppm in these spectra. Low physiological concentration of Mg (2 mM) was required to drive complex formation to completion. In addition, exchange between free and bound states is slow on the NMR time scale, as reflected in doubling of resonances from free and bound imino protons that were observed on addition of substoichiometric (0.5 equivalents) of added adenine or guanine, characteristic of tight complex formation.
Figure 3. Imino Proton NMR Spectra of Purine Binding to Aptamer Domains of Purine Riboswitches.
Imino proton NMR spectra (10–15 ppm) of the 71-mer add A-riboswitch in the absence (A) and presence (B) of one equivalent of adenine and of the 69-mer (with GGC and GUC sequences on the 5′ and 3′ ends, respectively) xpt G-riboswitch in the absence (C) and presence (D) of one equivalent of guanine. The spectra were recorded in 50 mM potassium acetate (pH 6.8) at 25°C. For both riboswitches, 2 mM Mg was added to drive complex formation to completion.
We have also investigated the binding of adenine and its analogs to the aptamer domain of the adenine-sensing ydhL mRNA. The imino proton spectra establish that this A-riboswitch forms 1:1 tight complexes not only with adenine, but also with 2-aminopurine and 2,6-diaminopurine (see Figure S1 in the Supplemental Data available with this article online). Complex formation is in slow exchange, with similar additional imino proton markers, characteristic of the bound state, observed in all three complexes.
Crystallization and Structure Determination
Despite sequence similarities, crystals of the A-riboswitch-adenine and G-riboswitch-guanine complexes were obtained under very different conditions. Crystals of the A-riboswitch complex were grown under high (200 mM) Mg concentration, pH 9.0 buffer, whereas the crystals of the G-riboswitch complex were grown under lower (20 mM) Mg concentration, pH 5.2 buffer. The A-riboswitch complex crystals belonged to space group P21212 and diffracted to 2.1 Å, whereas the crystals of the G-riboswitch complex belonged to space group C2221 and diffracted to 2.4 Å resolution.
We first solved the structure of the A-riboswitch-adenine complex. The native crystals of this complex were soaked in 60 mM BaCl2 solution, and the structure was determined by using the anomalous properties of Ba to solve the phase problem (see Experimental Procedures for details). The structure contains all RNA residues, 5 Mg cations, and 66 water molecules in the asymmetric unit and was refined to final R-factor/R-free values of 23.1/29.7 (Table S1). The G-riboswitch-guanine complex structure was solved by molecular replacement with the structure of the A-riboswitch-adenine complex as a model. In addition to RNA residues, the structure contains 18 water molecules per RNA molecule and was refined to final R-factor/R-free values of 23.2/26.4 (Table S1).
add A-Riboswitch-Adenine Complex Structure
Overall Topology
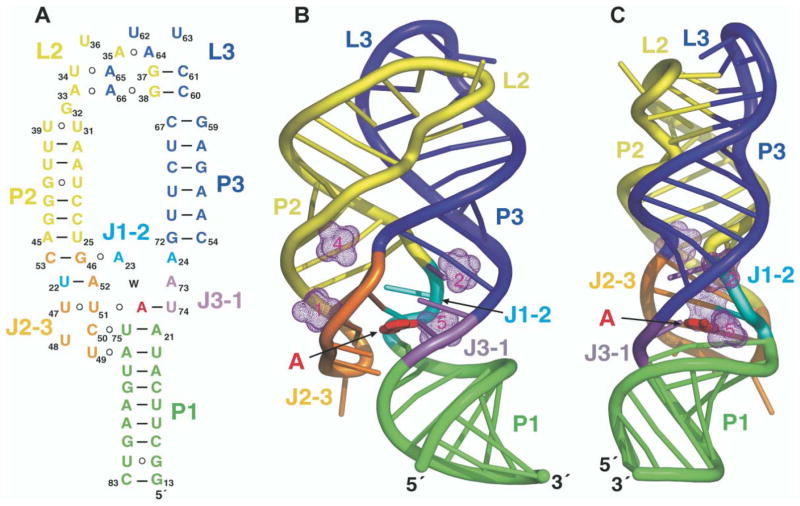
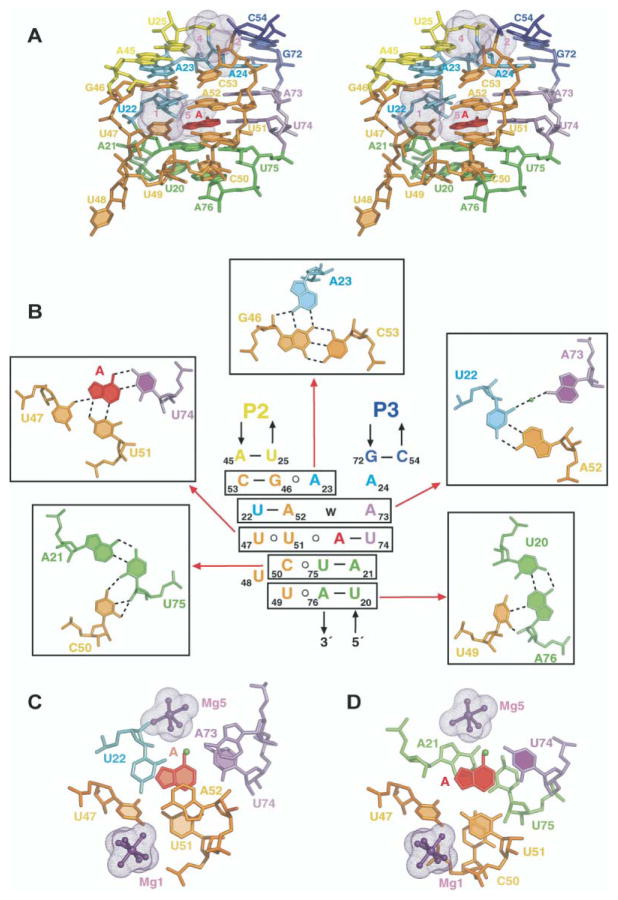
The adenine bound A-riboswitch complex adopts a tuning fork-like compact fold (schematic in Figure 4A; structures in Figures 4B and 4C), where stem P1 forms the handle of the tuning fork, and stems P2 and P3, which form the prongs, are aligned parallel to each other and anchored at the tips through extensive interaction between their hairpin loops L2 and L3. The central internal bubble zippers up through stacked base triple alignments between the three junction-connecting segments, J1-2, J2-3, and J3-1, and two junctional base pairs of stem P1, thereby generating an adenine-sensing pocket within the resulting core segment of the RNA scaffold.
Figure 4. Schematic and Structure of the A-Riboswitch-Adenine Complex.
(A) Schematic highlighting tertiary interactions in the folded structure of the A-riboswitch-adenine complex. The color-coding is as outlined in the caption to Figure 2.
(B and C) Ribbon representations (rotated by 90° along the vertical axis) of the A-riboswitch-adenine complex with the same color-coding scheme. The bound adenine is shown in red in a stick representation. Four of the five hydrated Mg cations are shown as dotted surfaces (remaining Mg is involved in packing interactions).
Zippering Up of Internal Bubble
The junctional bubble of the A-riboswitch contains tri-nucleotide J1-2, octanucleotide J2-3, and dinucleotide J3-1 junction-connecting segments (Figure 2A). Alignment of residues from all three junction-connecting segments on complex formation with adenine results in formation of a central core scaffold centered about the adenine binding site (stereo view in Figure 5A). Two triples, A23•(G46-C53) and water-mediated A73•(A52-U22), involving residues from J1-2, J2-3, and J3-1, are located above the adenine binding site, with adenines A23 and A73 positioned in the minor groove of their respective Watson-Crick base pairs (Figure 5B). Two additional triples, C50•(U75-A21) and U49•(A76-U20), involving residues in J2-3 and junctional base pairs of stem P1, are located below the adenine binding site, with C50 and U49 also positioned in the minor groove of their respective Watson-Crick base pairs. Finally, adenine recognition occurs through formation of a U51• (adenine-U74) triple, involving residues in J2-3 and J3-1, with U51 positioned in the minor groove of its Watson-Crick base pair. The stacking patterns of the adenine-containing U51•(adenine-U74) triple with the flanking A73•(A52-U22) and C50•(U75-A21) triples are shown in Figures 5C and 5D, respectively. Thus, the compact core is composed of a five-tiered triplex with extensive base stacking between tiers. Among the remaining J2-3 residues, U47 is positioned in the groove and pairs with the bound adenine and U51, whereas U48 is directed outwards from the stacked base triple architecture.
Figure 5. Details of the Tertiary Interactions within the Zippered Up Junctional Bubble in the Structure of the A-Riboswitch-Adenine Complex.
(A) Stereo pair of the junction-connecting J1-2, J2-3, and J-31 segments and two junctional stem P1 base pairs that constitute the core of the complex and include the adenine binding site. The color-coding is the same as in the caption to Figure 2, with the bound adenine shown in red.
(B) Schematic of the tertiary interactions involving a five-tiered arrangement of base triples. Individual base triples are boxed, and their pairing alignments (together with assignments) are shown on either side and above the schematic. Water molecule (w) is shown as a green ball. Hydrogen bonds are indicated by dotted lines.
(C) Stacking of the A73•(A52-U22) triple over the shaded U51•(adenine-U74) recognition triple.
(D) Stacking of the shaded U51•(adenine-U74) recognition triple over the C50•(U75-A21) triple. In (C) and (D), hydrated Mg ions, surrounded by the solvent-accessible surface in a mesh representation, are shown in ball-and-stick representation.
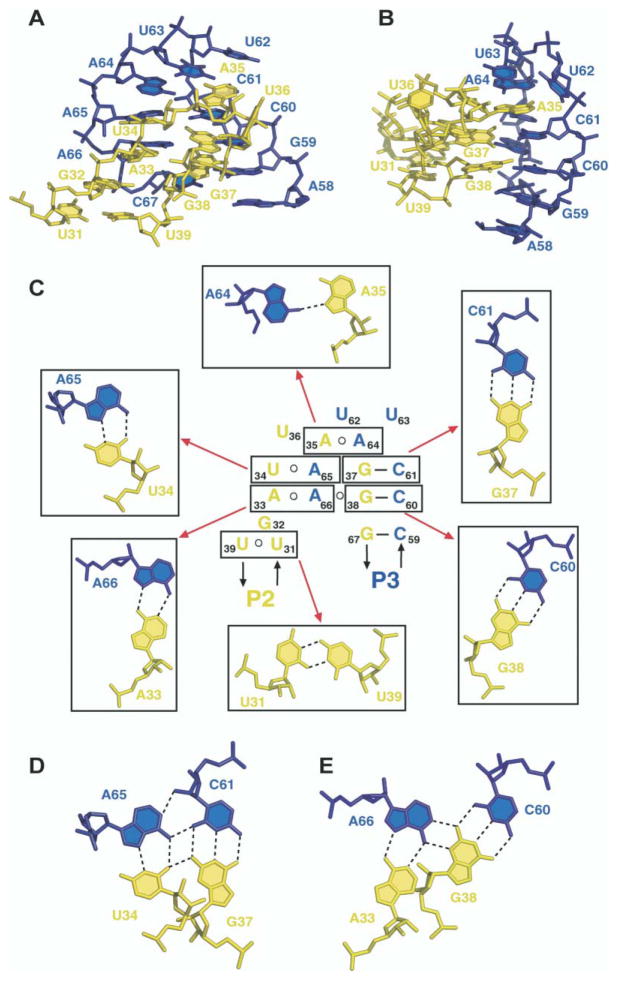
Kissing Interaction between Hairpin Loops
Hairpin loops L2 and L3, each of which contains seven nucleotides, are anchored together through formation of 5 bp in the structure of the A-riboswitch-adenine complex (Figures 6A and 6B). Two of these pairs, G38-C60 and G37-C61, are aligned through Watson-Crick pairing (Figure 6C), and three others form noncanonical pairs [34, 35]. The latter include the trans U34•A65, trans A33•A66 pair and trans A35•A64 pairs, with pairing alignments shown in Figure 6C. In addition, further alignment amongst the pairs results in formation of the A33•A66•C60-G38 tetrad (Figure 6E) and the U34• A65•C61-G37 tetrad (Figure 6D) platforms, thereby additionally anchoring the kissing interaction between the hairpin loops. Among the remaining loop bases, G32 is stacked between A33 and the U31•U39 junctional pair of P2, U36 is directed outwards from the stacked array of kissing loop interactions, and U62 and U63 form the tip of the kissing loop scaffold, with U62 stacked over the A35•A64 pair (Figure 6C). The extensive kissing interaction between hairpin loops results in a parallel alignment of stems P2 and P3.
Figure 6. Details of the Tertiary Interactions between the Kissing Hairpins in the Structure of the A-Riboswitch-Adenine Complex.
(A and B) Two views (rotated by 90° along the vertical axis) of the kissing interaction between loop L2, in yellow, and loop L3, in blue.
(C) Schematic of the tertiary interactions between kissing hairpin loops. Individual tertiary pairs are boxed, and their pairing alignments (together with assignments) are shown around the schematic. Pairing alignment of adjacent (D) U34•A65•C61-G37 and (E) A33• A66•C60-G38 tetrads.
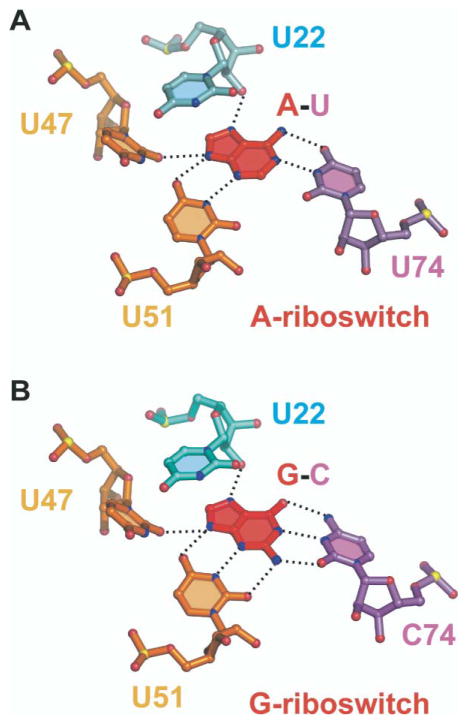
Adenine Recognition Specificity
The bound adenine moiety in the add A-riboswitch complex is held in position through formation of direct hydrogen bonds with three base edges and a sugar 2′-OH group (Figure 7A). The N1 and N6H2 atoms positioned along the Watson-Crick edge of the bound adenine form a pair of hydrogen bonds with the Watson-Crick edge of U74 of J3-1. The N3 and N9H atoms positioned along the minor groove edge of the bound adenine form a pair of hydrogen bonds with the Watson-Crick base edge of U51 of J2-3, with N9H also forming a hydrogen bond to O2 of U47 of J2-3. The N7 atom positioned along the major groove edge of the bound adenine forms a hydrogen bond with the 2′-OH of U22 of J1-2. Thus, all heteroatoms along the periphery of the bound adenine ring are recognized through hydrogen bond formation with residues from junction-connecting J1-2, J2-3, and J3-1 segments. Because the U51•(adenine-U74) base triple is sandwiched between the water-mediated A73•(A52-U22) and C50• (U75-A21) base triples (Figure 5A), the bound adenine is completely surrounded by RNA, both along its periphery and above and below its base plane.
Figure 7. Recognition of Bound Purines in Purine Riboswitches.
(A) Hydrogen-bonding alignments to bound adenine in the A-riboswitch. The bound adenine forms a Watson-Crick pair with U74. (B) Hydrogen-bonding alignments to bound guanine in the G-riboswitch. The bound guanine forms a Watson-Crick pair with C74. Hydrogen bonds involving 2′-OH of U22 and base edges of U47 and U51 are common to both riboswitches. Oxygen, nitrogen, and phosphorus atoms are shown, respectively, as red, blue, and yellow balls.
Divalent Cation Binding Sites
We observe five hydrated Mg binding sites in the A-riboswitch-adenine complex, one of which is on the surface and is involved in packing interactions. The remaining four Mg sites are positioned deep within grooves, primarily involving the junction-connecting segments in the core of the fold (Figures 4B and 4C). Mg1 and Mg4 are positioned within a deep groove made up of segments of J2-3 and the beginning of helix P2, and Mg2 and Mg5 are positioned within a deep groove made up of segments of J1-2, J3-1, and the beginning of helix P3. Hydrated Mg1 and Mg5 are closest to the adenine binding pocket (Figures 5C and 5D), with Mg5 located within hydrogen bonding distance of the N6 position of the bound adenine and thereby contributing to the locking-up of the binding pocket.
xpt G-Riboswitch-Guanine Complex Structure
Overall Topology and Tertiary Interactions
The global structure of the G-riboswitch-guanine complex is very similar to the global structure of the A-riboswitch-adenine complex (Figure 4). The same tertiary interactions hold the central core and kissing hairpins, and the purine binding pocket is sculptured by the same residue positions in both complexes. Because U62 in the A-riboswitch is replaced by G in the G-riboswitch (Figure 2), we observed formation of a G62•U63 base platform stabilized by a single hydrogen bond in the G-riboswitch.
Guanine Recognition Specificity
The bound guanine moiety in the G-riboswitch complex is held in position through the same set of hydrogen-bonding interactions observed for the bound adenine in the A-riboswitch (Figure 7), except that U74 in the A-riboswitch is replaced by C74 in the G-riboswitch. Thus, pyrimidine 74 is the specificity-determining residue in purine-sensing mRNAs. It aligns through Watson-Crick adenine-U74 pairing in the A-riboswitch and Watson-Crick guanine-C74 pairing in the G-riboswitch, as proposed earlier from mutagenesis data [24].
Discussion
Our research efforts address structural and functional issues related to folding and recognition by RNA-regulatory scaffolds associated with in vivo RNA aptamer modules. The primary challenge has been to understand the molecular recognition rules that govern ligand-mediated adaptive conformational transitions on RNA scaffolds that affect function. These principles underlie determinants of affinity and specificity, as well as global topology and allosteric transitions, thereby providing mechanistic insights into recognition processes associated with biological function.
Adaptive Conformational Transition on Complex Formation
The NMR results establish that the A-riboswitch binds adenine and G-riboswitch binds guanine selectively with 1:1 stoichiometry, with both complexes formed with high affinity (slow exchange on the NMR time scale). Complex formation occurs in the absence of divalent ions, but low (2 mM) Mg concentration was required to drive complex formation to completion.
We observe a large number of additional imino protons on proceeding from the free to the bound states of the A- and G-riboswitches (Figure 3). These additional imino protons on complex formation are detected in the 13.0–14.0 ppm region characteristic of imino protons involved in N-H••N hydrogen bonds and in the 10.0–11.0 ppm region characteristic of imino protons involved in N-H••O hydrogen bonds and/or those shielded from solvent. These results are indicative of stabilization of flexible regions within the riboswitches on ligand binding associated with an adaptive conformational transition, which results in a more compact tertiary fold on complex formation. These findings are consistent with the biochemical data published previously that indicated substantial structural stabilization of the highly conserved core of the aptamers [23, 24].
Global Architecture of Purine-Riboswitch Scaffolds
Both the A-riboswitch-adenine complex and the G-riboswitch-guanine complex adopt similar global scaffolds. The tuning-fork architecture of the three helical stems is achieved by the kissing interactions between hairpin loops L2 and L3, which forces stems P2 and P3 (the prongs of the tuning fork) to align in a parallel arrangement. The tuning fork architecture of the stems in turn defines the relative positioning of the junctional Watson-Crick base pairs A21-U75 of stem P1, U25-A45 of stem P2, and C54-G72 of stem P3. These base pairs serve as bookends bracketing the base triple interactions involving residues in J1-2, J2-3, and J3-1 junction-connecting segments that form the core of the architecture and are involved in the generation of the ligand binding pocket. It is striking that both riboswitch complexes adopt the same tertiary architectures despite different crystal packing interactions, pH, and Mg crystallization conditions.
Generation of a Central Core
A remarkable feature of the ligand bound A- and G-riboswitch scaffolds is the formation of a junctional core involving five stacked triples, with the bound purine constituting an integral part of the central triple. All five triples exhibit a common theme, in which the third base is positioned in the minor groove of a Watson-Crick base pair (Figure 5B) and is aligned with either the minor groove edge and/or sugar 2′-hydroxyl group, similar to what has been reported for a RNA pseudoknot scaffold [36], the P4-P5-P6 domain of the group I intron [37], and a vitamin B12-RNA aptamer complex [38]. It has been previously documented that this A-minor interaction strongly prefers Watson-Crick pairs over their noncanonical counterparts [37, 39] and that this plays a critical role in the specificity of RNA-RNA helix recognition [40].
There is a high degree of conservation of residues in the J1-2, J2-3, and J3-1 junction-connecting segments and the two junctional base pairs of stem P1 amongst the sequences of the purine-sensing mRNAs [24]. This conservation reflects the key roles these residues play in forming the stacked base triples array that defines the central core. Residue conservation is not maintained at position 24 in J1-2, position 48 in J2-3, and position 73 in J3-1, with the structures providing rational explanations for these observations. The central core architecture is anchored in place through formation of an A23-A24 platform [41], in which A23 and A24 are splayed out in the same plane, with A24 intercalating between an extended G72-A73 step. Thus, A24 acts as a spacer, while U48 loops out of the core scaffold. Residue A73 forms a water-mediated triple with the Watson-Crick A52-U22 pair (Figure 5B), and in the absence of direct interactions, the alignment is consistent with nonconservation of A at this position.
It is noteworthy that the junctional Watson-Crick A21-U75 and U20-A76 base pairs in stem P1 are highly conserved in the purine-riboswitches. This is now readily explainable because these base pairs are involved in C50•(U75-A21) and U49•(A76•U20) triple formation, with the former triple serving as the platform for stacking of the recognition triple. In contrast, the sequences of the remaining stem segments appear not to be important for riboswitch function.
Stitching Together the Ends
It was correctly anticipated that loops L2 and L3, given their complementary sequences, would pair with each other [23, 24]. Nevertheless, formation of two distinct types of trans A•A pairs and one trans A•U noncanonical pair was unexpected, as was their further alignment together with two tertiary Watson-Crick base pairs to form an adjacent pair of stacked mixed-tetrad alignments. Mixed tetrads have been reported previously in the structure of a viral pseudoknot [36] and facilitate the generation of a compact and maximally aligned kissing loop scaffold. The majority of the bases of loops L2 and L3 are involved in pairing between loops, and some of the remaining loop residues play distinct architectural roles. Hence, G32 is stacked into the scaffold as a spacer element, whereas U62 and U63 cap the interacting loop ends, and only U36 is looped out of the kissing scaffold.
The structure of the complex readily explains why the lengths of stems P2 (7 bp) and P3 (6 bp) are conserved. The observed lengths of stems P2 and P3 appear to be optimal for facilitating the observed kissing loop interactions. Such an establishment of distance and orientation constraints of stem-loop sequences P2-L2 and P3-L3 relative to the three-stem junction emphasizes the importance of peripheral tertiary kissing interactions in sculpting the topology of the ligand binding pocket, despite the large distance separating these two motifs.
Pyrimidine 74 as a Specificity Determinant
A systematic comparison between the A- and G-riboswitch sequences correctly led to the prediction that pyrimidine residue 74 is the key specificity determinant [24]. Thus, it was predicted that bound adenine would pair with U74 in the sequence of the A-riboswitch and that bound guanine would pair with C74 in the sequence of the G-riboswitch, presumably in each case through Watson-Crick pairing. Such an alignment would explain why the A-riboswitch containing U74 binds adenine and discriminates against guanine by greater than four orders of magnitude [24] and why the G-riboswitch containing C74 binds guanine and discriminates against adenine by six orders of magnitude [23]. Indeed, an A-riboswitch can be changed to a G-riboswitch simply by converting U74 to C74, and a G-riboswitch can be changed to an A-riboswitch by converting C74 to U74 [24].
Our structures of the A-riboswitch-adenine complex and the G-riboswitch-guanine complex indeed identify a Watson-Crick adenine-U74 alignment in the former complex compared to a Watson-Crick guanine-C74 alignment in the latter complex (Figure 7). These structures unambiguously demonstrate that the identity of a single nucleotide out of approximately 70 residues is sufficient for switching the specificity of a purine-sensing mRNA.
Purine Binding Pocket
Both adenine and guanine in their respective riboswitch complexes are completely surrounded by their common RNA scaffolds. Thus, constrained folding of junction-connecting segments juxtaposes structural elements to create a complementary surface for molecular recognition and discrimination against closely related analogs. Three pyrimidine rings and a sugar ring that line the binding cavity target the base edges of the bound adenine and guanine. Two of these pyrimidines, U47 and U51 from J2-3, are common to both complexes, and their Watson-Crick base edges align with the N3-N9H minor groove edge of both bound adenine and guanine (Figure 7). The 2′-OH group on the sugar ring of U22 from J1-2 targets the N7 of both adenine and guanine in both complexes. The specificity determinant residue is located at pyrimidine 74, with U74 and C74 targeting the Watson-Crick edge of bound adenine and guanine, respectively. It should be noted that both amino protons of the bound guanine are hydrogen bonded, consistent with a pronounced loss of binding affinity on substitution of guanine ligand by N-methyl-guanine [24].
The recognition base triples involving bound adenine or guanine are sandwiched between the A73•(A52-U22) and C50•(U75-A21) base triples (Figure 5A). The bound adenine or guanine was found to stack over both flanking triples (Figures 5C and Figures 5D). Such stacking interactions of planar purines sandwiched between base triple platforms have been previously reported for in vitro selected RNA aptamer complexes with bound planar ligands.
The bound purine ring systems are surrounded by RNA along their periphery as well as above and below their planar ring systems. Such an encapsulated recognition architecture readily explains why every functionalized position on the adenine/guanine heterocycle, even for closely related analogs, results in a substantial loss in binding affinity [23, 24]. The dramatic decrease in the majority of the analog binding affinities can be attributed to disruption of intermolecular hydrogen bonding interactions, and the remainder can be attributed to the consequences of steric occlusion.
Comparison of Adenine, Guanine, and Hypoxanthine Bound Riboswitches
The crystal structure of the hypoxanthine bound riboswitch [33] and our crystal structures of the adenine bound and guanine bound riboswitches are in excellent agreement. The crystallographic phases of the hypoxanthine complex were obtained through anomalous scattering of Co(NH3)6, whereas the phases in our structure of the adenine complex were solved through anomalous scattering properties of BaCl2. Hypoxanthine lacks the 2-amino group present in guanine, and, hence, the crystal structure of the hypoxanthine bound G-riboswitch [33] differs from its guanine bound G-riboswitch counterpart in the absence of two hydrogen bonds associated with 2-amino group recognition. The uniqueness of our contribution comparing the structures of bound A- and G-riboswitches reflects our ability to definitively establish the recognition principles by which a single specificity-determining residue permits discrimination between closely related metabolites.
Divalent Cation-Mediated Stabilization
There is ample documentation in the RNA literature that Mg cations can mediate interactions between structural domains, especially in regions where different segments of the phosphodiester backbone are brought in close proximity [42–44]. Such a stabilization effect of bound Mg cations is also observed in the crystal structure of the A-riboswitch-adenine complex (Figures 4B and Figures 4C) for crystals grown under high (200 mM) Mg concentration. In this structure, four Mg cations are positioned deep in grooves formed by junction-connecting segments in the core of the complex.
We have not been able to detect any bound hydrated Mg cations in the crystal structure of the G-riboswitch-guanine complex for crystals grown under lower (20 mM) Mg concentration. Our imino proton NMR spectra indicate that complex formation can occur in the absence of added Mg, but adding 2 mM Mg drives complex formation to completion.
Significance
Riboswitches are an important form of genetic control in certain bacteria, where they modulate the expression of numerous metabolic and transport proteins. Each riboswitch class carries a distinctive sequence and structure that is highly conserved among the organisms that use this form of regulatory system. With X-ray crystallography, we have determined high-resolution structures of the related purine-specific riboswitches that selectively bind adenine or guanine, complementing a parallel structure determination of a hypoxanthine bound riboswitch [33]. Although the A- and G-riboswitches only share 60% sequence identity, they form nearly identical binding pockets for their corresponding ligands. A single nucleotide within the core forms a Watson-Crick base pair with the ligand and, thus, serves as the main selectivity determinant between these two natural ligands. The structures of the A- and G-riboswitches bound to their respective ligands establish RNA’s ability to utilize A-minor motifs and base tetrads to facilitate folding of junctional architectures and thereby generate occluded binding pockets. Such scaffolds provide the infrastructure for targeting and discrimination between closely related metabolites, on the basis of precise hydrogen bonding and shape complementarity. In addition, the kissing interaction between the hairpin loops in the purine-riboswitch appears to be critical for both global scaffold formation and binding pocket architecture and thereby mediates long-range effects on ligand binding and release.
Experimental Procedures
RNA Preparation
The DNA fragments for riboswitch production were prepared by the annealing of chemically synthesized oligonucleotides. The DNA fragments were placed under control of the T7 promoter by cloning into StuI and HindIII sites of the pUT7 vector [45]. The add and xpt mRNAs were synthesized by in vitro transcription with T7 RNA polymerase with linearized plasmid DNA as templates. Self-cleaving hammerhead ribozyme sequence was added at the 3′-end of each RNA for subsequent cleavage to ensure length homogeneity. Purification of the transcripts was performed on 15% denaturing gels and was followed by cation exchange chromatography on MonoQ column (Amersham) and ethanol precipitation.
Complex Formation and Crystallization
The add A-riboswitch-adenine complex was prepared at 0.7 mM concentration in 50 mM potassium acetate buffer (pH 6.8) in the presence of 2 mM MgCl2. Because of the low solubility of guanine, the xpt G-riboswitch-guanine complex was first prepared at 10 μM concentration and then lyophilized to reduce the volume 100-fold. Each sample was combined with the same volume of the reservoir solution, and crystals were grown by the hanging-drop vapor diffusion method. Reservoir solutions were: (1) A-riboswitch-adenine complex, 3 M 1,6-hexanediol, 0.1 M Tris-HCl (pH 9.0), and 200 mM MgCl2; (2) G-riboswitch-guanine complex, 28% PEG400, 100 mM sodium citrate, 300 mM ammonium acetate (pH 5.2), 20 mM MgCl2, and 1% spermine. Crystals of the complexes grew to a maximal size of 300 × 50 × 50 μm in approximately 7 days at +4°C.
Structure Determination
The barium derivative was obtained by soaking the A-riboswitch-adenine complex crystals in stabilizing solution supplemented with 60 mM BaCl2 for 3 days. Crystals were flash-frozen in liquid nitrogen. The 2.7 Å resolution native and 2.95 Å resolution Ba-derivative data sets for the A-riboswitch-adenine complex were collected on the in-house Rigaku diffractometer at 1.54 Å wavelength. Other X-ray data (2.1 Å A-riboswitch-adenine complex and 2.4 Å G-riboswitch-guanine complex) were collected at beamline X25 at the National Synchrotron Light Source (NSLS). Data were processed with HKL2000 (HKL Research, Charlottesville, VA).
To determine the structure of the A-riboswitch-adenine complex, we first located six barium sites by single isomorphous replacement and anomalous scattering technique (SIRAS). The sites were found with SHELXD software [46] with the isomorphous 2.95 Å derivative and 2.7 Å native data. The position of the sites was refined with Mlphare [47]. The SIRAS phasing was performed with SHARP [48] and included the density modification procedure with SOLO-MON [49] assuming the optimized solvent content 40%. The initial model was built manually with the SIRAS electron density map and program O [50]. The model was then refined for the resolution range 20.0–2.1 Å with the higher resolution 2.1 Å native data set, as processed by CNS [51] and REFMAC [52] programs. Electron density was of sufficient quality to unambiguously interpret the entire structure except for the 5′-triphosphate and 3′-cyclic phosphate. Data collection, phasing, and refinement statistics are listed in Table S1. Adenine, hydrated Mg2+ cations, and water molecules were added to the model at the final stage of the refinement on the basis of analysis of the 2Fo-Fc and the difference Fo-Fc electron density maps.
The structure of the G-riboswitch-guanine complex was determined by molecular replacement for the resolution range 10.0–4.0 Å with Molrep software [47] with the A-riboswitch-adenine complex structure as a search model. The model was refined with the 2.4 Å resolution data set with protocols similar to those used to solve the A-riboswitch-adenine complex structure. The model contains all residues including 5′-triphosphate and 3′-cyclic phosphate.
NMR Experiments
Imino proton NMR spectra were recorded with Varian and Bruker NMR spectrometers at 25°C, with jump-and-return (JR) water suppression for detection [53]. The NMR sample conditions are listed in the caption to Figure 3.
Graphics
The figures were prepared with PyMOL (http://pymol.sourceforge.net/) and nuccyl (http://www.mssm.edu/students/jovinl02/research/nuccyl.html) software.
Supplementary Material
Acknowledgments
This research was supported by National Institutes of Health grant GM73618, the DeWitt Wallace Foundation, and the Abby Rockefeller Mauze Trust to D.J.P., National Institutes of Health grant GM068819 and National Science Foundation grant EIA-0323510 to R.R.B., and the Austrian Science Fund (FWF) to R.M. We thank P. Saha for initial experiments, A. Teplov for help with data collection, and M. Becker and the staff of the X25 and X12C beam lines at the National Synchrotron Light Source for assistance with data collection. D.J.P. is a member of the New York Structural Biology Center, which is supported by National Institutes of Health grant GM66354.
Footnotes
Accession Numbers
Coordinates for the 2.1 Å complex of adenine bound to the adenine-sensing add mRNA and the 2.4 Å complex of guanine bound to the guanine-sensing xpt mRNA have been deposited in the Protein Data Bank under accession codes 1Y26 and 1Y27, respectively.
Crystallographic data for the riboswitch-ligand complexes, as well as NMR spectra for the ydhL A-riboswitch bound to various ligands, are available at http://www.chembiol.com/cgi/content/full/11/12/1729/DC1/.
References
- 1.Batey RT, Rambo RP, Doudna JA. Tertiary motifs in RNA structure and folding. Angew Chem Int Ed Engl. 1999;38:2326–2343. doi: 10.1002/(sici)1521-3773(19990816)38:16<2326::aid-anie2326>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 2.Hermann T, Patel DJ. Stitching together RNA tertiary architectures. J Mol Biol. 1999;294:829–849. doi: 10.1006/jmbi.1999.3312. [DOI] [PubMed] [Google Scholar]
- 3.Doherty EA, Doudna JA. Ribozyme structure and mechanisms. Annu Rev Biophys Biomol Struct. 2001;30:457–475. doi: 10.1146/annurev.biophys.30.1.457. [DOI] [PubMed] [Google Scholar]
- 4.Lilley DMJ. Structure, folding and catalysis of the small nucleolytic ribozymes. Curr Opin Struct Biol. 1999;9:330–338. doi: 10.1016/S0959-440X(99)80044-X. [DOI] [PubMed] [Google Scholar]
- 5.Jaschke A. Artificial ribozymes and deoxyribozymes. Curr Opin Struct Biol. 2001;11:321–326. doi: 10.1016/s0959-440x(00)00208-6. [DOI] [PubMed] [Google Scholar]
- 6.Nowakowski J, Tinoco I., Jr . RNA structure in solution. In: Neidle S, editor. Oxford Handbook of Nucleic Acid Structure. New York: Oxford University Press; 1999. pp. 567–602. [Google Scholar]
- 7.Patel DJ. Adaptive recognition in RNA complexes with peptide and protein molecules. Curr Opin Struct Biol. 1999;9:74–87. doi: 10.1016/s0959-440x(99)80010-4. [DOI] [PubMed] [Google Scholar]
- 8.Leulliot N, Varani G. Current topics in RNA-protein recognition: Control of specificity and biological function through induced fit and conformational capture. Biochemistry. 2001;40:7947–7956. doi: 10.1021/bi010680y. [DOI] [PubMed] [Google Scholar]
- 9.Famulok M, Mayer G, Blind M. Nucleic acid ap-tamers: From selection in vitro to applications in vivo. Acc Chem Res. 2000;33:591–599. doi: 10.1021/ar960167q. [DOI] [PubMed] [Google Scholar]
- 10.Hermann T, Patel DJ. Adaptive recognition by nucleic acid aptamers. Science. 2000;287:820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- 11.Soukup GA, Breaker RR. Nucleic acid molecular switches. Trends Biotechnol. 1999;17:469–476. doi: 10.1016/s0167-7799(99)01383-9. [DOI] [PubMed] [Google Scholar]
- 12.Soukup GA, Breaker RR. Allosteric nucleic acid catalysts. Curr Opin Struct Biol. 2000;10:318–325. doi: 10.1016/s0959-440x(00)00090-7. [DOI] [PubMed] [Google Scholar]
- 13.Breaker RR. Engineered allosteric ribozymes as bio-sensor components. Curr Opin Biotechnol. 2002;13:31–39. doi: 10.1016/s0958-1669(02)00281-1. [DOI] [PubMed] [Google Scholar]
- 14.Soukup GA, Breaker RR. Engineering precision molecular switches. Proc Natl Acad Sci USA. 1999;96:3584–3589. doi: 10.1073/pnas.96.7.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaempfer R. RNA sensors: Novel regulators for gene expression. EMBO Rep. 2003;4:1043–1047. doi: 10.1038/sj.embor.7400005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR. Genetic control by a metabolite-binding RNA. Chem Biol. 2002;9:1043–1049. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- 17.Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E. Sensing small molecules by nascent RNA: A mechanism to control transcription in bacteria. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- 18.Winkler W, Cohen-Chalamish S, Breaker RR. An mRNA structure that controls gene expression by binding FMN. Proc Natl Acad Sci USA. 2002;99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind mRNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 20.McDaniel BAM, Grundy FJ, Artsimovitch I, Henkin TM. Transcription termination control of the S box system: Direct measurement of S-adenosylmethionine by the leader RNA. Proc Natl Acad Sci USA. 2003;100:3083–3088. doi: 10.1073/pnas.0630422100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Epshtein V, Mironov AS, Nudler E. The riboswitch-mediated control of sulfur metabolism in bacteria. Proc Natl Acad Sci USA. 2003;100:5052–5056. doi: 10.1073/pnas.0531307100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat Struct Biol. 2003;10:701–707. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- 23.Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. Riboswitches control fundamental biochemical pathways in Bacillus subtilisis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- 24.Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat Struct Mol Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- 25.Grundy FJ, Lehman SC, Henkin TM. The L box regulon: Lysine sensing by leader RNAs of bacterial lysine biosynthesis genes. Proc Natl Acad Sci USA. 2003;100:12057–12062. doi: 10.1073/pnas.2133705100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sudarsan N, Wickiser JK, Nakamura S, Ebert MS, Breaker RR. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev. 2003;17:2688–2697. doi: 10.1101/gad.1140003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR. A glycine-depedent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- 28.Nudler E, Mironov AS. The riboswitch control of bacterial metabolism. Trends Biochem Sci. 2004;29:11–17. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 29.Mandal M, Breaker RR. Gene regulation by ribo-switches. Nat Rev Mol Cell Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- 30.Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 31.Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sundarsan N, Jona I, et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci USA. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sudarsan N, Barrick JE, Breaker RR. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA. 2003;9:644–647. doi: 10.1261/rna.5090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Batey RB, Gilbert SD, Montagne RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- 34.Leontis NB, Stombaugh J, Westhof E. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leontis NB, Westhof E. Analysis of RNA motifs. Curr Opin Struct Biol. 2003;13:300–308. doi: 10.1016/s0959-440x(03)00076-9. [DOI] [PubMed] [Google Scholar]
- 36.Su L, Chen L, Egli M, Berger JM, Rich A. Minor groove RNA triplex in the crystal structure of a ribosomal frameshifting viral pseudoknot. Nat Struct Biol. 1999;6:285–292. doi: 10.1038/6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doherty EA, Batey RT, Masquida B, Doudna JA. A universal mode of helix packing in RNA. Nat Struct Biol. 2001;8:339–343. doi: 10.1038/86221. [DOI] [PubMed] [Google Scholar]
- 38.Sussman D, Nix JC, Wilson C. The structural basis for molecular recognition by the vitamin B12 RNA aptamer. Nat Struct Biol. 2000;7:53–57. doi: 10.1038/71253. [DOI] [PubMed] [Google Scholar]
- 39.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc Natl Acad Sci USA. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Battle DJ, Doudna JA. Specificity of RNA-RNA helix recognition. Proc Natl Acad Sci USA. 2002;99:11676–11681. doi: 10.1073/pnas.182221799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Szewczak AA, Kundrot CE, Cech TR, Doudna JA. RNA tertiary structure mediation by adenosine platforms. Science. 1996;20:1696–1699. doi: 10.1126/science.273.5282.1696. [DOI] [PubMed] [Google Scholar]
- 42.Hanna R, Doudna JA. Metal ions in ribozyme folding and catalysis. Curr Opin Chem Biol. 2000;4:166–170. doi: 10.1016/s1367-5931(99)00071-x. [DOI] [PubMed] [Google Scholar]
- 43.Egli M, Minasov G, Su L, Rich A. Metal ions and flexibility in a viral pseudoknot at atomic resolution. Proc Natl Acad Sci USA. 2002;99:4302–4307. doi: 10.1073/pnas.062055599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klein DJ, Moore PB, Steitz TA. The contribution of metal ions to the structural stability of the large ribosomal subunit. RNA. 2004;10:1366–1379. doi: 10.1261/rna.7390804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Serganov A, Rak A, Garber M, Reinbolt J, Ehresmann B, Ehresmann C, Grunberg-Manago M, Portier C. Ribosomal protein S15 from Thermus thermophilus Cloning, sequencing, overexpression of the gene and RNA-binding properties of the protein. Eur J Biochem. 1997;246:291–300. doi: 10.1111/j.1432-1033.1997.00291.x. [DOI] [PubMed] [Google Scholar]
- 46.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 47.CCP4 (Collaborative Computational Project Number 4) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 48.De La Fortelle E, Bricogne G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous and multiwavelength anomalous diffraction methods. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 49.Abrahams JP, Leslie AGW. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr D Biol Crystallogr. 1996;52:30–42. doi: 10.1107/S0907444995008754. [DOI] [PubMed] [Google Scholar]
- 50.Jones TA, Zou JY, Cowan SW, Kjeldgaard GJ. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 51.Brunger AT, Adams PD, Clore GM, De Lano WL, Gros P, Grosse-Kuntsleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;5:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 52.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–245. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 53.Phan AT, Gueron M, Leroy JL. Investigation of unusual DNA motifs. Methods Enzymol. 2001;338:341–371. doi: 10.1016/s0076-6879(02)38228-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.