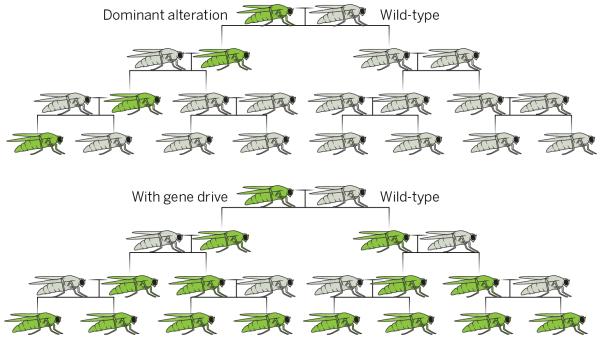
Gene drive systems promote the spread of genetic elements through populations by assuring they are inherited more often than Mendelian segregation would predict(see the figure). Natural examples of gene drive from Drosophila include sex-ratio meiotic drive, segregation distortion, and replicative transposition. Synthetic drive systems based on selective embryonic lethality or homing endonucleases have been described previously in Drosophila melanogaster (1–3), but they are difficult to build or are limited to transgenic populations. In contrast, RNA-guided gene drives based on the CRISPR/Cas9 nuclease can, in principle, be constructed by any laboratory capable of making transgenic organisms (4). They have tremendous potential to address global problems in health, agriculture, and conservation, but their capacity to alter wild populations outside the laboratory demands caution (4-7).Just as researchers working with self-propagating pathogens must ensure that these agents do not escape to the outside world, scientists working in the laboratory with gene drive constructs are responsible for keeping them confined (4,6,7).
This layout is not final
Two of us recently used a CRISPR/Cas9-based gene drive system to generate a Drosophilastrain homozygous for a loss-of-function mutation [the mutagenic chain reaction (6)](see the figure). Even though D. melanogaster ordinarily poses no threat to human health or agriculture, the accidental release of flies carrying gene drive constructs from the laboratorycould have unpredictable ecological consequences. This study therefore used institutionally approved stringent barrier methods. Only one experimenter handled the flies, inside an Arthropod Containment Level 2 insectary suitable for work with mosquitoes carrying human pathogens. Because barrier protocols can be vulnerable to human error (8), these authorssuggested (6) that additional molecular confinement methods described (4) and used by others of us in budding yeast (9) could further reduce risks. That these studies documented highly efficient RNA-guided gene drive in flies and yeast underscores the potential of the technology and the risk resulting from an accidental release.
As concerned scientists working in related areas, we engaged in collective discussionsto identify and publicize interim safety recommendations for laboratory research involving potential gene drive systems while formal national guidelines are developed. Although we cannot claim to represent all researchers, we share a commitment to the safe and responsible development of gene drive technology. Although we differ in our assessments of the types of precaution needed, we recognize that any single confinement strategy could fail. We therefore unanimously recommend that future studies usea combination of stringent confinement strategies (see the table) whenever possible and always use safeguards adequate for preventing the unintentional release of synthetic gene drive systems into natural populations.
Potentially stringent confinement strategies for gene drive research
| TYPE | STRINGENT CONFINEMENT STRATEGY | EXAMPLES |
|---|---|---|
| Molecular | Separate components required for genetic drive Target synthetic sequences absent from wild organisms |
sgRNA and Cas9 in separate loci (8) Drive targets a sequence unique to laboratory organisms (3,4,8) |
|
| ||
| Ecological | Perform experiments outside the habitable range of the organism Perform experiments in areas without potential wild mates |
Anopheles mosquitoes in Boston Anopheles mosquitoes in Los Angeles |
|
| ||
| Reproductive | Use a laboratory strain that cannot reproduce with wild organisms |
Drosophila with compound autosomes* |
|
| ||
| Barrier | Physical barriers between organisms and the environment
|
Triply nested containers, >3 doors (6) Anesthetize before opening (6) Low-temperature room, air-blast fans Keep careful records of organisms, one investigator performs all experiments (6) |
An example of reproductive confinement would be Drosophila laboratory strains with a compound autosome, where both copies of a large autosome are conjoined at a single centromere. These strains are fertile when crossed inter se but are sterile when outcrossed to any normal or wild-type strain because all progeny are monosomic or trisomic and die early in development.
RECOMMENDATIONS
RNA-guided gene drive systems are created by delivering into the germline a DNA cassette encoding Cas9 and a single synthetic guide RNA (sgRNA)that isflanked by sequences matchingthose on either side of the sgRNA target site (4). Cas9 nuclease-stimulated copying of the cassette into the target allele leads to continued Cas9+sgRNA expression and subsequent copying of the cassette into the other allele (6,9). The recurrent conversion of heterozygotes into homozygotes permits spread through populations(see the figure).
The vast majority of recent genome engineering approaches developed in model organisms neither involve nor risk the creation of gene drive systems. For example, Drosophila mutants can be readily generated by injecting sgRNAs or sgRNA-encoding plasmids into transgenic embryos expressing Cas9 (10–13) or by crossing sgRNA-expressing strains to Cas9-expressing strains (12–14).These approaches do not risk creating a gene drive system because cassettes encoding Cas9 and sgRNA are not inserted into the cut site or located adjacent to one another in the genome and can thus be safely used by researchers without additional precautions.Given the availability of efficient alternatives and the potential risks, we recommend that gene drive approaches to genome engineering be strictly reserved for cases that require their use.
The safest approach for using gene drives creates biallelic mutations with ansgRNA-only cassette that can spread only when combined with an unlinked Cas9 transgene (4). In such a “split gene drive system,” homozygous individuals lacking the Cas9 gene can be easily isolated in subsequent generations. The efficiency of gene drive exhibited by a split system in yeast is equivalent to that of a construct encoding both Cas9 and sgRNA (9). Split drive systems present a much lower risk if organisms are accidentally released because the population frequency of the Cas9 gene will be determined by normal, nondrive dynamics, consequently limiting the spread of the sgRNA cassette.
Nevertheless, any mutational event that moves the Cas9 gene into or directly adjacent to the sgRNA cassette could create an autonomous Cas9+sgRNA drive system by allowing the Cas9 gene to be copied into the target locus along with the sgRNA cassette upon repair of Cas9-induced DNA cleavage. Although the probability of such an event is extremely low, we recommend that at least one additional form of stringent confinement be used (see the table) and that the strains be continually monitored.
Other forms of stringent confinement include performing experiments in an area lacking wild populations (4) and, when the goal is to study gene drive systems in the laboratory, exclusively targeting synthetic sequences not found in natural populations (3,4,9). Because these strategies suffer from independent vulnerabilities, the safety improvements afforded by combining them will be multiplicative. Thus, the great majority of gene drive experiments can be performed with minimal risk of altering wild populations. Accordingly, we strongly recommend that
1) All work involving potential gene drive systems should be preceded by a thorough assessment by the relevant biosafety authorities of the risk of unwanted release from the laboratory. We encourage these authorities to seek guidance from external experts and make their evaluation available to others.
2) All laboratory gene drive experiments should employ at least two stringent confinement strategies (see the table) whenever possible to minimize the risk of altering wild populations. Using one form of confinement may be justified only if relevant biosafety authorities determine that it will reduce the probability of release to a level that is acceptably low. This probability must be defined on a case-by-case basis. The analyses necessary to confidently predict the efficacy of confinement strategies for gene drive systems are in a nascent form.Therefore, any proposal to use one rather than multiple forms of confinement requires even greater scrutiny and extensivedeliberation between regulatory authorities and scientists.
3) Organisms carrying gene drive constructs that could spread if the reproductivelycapable life stages were to escape in transit should not be distributed to other institutions until formal biosafety guidelines are established. Whenever possible, laboratories should instead send DNA constructs or information sufficient to reconstruct the gene drive. Protocols for distributing materials should be established in discussion with the wider research community and other relevant stakeholders.
Broadly inclusive and ongoing discussions among diverse groups concerning safeguards, transparency, proper use, and public involvement should inform expert bodies as they develop formal research guidelines for gene drive research in the laboratory and potential transitions to open field trials. We applaud the U.S. National Academy of Sciences for committing to provide recommendations for responsible gene drive research (15). By recommending strong safeguards and encouraging discussion of this technology, we hope to build a foundation of public trust for potential future applications in public health, sustainable agriculture, and ecological conservation.
ACKNOWLEDGMENTS
The authors are grateful for conversations with T. Wu, J. Lunshof, and A. Birnbaum. V.G., E.B., G.C., and K.E. are inventors on relevant provisional and nonprovisional patents filed by the University of California and Harvard University.
Footnotes
The spread of RNA-guided gene drive systems. Unlike the population dynamics of normal genomic alterations, gene drive systems can spread changes through wild populations by converting heterozygotes into homozygotes in each generation.
REFERENCES
- 1.Chen C-H, et al. Science. 2007;316597 doi: 10.1126/science.1138595. [DOI] [PubMed] [Google Scholar]
- 2.Akbari OS, et al. Curr. Biol. 2013;23671 doi: 10.1016/j.cub.2013.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan Y-S, Naujoks DA, Huen DS, Russell S. Genetics. 2011;18833 doi: 10.1534/genetics.111.127506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esvelt KM, Smidler AL, Catteruccia F, Church GM. eLife. 20142014:e03401. doi: 10.7554/eLife.03401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oye KA, et al. Science. 2014;345626 doi: 10.1126/science.345.6200.1010-c. [DOI] [PubMed] [Google Scholar]
- 6.Gantz VM, Bier E. Science. 2015;348442 doi: 10.1126/science.aaa5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burt A. Proc. Roy. Soc. London B. 2003;270921 [Google Scholar]
- 8.Henkel RD, Miller T, Weyant RS. Appl. Biosafety. 2012;17171 [Google Scholar]
- 9.DiCarlo JE, et al. bioRxiv. 2015:013896. [Google Scholar]
- 10.Ren X, et al. Proc. Natl. Acad. Sci. U.S.A. 2013;11019012 [Google Scholar]
- 11.Gratz SJ, et al. Genetics. 2014;196961 doi: 10.1534/genetics.113.160713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Port F, Chen H-M, Lee T, Bullock SL. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E2967. doi: 10.1073/pnas.1405500111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Port F, Muschalik N, Bullock SL. G3. 2015 doi: 10.1534/g3.115.019083. g3.115.019083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kondo S, Ueda R. Genetics. 2013;195715 doi: 10.1534/genetics.113.156737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.National Research Council Gene Drive Research in Non-Human Organisms: Recommendations for Responsible Conduct. (DELS-BLS-15-06, National Academy of Sciences, Washington, DC, 2015); http://bit.ly/CurrProjects-regul.