Abstract
The effects of divalent Mg2+ on the conformation and dynamics of the stem-loop transactivation response element (TAR) RNA from HIV-1 have been characterized using NMR residual dipolar couplings (RDCs). Order matrix analysis of one bond 13C–1H RDCs measured in TAR at [Mg2+]:[TAR] stoichiometric ratios of ~3:1 (TAR(3.0 Mg)) and ~4.5:1 (TAR(4.5 Mg)) revealed that Mg2+ reduces the average inter-helical angle from 47(±5)° in TAR(free) to 5(±7)° in TAR(4.5 Mg). In contrast to the TAR(free) state, the generalized degree of order for the two stems in TAR(4.5 Mg) is found to be identical within experimental uncertainty, indicating that binding of Mg2+ leads to an arrest of inter-helical motions in TAR(free). Results demonstrate that RDC-NMR methodology can provide new insight into the effects of Mg2+ on both the conformation and dynamics of RNA.
Keywords: transactivation response element, RNA folding, collective motions, divalent metal ions, nucleic acids
By neutralizing backbone electrostatic repulsions, divalent ions such as Mg2+ can profoundly affect RNA average conformation and plasticity, and thus have important consequences on folding,1–4 catalysis,5,6 and recognition.7 Understanding how divalent ions affect RNA conformation and function at a molecular level requires not only the atomic characterization of typically folded divalent ion bound conformations, but also what is more often an ensemble of partially unfolded divalent ion free conformations.1–4 However, elucidating high-resolution structures of dynamically inter-converting conformational substates poses significant challenges to techniques such as X-ray crystallography and NMR spectroscopy, which remain by and large, limited in applicability to “static” average structure determination of well folded conformations. While NMR relaxation measurements can probe “slow” (microseconds–milliseconds timescales) internal motions,8–10 these methods provide limited information about motional amplitudes and directions. Fluorescence resonance energy transfer (FRET)11 has emerged as a powerful alternative technique for measuring time-dependent changes in RNA global conformation,12 but structural resolution also remains limited to distances separating pairs of donor and acceptor dye molecules. These experimental limitations have made it difficult to more directly investigate the effect of divalent ions on RNA structural dynamics.
By providing long-range angular sensitivity to both structure and dynamic fluctuations over a wide range of timescales (<ms), the measurement of residual dipolar couplings (RDCs) in partially oriented systems13,14 has opened a new avenue for characterizing the conformational dynamics of flexible biomolecules using NMR.15–20 We recently reported the RDC-NMR characterization of a divalent cation free transactivation response element (TAR) RNA variant (TAR(free)) (Figure 1(a)).21 The TAR domain has been the subject of intense structural studies,22–25 both because its interaction with trans-activator protein Tat is critical for HIV-1 viral replication,26 making it an attractive drug target,27 and because it is a paradigm for studies of RNA–protein adaptive recognition.28,29 Our RDC-NMR investigation unveiled a highly dynamic global TAR conformation, executing directionally unbiased collective helical excursions with amplitudes of 46(±4)° about an average inter-helical angle of 47(±5)°.21
Figure 1.
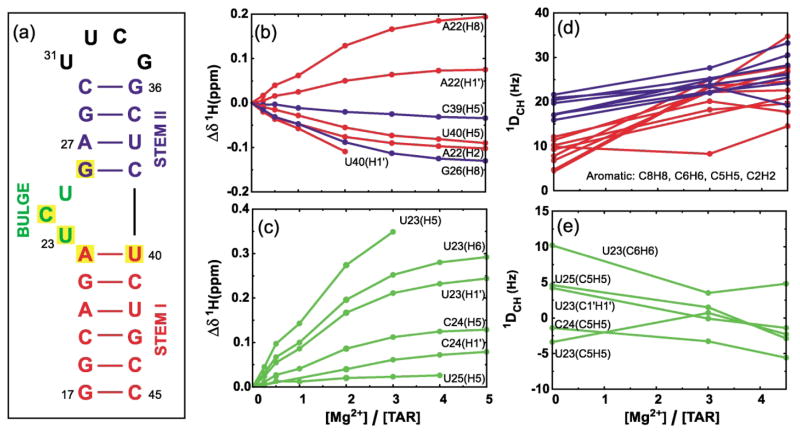
Secondary structure of the TAR RNA domain and variations in chemical shift and RDCs as a function of [Mg]:[TAR] stoichiometry. (a) The HIV-1 TAR analog used in this study. The six-residue hairpin loop in wt-TAR (CUGGGA) has been replaced with the more stable tetraloop (UUCG). Yellow boxes indicate residues having the largest Mg2+-induced 1H chemical shift perturbations[(Δδ (ppm) = δTAR(Mg) – δTAR(free)) > 0.1 ppm, positive values imply downfield shifts]. Perturbations in 1H chemical shifts for (b) residues in stem I (red) and II (blue) and, (c) bulge residues. RDC values measured between directly bonding C–H nuclei in (d) base moieties of residues in stems I (red) and II (blue) and (e) base and sugar moieties of bulge residues. Uniformly labeled 15N/13C-labeled TAR was prepared using standard procedures as described.21 NMR samples contained ~1.0 mM 15N/13C-labeled TAR, 15 mM sodium phosphate (pH 6.0–6.2), 25 mM sodium sulfate, 0.1 mM EDTA, and 3.0 and 4.5 mM Mg2+ for TAR(3.0 Mg) and TAR(4.5 Mg), respectively. An identical NMR sample also contained ~21 mg/ml of Pf1 phage.34,35 All NMR data were acquired on Varian Inova spectrometers operating at 1H frequencies of 500 and 600 MHz at 25 °C. Resonances were assigned by following incremental shifts upon addition of Mg2+. As described,21 splittings between [C8–H8, C6–H6, C2–H2], [C5–H5], [C1′ –H1′] were measured using 1H and 13C IPAP-HSQC experiments40 and between [C2′ –H2′, C3′ –H3′, C4′ –H4′] using the 1JCH-CT-CE-HSQC experiment.16 RDCs were calculated as the differences between splittings measured in the presence and absence of phage. The uncertainty in RDCs, estimated from measuring splittings in the 1H and 13C dimension, was <3.5 Hz and <4.5 Hz for TAR(3.0 Mg) and TAR(4.5 Mg), respectively. The greater uncertainty in RDCs measured in the presence of Mg2+ relative to TAR(free)21 is attributed to increased line widths arising from greater degree of alignment as well as exchange broadening due to non-specific Mg2+ binding.
Although several studies have reported evidence for binding of divalent metal ions to TAR, discrepancies persist regarding the effects of divalent ions on the TAR average conformation while little is also known about the effects of divalent ions on TAR dynamics. Using electric birefringence, Zacharias and Hageraman previously reported that Mg2+ (~2 mM) induces ~50% reduction in the inter-helical angle of a 120 nt long RNA containing the bulge region of TAR. In contrast, a 1.3 Å X-ray structure of an RNA duplex comprising the bulge region of TAR determined in the presence of much higher divalent ion concentrations (100 mM CaCl2) shows a coaxially aligned conformation with four Ca2+ bound to TAR. Three of these ions stabilize a looped out bulge conformation that is also distinct from previous solution NMR structures of TAR determined in the absence of divalent ions.23,24,30 Fluorescence studies also provide evidence for Mg2+ destabilizing complexes of TAR with Tat-derived peptides.31 More recently, experiments employing 19F chemical shift perturbations in combination with Pb2+-induced RNA cleavage have more directly established that the bulge region of TAR binds , Mg2+, and Ca2+ with increasing specificity in solution.32 Here, we have employed RDC-NMR to investigate the effects of Mg2+ on both TAR average conformation and dynamics under solution conditions.
Binding of Mg2+ to TAR was initially examined by recording 2D 13C–1H correlation spectra of TAR in the presence of increasing Mg2+ concentrations. Monotonous variations in 1H and 13C chemical shifts were observed (Figure 1(b) and (c)), indicating rapid exchange (105–103 s−1) between free and Mg2+ “bound” TAR conformations. In agreement with previous 19F NMR studies,32 the largest chemical shift perturbations (>0.1 ppm in 1H) are observed in and around bulge residues (Figure 1(a), in yellow), while negligible shifts are observed for the UUCG tetraloop. Although it is not possible to distinguish contributions to chemical shift perturbations arising from conformational change and Mg2+ localization, these results indicate that Mg2+ primarily binds residues in and around the bulge, consistent with the previously reported X-ray structure of TAR bound to Ca2+.33 Interestingly, while most of the stem resonances move downfield (Figure 1(b)), bulge resonances exclusively move upfield (Figure 1(c)). Modest line broadening was also observed with increasing Mg2+ concentrations, especially for the C8H8 resonances of guanine residues. This was attributed in part to exchange broadening due to non-specific Mg2+binding to TAR.
To investigate the effects of Mg2+ on the structure and dynamics of TAR, RDCs were measured between directly bonded 1H–13C nuclei (1DCH) at [Mg2+]:[TAR] stoichiometric ratios of ~3:1 [TAR(3.0 Mg)] and ~4.5:1 [TAR(4.5 Mg)]. Based on a two-state interpretation of the chemical shift titrations (Figure 1(b) and (c)), the latter stoichiometries correspond to ~84% and ~94% of TAR existing in a Mg2+ bound conformation, respectively. Initial dissolution of TAR(3.0 Mg) in ~21 mg/ml Pf1 phage34,35 yielded NMR spectra identical to those recorded for TAR(free). This effect was attributed to sequestering of Mg2+ by phage as spectra of TAR(3.0 Mg) and TAR(4.5 Mg) could be restored by incremental addition of Mg2+ to final concentrations of ~6.0 and ~7.5 mM, respectively. In all cases, the 2H splittings, and hence the apparent degree of phage alignment remained unchanged at ~21 Hz. Greater line broadening was also observed for TAR(3.0 Mg) and TAR(4.5 Mg) relative to TAR(free) when dissolved in phage. Although the resulting uncertainty in RDCs, estimated from measuring splittings in the 1H and 13C dimension, was greater for TAR(3.0 Mg) (<3.5 Hz) and TAR(4.5 Mg) (<4.5 Hz) compared to TAR(free) (~1 Hz), this adverse effect was in part offset by an observed Mg2+-induced increase (~32%) in the total TAR alignment (see below). Hence, in addition to exchange broadening due to non-specific Mg2+ binding, the resonances in TAR(3.0 Mg) and TAR(4.5 Mg) likely experience additional line broadening relative to TAR(free) due to greater alignment and dipolar interactions. Due to the greater overlap observed in spectra of TAR in the presence of Mg2+, a smaller number of RDCs could on average be measured in TAR(3.0 Mg) and TAR(4.5 Mg) relative to TAR(free) (Table 1).
Table 1.
Statistics for order matrix analysis of RDCs measured in the two stems of TAR as a function of Mg2+ concentration
| Stem | No. RDCs | CN | RMSD (Hz) | RMSDN (Hz) × 103 | R |
|---|---|---|---|---|---|
| TAR(free)stem I | 18 | 2.5 | 1.9 | 3.7 | 0.99 |
| TAR(free)stem II | 22 | 2.3 | 2.6 | 3.0 | 0.98 |
| TAR(3.0 Mg)stem I | 17 | 2.5 | 3.9 | 3.7 | 0.94 |
| TAR(3.0 Mg)stem II | 11 | 3.3 | 2.8 | 2.5 | 0.98 |
| TAR(4.5 Mg)stem I | 16 | 5.9 | 4.5 | 3.9 | 0.94 |
| TAR(4.5 Mg)stem II | 16 | 2.8 | 4.0 | 3.4 | 0.97 |
Shown are the total number of RDCs employed in the order matrix analysis (No. RDCs), resulting condition numbers (CN) defined as the ratio of the largest to smallest singular value in the singular value decomposition, root mean square deviation between experimental RDCs and values calculated using best fit order tensor parameters (RMSD), RMSD values normalized for degree of alignment (RMSD (Hz)/ϑ), and correlation coefficient between measured and predicted RDCs (R). Parameters are reported using input coordinates from idealized A-form RNA duplexes. Note that variations in stem-specific RMSD values correlate with the observed degree of alignment, such that RMSDN values are more uniform across the stems.
The RDC values measured in TAR(3.0 Mg) and TAR(4.5 Mg) along with previous values measured for TAR(free)21 are compared in Figure 1 separately for stem (Figure 1(d)) and bulge residues (Figure 1(e)). Although Mg2+ leads to changes in almost all of the RDCs measured in TAR (Figure 1(d) and (e)), two variations are noteworthy. First, the pronounced difference between the magnitudes of RDCs measured in stems I and II in TAR(free), which was previously attributed to inter-helical kinking and dynamics,21 is diminished in TAR(3.0 Mg), and indiscernible in TAR(4.5 Mg) (Figure 1(d)). Second, whereas Mg2+ induces an increase in the absolute magnitude of stem RDCs (Figure 1(d)), a decrease is observed for RDCs measured in juxtaposed bulge residues (Figure 1(e)). This attenuation is particularly pronounced and uniform across three RDCs measured in residue U23. These results indicate the Mg2+ changes both the global inter-helical and local bulge structure and/or dynamics of TAR.
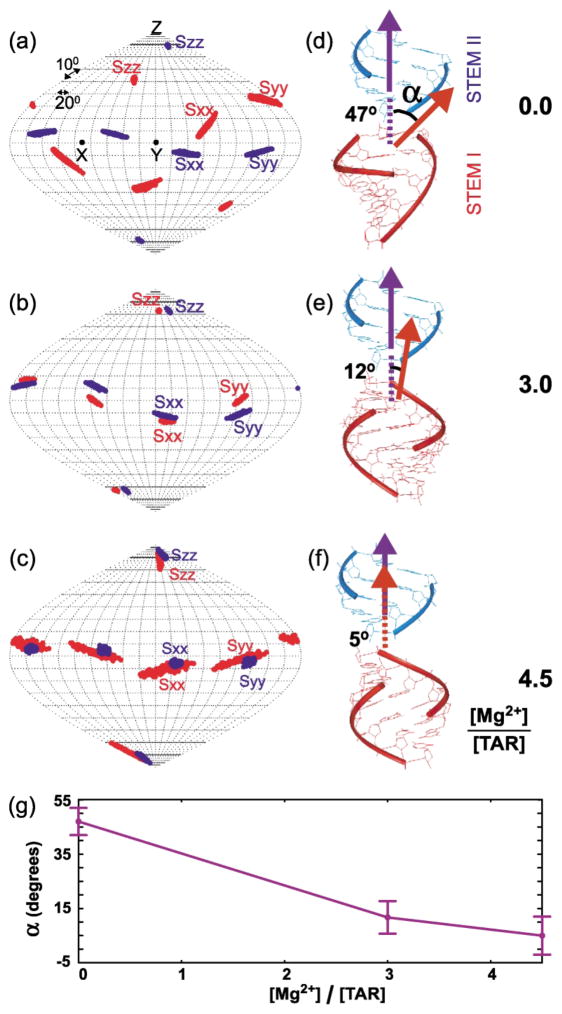
To examine the effects of Mg2+ on the global conformation of TAR, RDCs measured in Watson–Crick regions of stems I and II were used to solve for stem-specific order matrix elements using idealized A-form geometries as input coordinates.36,37 This analysis revealed that Mg2+ induces a change in the global conformation of TAR. As now reported in a number of RDC-NMR studies of RNA,19–21,38,39 measured RDCs were found to agree within experimental uncertainty with local geometries for the stems derived from idealized A-form helices in all of TAR(free),21 TAR(3.0 Mg) and TAR(4.5 Mg) (Table 1). However, a change in the relative orientations of stem principal axes (Szz, Syy, Sxx), and hence stem alignment is observed with increasing Mg2+ concentrations. This can be visualized when stem-specific principal orientations (Szz, Syy, Sxx) are mapped relative to a continuous, coaxially aligned A-form helix (Figure 2(a)–(c)). Greater overlap between stem solutions and hence inter-helical straightening is observed in going from TAR(free) (Figure 2(a)),21 to TAR(3.0 Mg) (Figure 2(b)) and TAR(4.5 Mg) (Figure 2(c)). Correspondingly, TAR conformations determined by superimposing stem-centered principal axes exhibit a significant decrease in the average inter-helical angle (a) from 47(±5)° in TAR (free)21 (Figure 2(d)), to 12(±6)° in TAR(3.0 Mg) (Figure 2(e)), and 5(±7)° in TAR(4.5 Mg) (Figure 2(f)). Note that the conformation of TAR(3.0 Mg) should be interpreted with caution. In particular, it is difficult to decipher if the observed conformation corresponds to a population-weighted average of free and Mg2+-bound states or a state in which Mg2+ binding sites in TAR have not been completely occupied. It is nevertheless interesting to note that the observed inter-helical angle for TAR(3.0 Mg) can be reproduced based on a population weighted average of TAR(free) and TAR(4.5 Mg) states (84% × 5°(bound) + 16% × 47°(free) ≈12°). This is consistent with exchange of TAR between Mg2+ free and bound forms and hence cooperative Mg2+ binding. Although the relatively small number of RDCs measured in TAR(3.0 Mg)stemII and TAR(4.5 Mg)stemI resulted in higher condition number and consequently higher uncertainty in derived order matrix elements, the precision with which the TAR inter-helical angle is determined and the overall trend observed across TAR(free), TAR(3.0 Mg) and TAR(4.5 Mg) strongly suggest that Mg2+ induces complete coaxial alignment of the TAR conformation. The accuracy of derived principal orientational solutions is also supported by the fact that as expected for phage-induced alignment,35 co-axial alignment of helices is accompanied by greater coincidence between the principal (Szz) and helix (“z”) axes of TAR (Figure 2(a)–(c)). The effects of Mg2+ on the TAR global conformation observed here using RDC-NMR (Figure 2(g)) can reconcile the standing discrepancy between bent and linear TAR conformations determined by NMR23,24,30 and X-ray crystallography,33 respectively, which can be attributed to the absence of divalent ions in the former rather than crystal packing forces in the latter. Our results are also in qualitative agreement with transient electric birefringence studies,25 which report ~50% reduction in the TAR inter-helical angle at lower Mg2+ concentrations (~2 mM), noting that the authors did not preclude the possibility for further inter-helical straightening at higher Mg2+ concentrations.
Figure 2.
TAR inter-helical conformation as a function of [Mg]:[TAR] stoichiometry. Order tensor orientations (Sxx, Syy, Szz) for stem I (red) and II (blue) in (a) TAR(free) (b) TAR(3.0 Mg) and (c) TAR(4.5 Mg). Ribbon representation of the relative orientation of stem I (in red) and II (in blue) determined by superimposing stem-specific principal axes for (d) TAR(free), (e) TAR(3.0 Mg) and (f) TAR(4.5 Mg), where the helix axis of stem I is oriented towards the reader. The helix axis of stem II is superimposed for all three conformations along the molecular z direction. (g) The average and allowed range of determined inter-helical angles (a). Order matrix analysis36,37 was carried out independently for stems I and II for both TAR(3.0 Mg) and TAR(4.5 Mg), closely following the procedure previously described for TAR(free)21(see Table 1). In all cases, repeating order matrix analyses when excluding data measured in terminal residues in various permutations had a negligible effect on the average values of derived structural and dynamic parameters. Stems I and II were rotated into the principal axis system (PAS) using the program Insight II, and one orientation among four degenerate solutions could be selected based on linkage geometry considerations.21 In particular, the translational disposition of the two stems was determined by fixing the distance between atoms U40 (P) and C39(O3′) at 1.59 Å . With the latter translational constraint, two of the four inter-stem orientations generated through 180° rotations about the Sxx and Syy directions could be discarded because they lead to anti-parallel stem alignments that is inconsistent with the secondary structure and sequence (terminal residues G17–C45 in stem I are oriented towards residues G26– C39 in stem II). Among the two remaining solutions involving180° rotation about Szz, one of the solutions could be discarded because it lead to a considerable distance between A22(O3′) and G26(P) (~35 Å and 33 Å for TAR(3.5 Mg) and TAR(4.5 Mg), respectively) which cannot be connected using a three-residue bulge (the same distance in A-form helices is only ~17 Å).
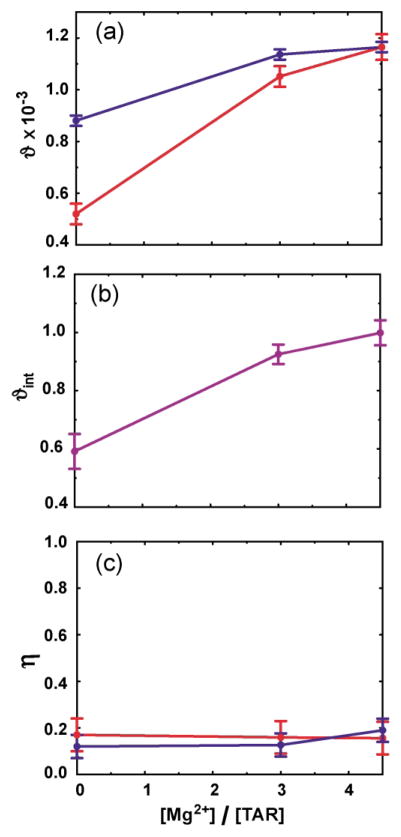
The generalized degree of order (ϑ)17 and asymmetry (η) are two additional stem-specific principal order parameters derived from the order matrix analysis,36 which describe the degree and asymmetry of molecular alignment, respectively. While these values should be identical for rigidly connected helices, inter-helical motions can lead to differences in a manner dependent on both motional amplitudes and directions.17,21 For TAR(free), inter-helical motions result in significant attenuation of ϑstemI relative to ϑstemII21 (Figure 3(a)). Remarkably, this difference, and hence the amount of inter-helical motions, is significantly diminished with increasing Mg2+ concentrations (Figure 3(a)). As can be seen in Figure 3(b), the internal generalized degree of order (ϑint = ϑstemI/ϑstemII) increases from 0.59(±0.06) in TAR(free) to 0.93(±0.03) in TAR(3.0 Mg), and 0.99(±0.03) in TAR(4.5 Mg) (Figure 3(b)). In contrast to the ϑ values, the computed stem-specific η values remain similar and only marginally change with increasing Mg2+ concentrations (Figure 3(c)). Once again, this overall trend provides strong evidence for a dramatic Mg2+-induced arrest of inter-helical motions in TAR(free).
Figure 3.
Principal order parameters and inter-helical dynamics in TAR as a function of [Mg]:[TAR] stoichiometry. (a) The generalized degree of order (ϑ) for stem I (red) and II (blue), (b) internal generalized degree of order (ϑint = ϑstem I/ϑstem II), (c) asymmetry parameter (η = (Syy − Sxx)/Szz).
As noted earlier, despite having almost identical 2H splittings and hence phage order in all of TAR(free), TAR(3.0 Mg) and TAR(4.5 Mg), Mg2+ induces an apparent ~32% increase in the total alignment of TAR. This can be seen as a Mg2+-induced increase in the ϑ value for the more ordered stem II (Figure 3(a)). A number of factors can give rise to this behavior. First, increase in the structural anisotropy of “linear” TAR(4.5 Mg) versus “bent” TAR(free) can result in an increase in molecular alignment. Second, and as discussed previously,21 motions of stem I in TAR(free) could in principle reduce the absolute alignment of stem II, an effect that is attenuated in the more rigid TAR(4.5 Mg). Finally, it is possible that quenching of electrostatic repulsions between TAR and phage through the observed Mg2+ binding to both phage and TAR allows for greater intermolecular proximity and hence greater TAR alignment.
In conclusion, we have described large Mg2+-induced variations in the structure and dynamics of TAR. Although the total number and coordination of bound Mg2+ along with the conformation of bulge residues remains to be investigated, our results show that Mg2+ significantly affects both the global conformation and dynamics of TAR, stabilizing a coaxially aligned conformation. It is noteworthy that based on our previous RDC study of TAR(free),21 this coaxially aligned conformation falls within the envelope of inter-helical conformations that are apparently sampled dynamically in TAR(free) (Figure 3(e)). We are now examining whether the Mg2+-induced changes in RDCs measured in the bulge residues correspond to changes in structure, dynamics, or both. The RDC-NMR approach outlined here should continue to illuminate the conformational transitions associated with RNA folding, catalysis, and recognition.
Acknowledgments
We thank Yuying Gosser for NOE spectra of TAR in the presence of Mg2+ and Maurice Gueron for useful comments. This research was supported by NIH CA46778.
Abbreviations used
- RDC
residual dipolar couplings
- TAR
transactivation response element
References
- 1.Misra VK, Draper DE. On the role of magnesium ions in RNA stability. Biopolymers. 1998;48:113–135. doi: 10.1002/(SICI)1097-0282(1998)48:2<113::AID-BIP3>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 2.Brion P, Westhof E. Hierarchy and dynamics of RNA folding. Annu Rev Biophys Biomol Struct. 1997;26:113–137. doi: 10.1146/annurev.biophys.26.1.113. [DOI] [PubMed] [Google Scholar]
- 3.Chen SJ, Dill KA. RNA folding energy landscapes. Proc Natl Acad Sci USA. 2000;97:646–651. doi: 10.1073/pnas.97.2.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Treiber DK, Williamson JR. Beyond kinetic traps in RNA folding. Curr Opin Struct Biol. 2001;11:309–314. doi: 10.1016/s0959-440x(00)00206-2. [DOI] [PubMed] [Google Scholar]
- 5.Hanna R, Doudna JA. Metal ions in ribozyme folding and catalysis. Curr Opin Chem Biol. 2000;4:166–170. doi: 10.1016/s1367-5931(99)00071-x. [DOI] [PubMed] [Google Scholar]
- 6.Scott WG. RNA structure, metal ions, and catalysis. Curr Opin Chem Biol. 1999;3:705–709. doi: 10.1016/s1367-5931(99)00029-0. [DOI] [PubMed] [Google Scholar]
- 7.Hermann T, Westhof E. Simulations of the dynamics at an RNA–protein interface. Nature Struct Biol. 1999;6:540–544. doi: 10.1038/9310. [DOI] [PubMed] [Google Scholar]
- 8.Hoogstraten CG, Wank JR, Pardi A. Active site dynamics in the lead-dependent ribozyme. Biochemistry. 2000;39:9951–9958. doi: 10.1021/bi0007627. [DOI] [PubMed] [Google Scholar]
- 9.Palmer AG, III, Kroenke CD, Loria JP. Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Methods Enzymol. 2001;339:204–238. doi: 10.1016/s0076-6879(01)39315-1. [DOI] [PubMed] [Google Scholar]
- 10.Dayie KT, Brodsky AS, Williamson JR. Base flexibility in HIV-2 TAR RNA mapped by solution (15)N, (13)C NMR relaxation. J Mol Biol. 2002;317:263–278. doi: 10.1006/jmbi.2001.5424. [DOI] [PubMed] [Google Scholar]
- 11.Stryer L, Haugland RP. Energy transfer: a spectroscopic ruler. Proc Natl Acad Sci USA. 1967;58:719–726. doi: 10.1073/pnas.58.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weiss S. Fluorescence spectroscopy of single biomolecules. Science. 1999;283:1676–1683. doi: 10.1126/science.283.5408.1676. [DOI] [PubMed] [Google Scholar]
- 13.Tolman JR, Flanagan JM, Kennedy MA, Prestegard JH. Nuclear magnetic dipole interactions in field-oriented proteins—information for structure determination in solution. Proc Natl Acad Sci USA. 1995;92:9279–9283. doi: 10.1073/pnas.92.20.9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tjandra N, Bax A. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science. 1997;278:1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- 15.Tolman JR, Flanagan JM, Kennedy MA, Prestegard JH. NMR evidence for slow collective motions in cyanometmyoglobin. Nature Struct Biol. 1997;4:292–297. doi: 10.1038/nsb0497-292. [DOI] [PubMed] [Google Scholar]
- 16.Prestegard JH, Al-Hashimi HM, Tolman JR. NMR structures of biomolecules using field oriented media and residual dipolar couplings. Q Rev Biophys. 2000;33:371–424. doi: 10.1017/s0033583500003656. [DOI] [PubMed] [Google Scholar]
- 17.Tolman JR, Al-Hashimi HM, Kay LE, Prestegard JH. Structural and dynamic analysis of residual dipolar coupling data for proteins. J Am Chem Soc. 2001;123:1416–1424. doi: 10.1021/ja002500y. [DOI] [PubMed] [Google Scholar]
- 18.Meiler J, Prompers JJ, Peti W, Griesinger C, Bruschweiler R. Model-free approach to the dynamic interpretation of residual dipolar couplings in globular proteins. J Am Chem Soc. 2001;123:6098–6107. doi: 10.1021/ja010002z. [DOI] [PubMed] [Google Scholar]
- 19.Bondensgaard K, Mollova ET, Pardi A. The global conformation of the hammerhead ribozyme determined using residual dipolar couplings. Biochemistry. 2002;41:11532–11542. doi: 10.1021/bi012167q. [DOI] [PubMed] [Google Scholar]
- 20.Sibille N, Pardi A, Simorre JP, Blackledge M. Refinement of local and long-range structural order in theophylline-binding RNA using C-13–H-1 residual dipolar couplings and restrained molecular dynamics. J Am Chem Soc. 2001;123:12135–12146. doi: 10.1021/ja011646+. [DOI] [PubMed] [Google Scholar]
- 21.Al-Hashimi HM, Gosser Y, Gorin A, Hu W, Majumdar A, Patel DJ. Concerted motions in HIV-1 TAR RNA may allow access to bound state conformations: RNA dynamics from NMR residual dipolar couplings. J Mol Biol. 2002;315:95–102. doi: 10.1006/jmbi.2001.5235. [DOI] [PubMed] [Google Scholar]
- 22.Puglisi JD, Tan R, Calnan BJ, Frankel AD, Williamson JR. Conformation of the TAR RNA–arginine complex by NMR spectroscopy. Science. 1992:257. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]
- 23.Aboul-ela F, Karn J, Varani G. Structure of HIV-1 TAR RNA in the absence of ligands reveals a novel conformation of the trinucleotide bulge. Nucl Acids Res. 1996;24:3974–3981. doi: 10.1093/nar/24.20.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Long KS, Crothers DM. Characterization of the solution conformations of unbound and Tat peptide-bound forms of HIV-1 TAR RNA. Biochemistry. 1999;38:10059–10069. doi: 10.1021/bi990590h. [DOI] [PubMed] [Google Scholar]
- 25.Zacharias M, Hagerman PJ. The bend in RNA created by the transactivation response element bulge of human-immunodeficiency-virus is straightened by arginine and by tat-derived peptide. Proc Natl Acad Sci USA. 1995;92:6052–6056. doi: 10.1073/pnas.92.13.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cullen BR. Trans-activation of human immunodeficiency virus occurs via a bimodal mechanism. Cell. 1986;46:973–982. doi: 10.1016/0092-8674(86)90696-3. [DOI] [PubMed] [Google Scholar]
- 27.Gallego J, Varani G. Targeting RNA with small-molecule drugs: therapeutic promise and chemical challenges. Acc Chem Res. 2001;34:836–843. doi: 10.1021/ar000118k. [DOI] [PubMed] [Google Scholar]
- 28.Hermann T, Patel DJ. Adaptive recognition by nucleic acid aptamers. Science. 2000;287:820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- 29.Leulliot N, Varani G. Current topics in RNA–protein recognition: control of specificity and biological function through induced fit and conformational capture. Biochemistry. 2001;40:7947–7956. doi: 10.1021/bi010680y. [DOI] [PubMed] [Google Scholar]
- 30.Puglisi JD, Chen L, Frankel AD, Williamson JR. Role of RNA structure in arginine recognition of TAR RNA. Proc Natl Acad Sci USA. 1993;90:3680–3684. doi: 10.1073/pnas.90.8.3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arzumanov A, Godde F, Moreau S, Toulme JJ, Weeds A, Gait MJ. Use of the fluorescent nucleoside analogue benzo[g ]quinazoline 2′-O-methyl-beta-D-ribofuranoside to monitor the binding of the HIV Tat protein or of anitsense oligonucleotides to the TAR RNA stem-loop. Helv Chim Acta. 2000;83:1424–1436. [Google Scholar]
- 32.Olejniczak M, Gdaniec Z, Fischer A, Grabarkiewicz T, Bielecki L, Adamiak RW. The bulge region of HIV-1 TAR RNA binds metal ions in solution. Nucl Acids Res. 2002;30:4241–4249. doi: 10.1093/nar/gkf541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ippolito JA, Steitz TA. A 1.3-angstrom resolution crystal structure of the HIV-1 trans-activation response region RNA stem reveals a metal ion-dependent bulge conformation. Proc Natl Acad Sci USA. 1998;95:9819–9824. doi: 10.1073/pnas.95.17.9819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hansen MR, Mueller L, Pardi A. Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nature Struct Biol. 1998;5:1065–1074. doi: 10.1038/4176. [DOI] [PubMed] [Google Scholar]
- 35.Hansen MR, Hanson P, Pardi A. Filamentous bacteriophage for aligning RNA, DNA, and proteins for measurement of nuclear magnetic resonance dipolar coupling interactions. Method Enzymol. 2000;317:220–240. doi: 10.1016/s0076-6879(00)17017-x. [DOI] [PubMed] [Google Scholar]
- 36.Saupe A. Recent results in the field of liquid crystals. Angew Chem, Int Ed Engl. 1968;7:97–112. [Google Scholar]
- 37.Losonczi JA, Andrec M, Fischer MWF, Prestegard JH. Order matrix analysis of residual dipolar couplings using singular value decomposition. J Magn Reson. 1999;138:334–342. doi: 10.1006/jmre.1999.1754. [DOI] [PubMed] [Google Scholar]
- 38.Mollova ET, Hansen MR, Pardi A. Global structure of RNA determined with residual dipolar couplings. J Am Chem Soc. 2000;122:11561–11562. [Google Scholar]
- 39.Al-Hashimi HM, Gorin A, Majumdar A, Gosser Y, Patel DJ. Towards structural genomics of RNA: Rapid NMR resonance assignment and simultanous RNA tertiary structure determination using residual dipolar couplings. J Mol Biol. 2002;318:637–649. doi: 10.1016/S0022-2836(02)00160-2. [DOI] [PubMed] [Google Scholar]
- 40.Ottiger MDF, Bax A. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J Magn Reson. 1998;131:373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]