Abstract
It has now been nearly two decades since the first solution structures of DNA duplexes covalently damaged by metabolically activated polycyclic aromatic hydrocarbons and amines were determined by NMR. Dozens of such high-resolution structures are now available, and some broad structural themes have been uncovered. It has been hypothesized that the solution structures are relevant to the biochemical processing of the adducts. The structural features of the adducts are considered to determine their mutational properties in DNA polymerases and their repair susceptibilities. In recent years, a number of crystal structures of DNA adducts of interest to our work have been determined in DNA polymerases. Accordingly, it is now timely to consider how NMR solution structures relate to structures within DNA polymerases. The NMR solution structural themes for polycyclic aromatic adducts are often observed in polymerase crystal structures. While the polymerase interactions can on occasion override the solution preferences, intrinsic adduct conformations favored in solution are often manifested within polymerases and likely play a significant role in lesion processing.
1. Introduction
Polycyclic aromatic hydrocarbons (PAHs)1 and amines are well-known environmental chemicals present in tobacco smoke, automobile and diesel exhaust, and broiled fish and meats (1, 2). Metabolic activation of these substances can produce highly reactive products, which may subsequently react with DNA, yielding carcinogen–DNA adducts (1, 3, 4). These adducts, if not accurately repaired by the nucleotide excision repair machinery (5), may produce mutations during DNA replication. Such mutations, when present in DNA sequences that function in regulating the cell cycle, may lead to the initiation of cancer (1, 6).
It has now been nearly two decades since the first structures of DNA duplexes covalently damaged by metabolically activated PAHs and amines were determined by NMR solution methods. Dozens of such high-resolution structures are now available, and some broad structural themes have been uncovered. It has been hypothesized that the solution structures are relevant to the biochemical functioning of the adducts. The structural features of the adducts are considered to determine their mutational properties in DNA polymerases and their repair susceptibilities. Extensive reviews of NMR solution structures of damaged DNA have been presented by Geacintov et al. (7) and Patel et al. (8); recently, Cho has reviewed evidence for conformational heterogeneity in adduct structures (9), and Lukin and de Los Santos have presented a comprehensive survey of NMR structures of damaged DNA (10). In recent years, a number of crystal structures of DNA adducts of interest to our work have been determined in DNA polymerases (11–15). Accordingly, it now seems timely to consider how NMR solution structures relate to structures within DNA polymerases.
Of particular interest are adducts with available NMR solution structures as well as X-ray crystal structures within polymerases. Among the PAHs, we focus on benzo[a]pyrene (B[a]P) and benzo[c]phenanthrene (B[c]Ph); among the aromatic amines, we focus on 2-aminofluorene (AF) and N-2-acetyl-aminofluorene (AAF), with some selected consideration of other aromatic amine adducts for which NMR data are at hand. We first review conformational themes defined in NMR studies.
2. NMR Structural Themes
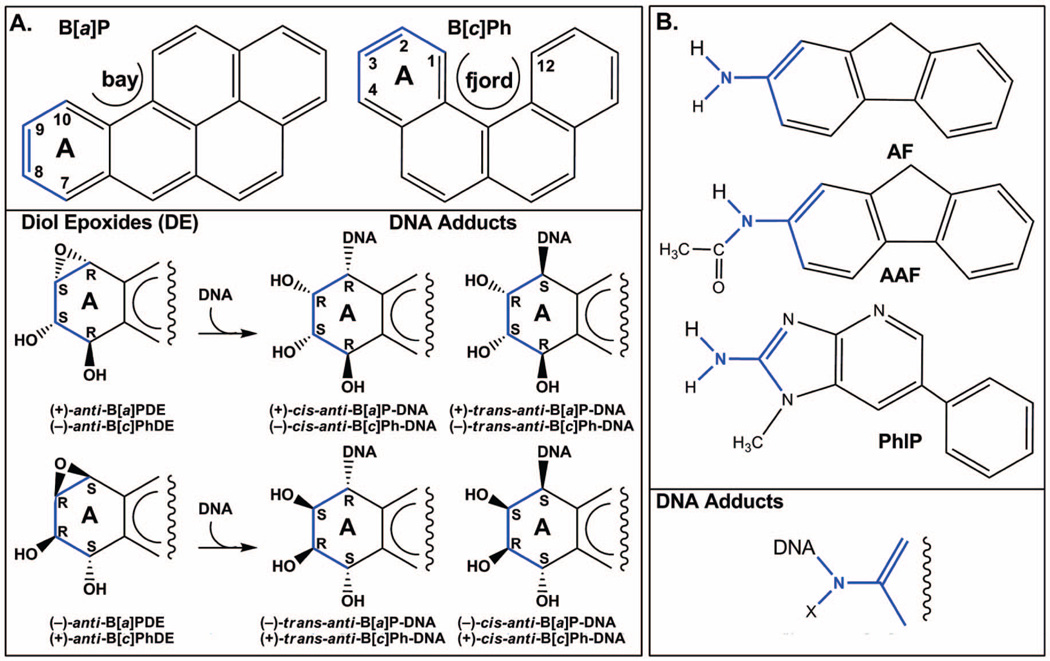
B[a]P and B[c]Ph represent two model PAHs with uniquely different topological features (Figure 1A, upper panel). The planar B[a]P has a characteristic bay region, while B[c]Ph, one of the most tumorigenic PAHs (16), has a sterically hindered fjord region, which causes nonplanarity in the aromatic ring system (17). In addition, the B[a]P ring system is extended while that of B[c]Ph is curved. Metabolic activation of B[a]P and B[c]Ph produces, among others, a pair of mirror image diol epoxides in each case (Figure 1A, lower panel); these can react with the amino groups of adenine and guanine by trans- and cis-epoxide opening to produce adducts differing in base-linkage site, stereochemistry, and ring topology (18).
Figure 1.
Structures of representative PAHs (A) and polycyclic aromatic amines (B), and their DNA adducts. PhIP is a heterocyclic aromatic amine. PAHs are adducted to DNA via the N2 of guanine or the N6 of adenine. Polycyclic aromatic amines are adducted to DNA via the C8 of G. Chiral centers in the A ring of the PAH adducts and the linkage site of the polycyclic aromatic amine adducts are shown in blue. X is H or COCH3.
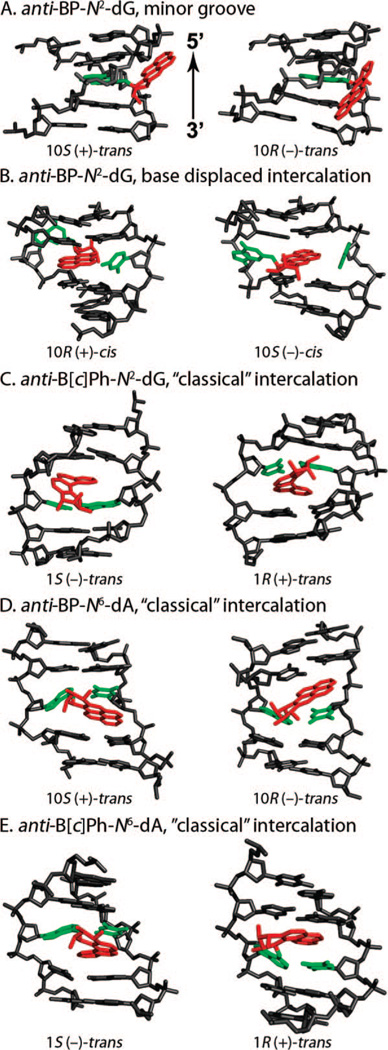
NMR conformational themes that have been defined for these adducts (7, 10) are shown in Figure 2. For guanine (G) adducts derived from B[a]P, trans adducts in duplexes prefer to reside in the B-DNA minor groove with Watson–Crick pairing at the lesion site retained. The 10S adduct is oriented in the 5′-direction along the modified strand, while the 10R adduct is 3′-oriented (Figure 2A). The cis adducts favor a base-displaced intercalated conformation with Watson–Crick pairing disrupted at the modification site and the pyrenyl ring system inserted in place of the modified G and its partner cytosine (C), both of which are displaced into the major groove (Figure 2B) in the (−)-cis case. However, the displaced G is in the minor groove in the case of the (+)-cis adduct. Here, opposite orientations in 10S and 10R stereoisomeric adducts are manifested by protrusion of the puckered benzylic ring into the minor groove with the pyrenyl rings directed towards the major groove in the 10S adduct case and visa versa for the 10R adduct. There is evidence that equilibria between these conformational families are governed by base-sequence context in both cis and trans adducts (7, 10, 19, 20). The B[c]Ph G adducts adopt a third conformational theme, “classical intercalation”, with perturbed Watson–Crick base pairing retained at the lesion site and the aromatic ring system intercalated on the 5′-side of the modified base pair in the 1S case and on the 3′-side in the 1R case (Figure 2C). The helix is stretched and unwound to permit insertion of the aromatic rings. In the case of the adenine adducts, classical intercalation themes are observed in lesions derived from both B[a]P (Figure 2D) and B[c]Ph (Figure 2E), with 3′-side intercalation for 10S-B[a]P and 1S-B[c]Ph adducts and 5′-side intercalation for the respective R stereoisomeric adducts. While the overall conformational family is the same in the B[a]P and B[c]Ph adducts, there are important differences in detail stemming from the topological differences of the bay and fjord region adducts, which profoundly affect their repair susceptibilies, as has been discussed previously (21–23).
Figure 2.
Conformational themes for B[a]P and B[c]Ph adducts in duplex DNA in solution. The 5′- to 3′-directionality of all modified strands is given in panel A.
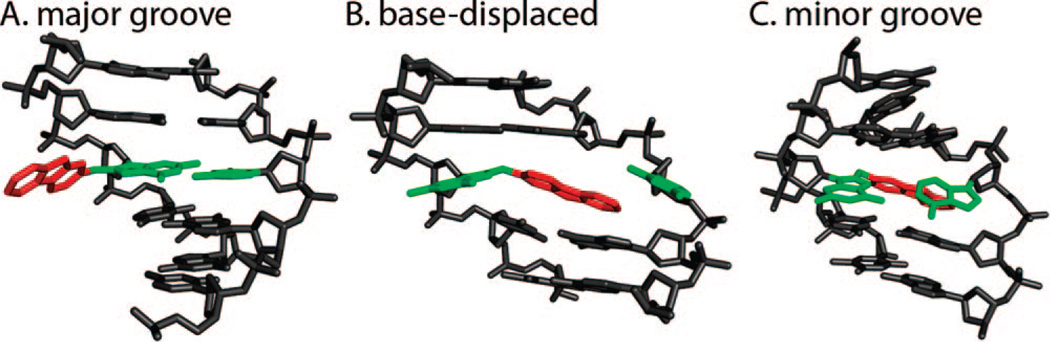
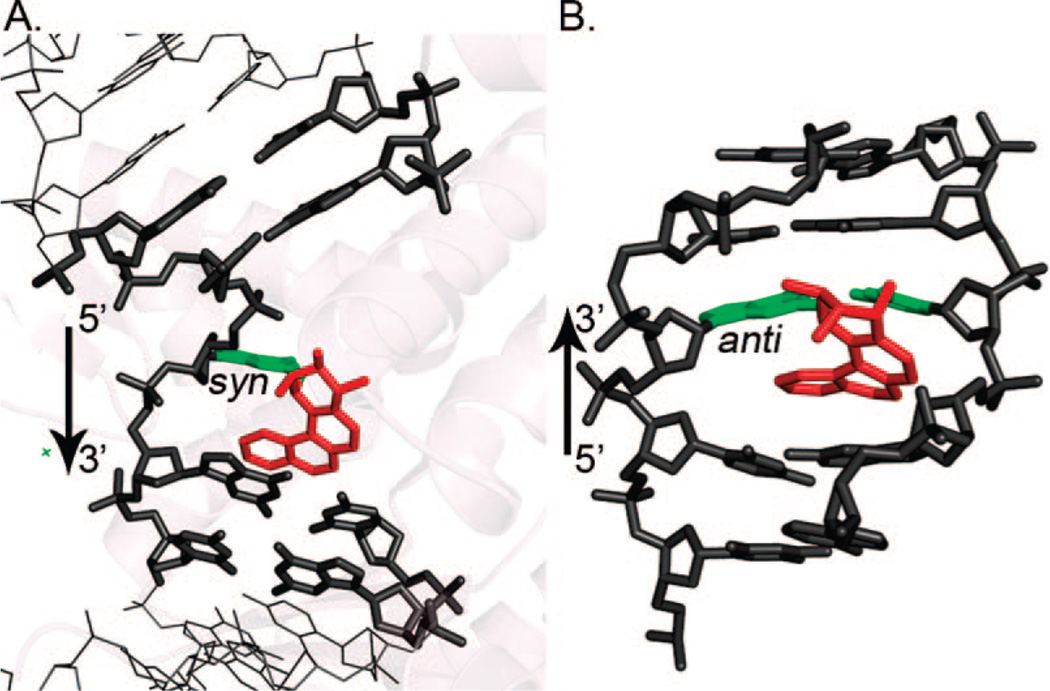
A second group of chemicals of interest in our structural studies are the polycyclic aromatic amines (Figure 1B, upper panel). Their metabolites form predominant adducts to the C8 of G (Figure 1B, lower panel), and additional minor products have also been observed (8, 10). The AF adducts have been investigated most extensively, and their representative structural themes have been defined (8, 9). Three conformational families have been observed as follows: (i) aromatic rings in the B-DNA major groove with Watson–Crick pairing intact and modified G anti (Figure 3A); (ii) base-displaced intercalated structures with Watson–Crick pairs disrupted at the lesion site, with aromatic rings inserted into the helix and the modified G in the syn conformation (Figure 3B); and (iii) adducts positioned in the B-DNA minor groove and with the modified G syn, characterized for the case of a G:A mismatch at the lesion site (Figure 3C). The equilibrium between major groove/anti and base-displaced intercalated/syn conformers is determined by base-sequence context (8), with enhanced carcinogen/base stacking likely accounting for preferred stabilization of the base-displaced structures in certain sequences. The conformational balance is dependent on ring topology as well. Thus, the 1-aminopyrene adduct with its multiple rings shows only a syn/base-displaced intercalated conformation (24). In the case of the 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) adduct, this conformation is about 90% of the population with the adduct in the remaining population solvent-exposed in a groove (25). For the 2-amino-3-methylimidazo[4,5-f]quinoline adduct, base-sequence context also plays a key role in determining the conformational balance between major groove and base-displaced intercalated conformations (26, 27). The flexible 4-aminobiphenyl adduct is predominantly in the major groove (28). The AAF adduct adopts a syn/base-displaced intercalated conformation in about 70% of the population; the conformations of the remaining adducts are uncharacterized (29). The syn conformation is particularly preferred by the AAF adduct due to crowding of the bulky acetyl group with adjacent sugar when the modified G is anti, as first noted by Grunberger et al. (30) and by Fuchs and Daune (31) in their base-displacement and insertion-denaturation models. However, molecular dynamics simulations show that modest sugar repuckering can alleviate the crowding and permit the anti conformation, thereby allowing Watson–Crick pairing (32). Observed preferential incorporation of dCTP opposite dG-AAF in DNA polymerases (33–37) is thus structurally explained (32).
Figure 3.
Conformational themes for AF adducts in duplex DNA in solution.
The variety of conformational themes adopted by these bulky aromatic adducts is governed by a subtle interplay of competing forces. To accommodate the adducted base, an energetic price is exacted in distorting the DNA through disturbed stacking, Watson–Crick pairing, and other perturbations such as stretching and unwinding. However, shielding of the adduct aromatic rings from solvent and/or stacking them with bases within the DNA helix is favorable. Van der Waals and electrostatic interactions between the adduct and the DNA backbone can also provide stabilizing forces. Thus, conformational preferences are a combined outcome of structural factors including adduct topology, nature of modified base (adenine or guanine), linkage site (e.g., C8 or N2 of G), and base-sequence context that together determine the minimum free energy conformation.
3. DNA Polymerases
Crystallographic analyses of a number of replicative DNA polymerases have yielded excellent structures that defined common features of these enzymes. They are shaped like a right hand with palm, finger, and thumb domains, with the active site located in the palm domain (38). Replicative DNA polymerases employ an induced-fit mechanism in which the polymerase closes largely through rotation of the finger domain when the Watson–Crick partner to the template enters the active site. An α-helix of the finger domain closes around the nascent base pair with a tight steric fit (39). The resulting active site is catalytically competent and ready for the nucleotidyl transfer reaction. Two metal ions, usually Mg2+, are critical to the catalytic mechanism, as are conserved carboxylate residues (Asp or Glu) (40). These polymerases have open pockets on the developing major groove side of the templating base. However, the minor groove side is tightly packed with amino acid residues; these protein–DNA interactions are essential for processivity and fidelity (41–43). The repair polymerase β (Pol β), a member of the X family, has overall structural and mechanistic features that are similar, although the domains differ (44).
When replicative polymerases encounter a lesion, they are frequently stalled and replaced by specialized lesion-bypass polymerases, many of which are from the Y family (45–48). One or more bypass polymerases are involved in transiting the lesion and extending several residues past it before the replicative polymerase resumes replication. Bypass polymerases are typically low fidelity on unmodified DNA and may be high or low fidelity in lesion bypass (43). Crystallographic characterizations of the Y family polymerase Dpo4, from the archaeon bacterium Sulfolobus solfataricus (14, 49–60), have provided structural understanding of this DinB homologue (37) of the human polymerase κ (61). Like replicative polymerases, Dpo4 is shaped like a right hand with an open pocket on the developing major groove side of the template and employs a two-metal ion mechanism for nucleotidyl transfer. However, the finger domain of Dpo4 is short with an additional flexible little finger domain holding the DNA backbone from the major groove side. The spacious active site, open and water-accessible on both the major and the minor groove side of the template, is capable of accommodating two templating residues. Entry of the correct dNTP partner to the template does not trigger an induced fit closing motion. Instead, a motion of translocation involving movement of the little finger and subsequently that of the thumb, relative to the DNA, is observed as the DNA is elongated by one residue in a replication cycle. This process produces a counter-clockwise rotation (when the ternary complex is viewed in the 5′- to 3′-direction of the template strand) and a concerted translation (in the 3′- to 5′-direction of the template) of the polymerase (57). A detailed chemical mechanism for nucleotidyl transfer in Dpo4 has recently been delineated (62). Recent crystal structures of human Y family polymerases Pol ι (63–66) and Pol κ (67) reveal significant similarities as well as individual structural features tailored to the specific enzyme function in nucleotide incorporation opposite lesions or extension beyond lesions.
4. Adduct Structures in DNA Polymerases
We now turn to structures of bulky adducts of interest within DNA polymerases. In this connection, the representative Pol I A family replicative polymerase Bacillus fragment (BF) from Bacillus stearothermophilus has provided valuable crystallographic insights. Crystal structures of BF without lesion have identified a series of positions in its active site region that the templating base traverses through a replicative cycle (68). The next-to-be replicated base occupies the preinsertion site, between the O and the O1 helices, in the open binary complex; Tyr 714 occupies the insertion position. At the next step, the template and its partner dNTP are placed in the insertion site in the closed ternary complex; the O and O1 helices have rotated, thus closing the active site of the polymerase, and Tyr 714 has vacated the active site. Following the nucleotidyl transfer reaction, the nascent base pair is translocated to the postinsertion site with the next base (5′ to the nascent base pair on the template strand) occupying the preinsertion site in the open polymerase binary complex, ready for another cycle of replication.
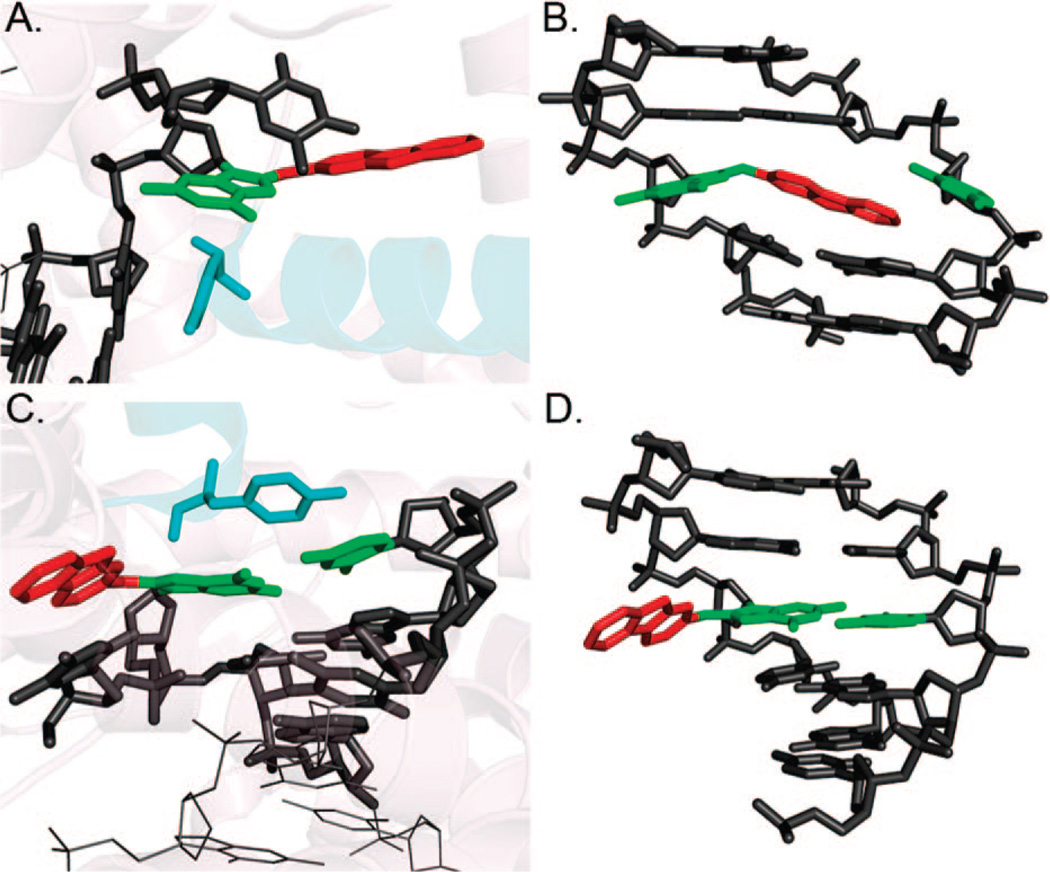
A crystal structure of the 10S-(+)-trans-anti-B[a]P-N2-dG adduct in the postinsertion site of BF (11) has been solved (PDB ID:1XC9). In this structure, the B[a]P rings are 5′-directed along the template strand on the minor groove side with the Watson–Crick pair intact at the lesion site (Figure 4A), as in the DNA duplex structure solved by NMR (Figure 2A, left panel), although the minor groove is not fully developed in the polymerase structure and detailed orientation of the B[a]P rings differs somewhat. However, an NMR solution structure of this adduct at a single-strand/double-strand junction with dC incorporated opposite the damaged guanine (PDB ID: 1AXO) (69) is very similar in orientational detail (Figure 4B). The B[a]P disrupts the critical polymerase minor groove contacts, hence accounting for the predominant blockage observed in primer extension studies of this adduct with BF. Small amounts of bypass are also experimentally noted and have been interpreted through modeling to entail a syn conformation of the damaged base with the B[a]P rings in the open pocket on the major groove side (70).
Figure 4.
Structures of the 10S-(+)-trans-anti-B[a]P-N2-dG adduct (A) in polymerase BF (PDB ID:1XC9) and (B) at a single-strand/double-strand junction in solution (PDB ID:1AXO).
Two different structures of dG–AF adducts in DNA–BF complexes have also been studied (12), with AF in the preinsertion site in one structure (PDB ID:1UA0) and in the postinsertion site in the other (PDB ID:1UA1). Interestingly, the adducted base is replicated by the polymerase, transiting from preinsertion to insertion and postinsertion sites. In the preinsertion site, the modified guanine is syn, and the AF rings are accommodated in the preinsertion site with extensive van der Waals interactions, with the syn-G stacked with the adjacent sugar (Figure 5A). While the O1 helix is disordered, the O helix remains ordered, open, and properly positioned for transfer of the adducted base to the insertion site. The polymerase is otherwise little disturbed. In transiting to the postinsertion site, however, the modified G is rotated to anti, and Watson–Crick pairing is intact with the AF rings in the major groove pocket (Figure 5C). The active site regions are markedly distorted; the next templating base is stacked with the fluorenyl rings and is blocked from entering the preinsertion site. The O and O1 helices are also distorted, and Tyr 714 is displaced. These structures explain replication of the modified G and also account for the observed predominant blockage of further primer extension. A surprising syn–anti rotation occurs in the polymerase, as was also observed in the NMR solution structures of this adduct. In the postinsertion site, the structure is very similar to the major groove NMR solution structure. The conformation in the preinsertion site where the modified G is single-stranded resembles the AF base-displaced intercalated and minor groove structures in that they share the syn conformation. The existence of a syn–anti glycosidic bond rotation in the polymerase, as in solution, is remarkable, as is the similarity between the NMR solution and the polymerase postinsertion site structures.
Figure 5.
Structures of dG-C8-AF in polymerase BF at (A) preinsertion (PDB ID:1UA0) and (C) postinsertion (PDB ID:1UA1) sites, respectively. The corresponding conformational themes in solution are shown in panels B and D, respectively. Tyr714 of the O helix is shown in cyan.
A crystal structure of a dG–AAF adduct in BF in the preinsertion site was also investigated and was found to have an open polymerase conformation with a well-ordered preinsertion site that was empty (12). However, the Pol I A family polymerase from bacteriophage T7 did yield a structure of the adduct (PDB ID: 1X9M) (13). The intent was to crystallize a ternary complex with templating dG–AAF and incoming ddCTP. However, no incoming nucleotide was observed in the crystal. The dG and adjacent sugar were located on the surface of the finger subdomain, flipped out of the polymerase active site in the syn conformation. The AAF rings were inserted into a hydrophobic pocket behind the O helix, which was rotated about 35° from the closed position that it adopts in the unmodified ternary complex. The site for binding the incoming nucleotide is blocked by the displaced O helix. This structure thus explains the predominant blockage one base before this lesion in T7 DNA polymerase. The strongly preferred syn conformation of this adduct in solution (29) is manifested in the polymerase.
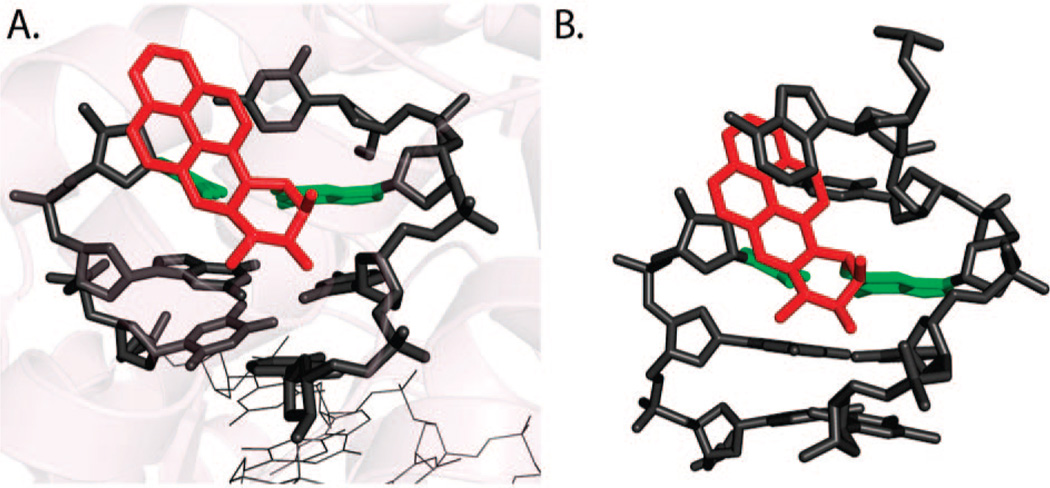
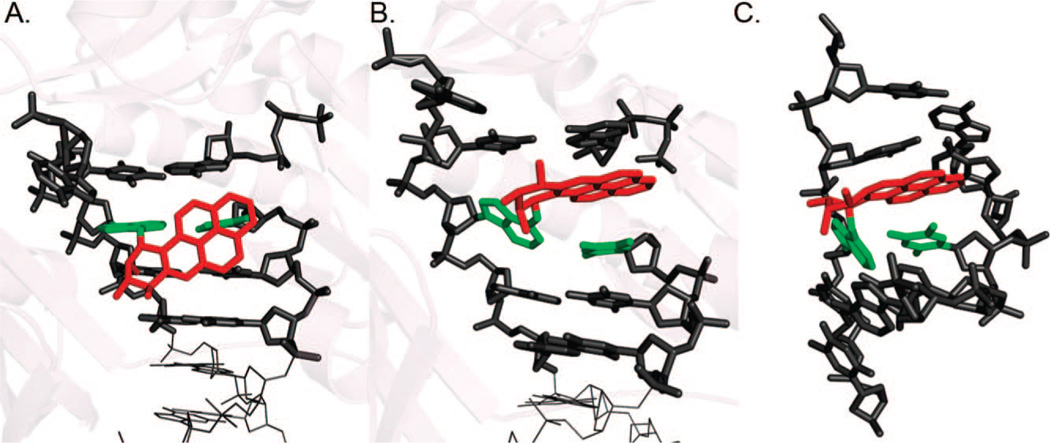
A recent crystal structure has been solved for Pol β, in which a 1S-(−)-trans-anti-B[c]Ph-N2-dG adduct is the templating base in a DNA substrate with a one nucleotide gap (PDB ID: 2I9G) (15). The phenanthrenyl ring system is stacked over the base pair immediately 3′ to the modified deoxyguanosine, which is in the syn conformation (Figure 6A). This base is displaced downstream and prevents the polymerase from closing to form a catalytically active complex. Nonetheless, inefficient and error-prone bypass was observed with preferred misinsertion of purine opposite the lesion; it was suggested that the bypass might be untemplated and favored by the enlarged binding site and purine stacking with the aromatic B[c]Ph rings. Interestingly, a similar carcinogen–base linkage geometry, defined by torsion angles α′ [N1(G)–C2(G)–N2(G)–C1(B[c]Ph)] and β′ [C2(G)–N2(G)–C1(B[c]Ph)–C2(B[c]Ph)], was found in the NMR solution structure of this adduct (PDB ID:1HWV), although the modified nucleoside adopted the anti conformation and Watson–Crick pairing was observed in the NMR structure (Figure 6B) (71).
Figure 6.
Structures of the 1S-(−)-trans-anti-B[c]Ph-N2-dG adduct (A) in Pol β (PDB ID:2I9G) and (B) in duplex DNA in solution (PDB ID:1HWV).
In the Y family bypass polymerase, Dpo4, crystal structures have been solved for the 10R-(+)-cis-anti-B[a]P-N6-dA adduct (PDB ID:1S0M) (14), which has also been characterized by high-resolution NMR (PDB ID:1AXV) (72). In the crystal, the modified anti-adenine was in the postinsertion site opposite T. Two different conformers were found. In one conformation, the pyrenyl rings occupied the major groove open pocket with little perturbation to the polymerase; this conformer appeared to be fairly near-reaction ready with a distance between primer terminal O3′ and Pα of the dNTP < 5 Å (Figure 7A). The second conformer was nearly identical to the NMR solution structure, with pyrenyl rings intercalated on the 5′-side of the damaged base whose Watson–Crick pairing was perturbed but not ruptured. In this case, the O3′–Pα distance was >10 Å, which would preclude the nucleotidyl transfer reaction (Figure 7B). Ling et al. (14) suggest that poor extension beyond the adduct in Dpo4 occurs because the major groove conformer that appears most catalytically competent is in an energetically unfavorable domain in solution (73) and would occur less frequently. However, in Dpo4, the energetic advantage derived from the comfortable housing in the Dpo4 major groove pocket offsets the energy penalty.
Figure 7.
Structures of the 10R-(+)-cis-anti-B[a]P-N6-dA adduct (A, B) in polymerase Dpo4 (PDB ID:1S0M) and (C) in duplex DNA in solution (PDB ID:1AXV).
A recent series of crystal structures of Dpo4 containing three steps in the replicative cycle (a short binary complex, a ternary complex with dNTP, and a long binary complex following reaction, which is elongated by one nucleotide) have revealed the mechanism of translocation in this enzyme (57). The consecutive motion of the little finger and thumb, relative to the DNA, that was revealed provided insights into how bulky adducts may affect translocation in this enzyme. Modeling studies for the PhIP-C8-dG, AAF-C8-dG, and 10S-(+)-transanti-B[a]P-N2-dG adducts suggested that translocation would be much more impeded by the multiringed C8–dG adducts on the major groove side of the modified template as compared to the N2–dG adduct on the minor groove side (27, 74). The key feature is the steric blockage of the little finger rotation by the C8–dG major groove adducts but not the minor groove N2–dG adduct (Figure 8A). Primer extension studies in DinB family polymerases have shown more facile bypass of N2–dG than C8–dG adducts (Figure 8B) (37, 75, 76), in line with these modeling studies. Future investigations of translocation mechanisms in the face of bulky adducts have been facilitated by these Dpo4 crystal structures.
Figure 8.
Models of translocation blockage in polymerase Dpo4: (A) major groove positioned anti-dG-C8-PhIP adduct and (B) minor groove positioned 10S-(+)-trans-anti-B[a]P-N2-dG adduct in Dpo4. The Dpo4 little finger domain is shown in purple.
5. Conclusion
In conclusion, it appears on the basis of present evidence that NMR solution structural themes for polycyclic aromatic adducts are often observed in crystal structures of DNA polymerases. Striking similarities are observed for B[a]P-, B[c]Ph-, AAF-, and AF-derived adducts. However, polymerase interactions may at times override the intrinsic preferences in solution, even permitting an unfavorable solution structure to occur within the polymerase (14, 73). Favorable steric factors and hydrophobic interactions involving the bulky adduct are important considerations in this connection. Solvent exposure of the polycyclic aromatic ring system may be minimized through interactions with specific niches of the polymerase, as has recently been observed for a 10S-(+)-trans-anti-B[a]P-N2-dG adduct in the postinsertion site in Dpo4 (77, 78). Interestingly, in this case, too, the carcinogen–base linkage site adopts conformational domains similar to those in the NMR solution structure (19). The adduct structures in polymerases have begun to provide structural understanding of polymerase blockage and bypass of bulky lesions. In turn, modeling studies are beginning to take advantage of this knowledge to evaluate structural properties producing blockage or bypass by these types of lesions (70, 74, 79). Future structural studies within polymerases are eagerly awaited to provide further insights.
Acknowledgments
This work is supported by NIH Grants CA28038 and CA75449 to S.B., CA99194 to N.E.G., and CA46533 to D.J.P. Molecular images were made with PyMOL (DeLano Scientific, LLC).
Footnotes
Abbreviations: PAHs, polycyclic aromatic hydrocarbons; B[a]P, benzo[a]pyrene; B[c]Ph, benzo[c]phenanthrene; G, guanine; C, cytosine; AF, 2-aminofluorene; AAF, 2-acetylaminofluorene; PhIP, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine; Pol β, DNA polymerase β; dG-C8-PhIP, N2-(2′-deoxyguanosin-8-yl)-PhIP; dG, 2′-deoxyguanosine; dG-C8-AF, N2-(2′-deoxyguanosin-8-yl)-2-aminofluorene; BF, Bacillus fragment; PDB, Protein Data Bank; dG-C8-AAF, N2-(2′-deoxyguanosin-8-yl)-2-acetylaminofluorene.
References
- 1.Luch A. Nature and nurture—Lessons from chemical carcinogenesis. Nat. Rev. Cancer. 2005;5:113–125. doi: 10.1038/nrc1546. [DOI] [PubMed] [Google Scholar]
- 2.Turesky RJ. Formation and biochemistry of carcinogenic heterocyclic aromatic amines in cooked meats. Toxicol. Lett. 2007;168:219–227. doi: 10.1016/j.toxlet.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 3.Xue W, Warshawsky D. Metabolic activation of polycyclic and heterocyclic aromatic hydrocarbons and DNA damage: A review. Toxicol. Appl. Pharmacol. 2005;206:73–93. doi: 10.1016/j.taap.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 4.Luch A. The mode of action of organic carcinogens on cellular structures. EXS. 2006:65–95. doi: 10.1007/3-7643-7378-4_4. [DOI] [PubMed] [Google Scholar]
- 5.Gillet LC, Schärer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem. Rev. 2006;106:253–276. doi: 10.1021/cr040483f. [DOI] [PubMed] [Google Scholar]
- 6.Poirier MC. Chemical-induced DNA damage and human cancer risk. Nat. Rev. Cancer. 2004;4:630–637. doi: 10.1038/nrc1410. [DOI] [PubMed] [Google Scholar]
- 7.Geacintov NE, Cosman M, Hingerty BE, Amin S, Broyde S, Patel DJ. NMR solution structures of stereoisometric covalent polycyclic aromatic carcinogen-DNA adduct: Principles, patterns, and diversity. Chem. Res. Toxicol. 1997;10:111–146. doi: 10.1021/tx9601418. [DOI] [PubMed] [Google Scholar]
- 8.Patel DJ, Mao B, Gu Z, Hingerty BE, Gorin A, Basu AK, Broyde S. Nuclear magnetic resonance solution structures of covalent aromatic amine-DNA adducts and their mutagenic relevance. Chem. Res. Toxicol. 1998;11:391–407. doi: 10.1021/tx9702143. [DOI] [PubMed] [Google Scholar]
- 9.Cho BP. Dynamic conformational heterogeneities of carcinogen-DNA adducts and their mutagenic relevance. J. Environ. Sci. Health, Part C. 2004;22:57–90. doi: 10.1081/LESC-200038217. [DOI] [PubMed] [Google Scholar]
- 10.Lukin M, de Los Santos C. NMR structures of damaged DNA. Chem. Rev. 2006;106:607–686. doi: 10.1021/cr0404646. [DOI] [PubMed] [Google Scholar]
- 11.Hsu GW, Huang X, Luneva NP, Geacintov NE, Beese LS. Structure of a high fidelity DNA polymerase bound to a benzo[a]pyrene adduct that blocks replication. J. Biol. Chem. 2005;280:3764–3770. doi: 10.1074/jbc.M411276200. [DOI] [PubMed] [Google Scholar]
- 12.Hsu GW, Kiefer JR, Burnouf D, Becherel OJ, Fuchs RP, Beese LS. Observing translesion synthesis of an aromatic amine DNA adduct by a high-fidelity DNA polymerase. J. Biol. Chem. 2004;279:50280–50285. doi: 10.1074/jbc.M409224200. [DOI] [PubMed] [Google Scholar]
- 13.Dutta S, Li Y, Johnson D, Dzantiev L, Richardson CC, Romano LJ, Ellenberger T. Crystal structures of 2-acetylaminofluorene and 2-aminofluorene in complex with T7 DNA polymerase reveal mechanisms of mutagenesis. Proc. Natl. Acad. Sci. U.S.A. 2004;101:16186–16191. doi: 10.1073/pnas.0406516101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ling H, Sayer JM, Plosky BS, Yagi H, Boudsocq F, Woodgate R, Jerina DM, Yang W. Crystal structure of a benzo[a]pyrene diol epoxide adduct in a ternary complex with a DNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 2004;101:2265–2269. doi: 10.1073/pnas.0308332100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Batra VK, Shock DD, Prasad R, Beard WA, Hou EW, Pedersen LC, Sayer JM, Yagi H, Kumar S, Jerina DM, Wilson SH. Structure of DNA polymerase β with a benzo[c]phenanthrene diol epoxide-adducted template exhibits mutagenic features. Proc. Natl. Acad. Sci. U.S.A. 2006;103:17231–17236. doi: 10.1073/pnas.0605069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levin W, Chang RL, Wood AW, Thakker DR, Yagi H, Jerina DM, Conney AH. Tumorigenicity of optical isomers of the diastereomeric bay-region 3,4-diol-1,2-epoxides of benzo(c)phenanthrene in murine tumor models. Cancer Res. 1986;46:2257–2261. [PubMed] [Google Scholar]
- 17.Hirshfeld FL. The structure of overcrowded aromatic compounds. Part VII. Out-of-plane deformation in benzo[c]phenanthrene and 1,12-dimethyl-benzo[c]phenanthrene. J. Chem. Soc. 1963:2126–2135. [Google Scholar]
- 18.Conney AH. Induction of microsomal enzymes by foreign chemicals and carcinogenesis by polycyclic aromatic hydrocarbons: G. H. A. Clowes Memorial Lecture. Cancer Res. 1982;42:4875–4917. [PubMed] [Google Scholar]
- 19.Cosman M, de los Santos C, Fiala R, Hingerty BE, Singh SB, Ibanez V, Margulis LA, Live D, Geacintov NE, Broyde S, et al. Solution conformation of the major adduct between the carcinogen (+)-anti-benzo[a]pyrene diol epoxide and DNA. Proc. Natl. Acad. Sci. U.S.A. 1992;89:1914–1918. doi: 10.1073/pnas.89.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fountain MA, Krugh TR. Structural characterization of a (+)-trans-anti-benzo[a]pyrene-DNA adduct using NMR, restrained energy minimization, and molecular dynamics. Biochemistry. 1995;34:3152–3161. doi: 10.1021/bi00010a004. [DOI] [PubMed] [Google Scholar]
- 21.Geacintov NE, Broyde S, Buterin T, Naegeli H, Wu M, Yan S, Patel DJ. Thermodynamic and structural factors in the removal of bulky DNA adducts by the nucleotide excision repair machinery. Biopolymers. 2002;65:202–210. doi: 10.1002/bip.10239. [DOI] [PubMed] [Google Scholar]
- 22.Geacintov NE, Naegeli H, Patel DJ, Broyde S. Structural aspects of polycyclic aromatic carcinogen-damaged DNA and its recognition by NER proteins. In: Siede W, Kow Y, Doetsch P, editors. DNA Damage Recognition. New York: Taylor and Francis; 2005. pp. 263–296. [Google Scholar]
- 23.Wu M, Yan S, Patel DJ, Geacintov NE, Broyde S. Relating repair susceptibility of carcinogen-damaged DNA with structural distortion and thermodynamic stability. Nucleic Acids Res. 2002;30:3422–3432. doi: 10.1093/nar/gkf427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mao B, Vyas RR, Hingerty BE, Broyde S, Basu AK, Patel DJ. Solution conformation of the N-(deoxyguanosin-8-yl)-1-aminopyrene ([AP]dG) adduct opposite dC in a DNA duplex. Biochemistry. 1996;35:12659–12670. doi: 10.1021/bi961078o. [DOI] [PubMed] [Google Scholar]
- 25.Brown K, Hingerty BE, Guenther EA, Krishnan VV, Broyde S, Turteltaub KW, Cosman M. Solution structure of the 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine C8-deoxyguanosine adduct in duplex DNA. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8507–8512. doi: 10.1073/pnas.151251898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elmquist CE, Wang F, Stover JS, Stone MP, Rizzo CJ. Conformational differences of the C8-deoxyguanosine adduct of 2-amino-3-methylimidazo[4,5-f]quinoline (IQ) within the NarI recognition sequence. Chem. Res. Toxicol. 2007;20:445–454. doi: 10.1021/tx060229d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang F, DeMuro NE, Elmquist CE, Stover JS, Rizzo CJ, Stone MP. Base-displaced intercalated structure of the food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in the recognition sequence of the NarI restriction enzyme, a hotspot for −2 bp deletions. J. Am. Chem. Soc. 2006;128:10085–10095. doi: 10.1021/ja062004v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho BP, Beland FA, Marques MM. NMR structural studies of a 15-mer DNA sequence from a ras protooncogene, modified at the first base of codon 61 with the carcinogen 4-aminobiphenyl. Biochemistry. 1992;31:9587–9602. doi: 10.1021/bi00155a011. [DOI] [PubMed] [Google Scholar]
- 29.O’Handley SF, Sanford DG, Xu R, Lester CC, Hingerty BE, Broyde S, Krugh TR. Structural characterization of an N-acetyl-2-aminofluorene (AAF) modified DNA oligomer by NMR, energy minimization, and molecular dynamics. Biochemistry. 1993;32:2481–2497. doi: 10.1021/bi00061a005. [DOI] [PubMed] [Google Scholar]
- 30.Grunberger D, Nelson JH, Cantor CR, Weinstein IB. Coding and conformational properties of oligonucleotides modified with the carcinogen N-2-acetylaminofluorene. Proc. Natl. Acad. Sci. U.S.A. 1970;66:488–494. doi: 10.1073/pnas.66.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuchs R, Daune M. Changes of stability and conformation of DNA following the covalent binding of a carcinogen. FEBS Lett. 1971;14:206–208. doi: 10.1016/0014-5793(71)80618-x. [DOI] [PubMed] [Google Scholar]
- 32.Wang L, Broyde S. A new anti conformation for N-(deoxyguanosin-8-yl)-2-acetylaminofluorene (AAF-dG) allows Watson-Crick pairing in the Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4) Nucleic Acids Res. 2006;34:785–795. doi: 10.1093/nar/gkj479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuan F, Zhang Y, Rajpal DK, Wu X, Guo D, Wang M, Taylor JS, Wang Z. Specificity of DNA lesion bypass by the yeast DNA polymerase η. J. Biol. Chem. 2000;275:8233–8239. doi: 10.1074/jbc.275.11.8233. [DOI] [PubMed] [Google Scholar]
- 34.Rabkin SD, Strauss BS. A role for DNA polymerase in the specificity of nucleotide incorporation opposite N-acetyl-2-aminofluorene adducts. J. Mol. Biol. 1984;178:569–594. doi: 10.1016/0022-2836(84)90239-0. [DOI] [PubMed] [Google Scholar]
- 35.Masutani C, Kusumoto R, Iwai S, Hanaoka F. Mechanisms of accurate translesion synthesis by human DNA polymerase η. EMBO J. 2000;19:3100–3109. doi: 10.1093/emboj/19.12.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Yuan F, Wu X, Taylor JS, Wang Z. Response of human DNA polymerase ι to DNA lesions. Nucleic Acids Res. 2001;29:928–935. doi: 10.1093/nar/29.4.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boudsocq F, Iwai S, Hanaoka F, Woodgate R. Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4): an archaeal DinB-like DNA polymerase with lesion-bypass properties akin to eukaryotic pol η. Nucleic Acids Res. 2001;29:4607–4616. doi: 10.1093/nar/29.22.4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rothwell PJ, Waksman G. Structure and mechanism of DNA polymerases. Adv. Protein Chem. 2005;71:401–440. doi: 10.1016/S0065-3233(04)71011-6. [DOI] [PubMed] [Google Scholar]
- 39.Beard WA, Wilson SH. Structural insights into DNA polymerase β fidelity: Hold tight if you want it right. Chem. Biol. 1998;5:R7–R13. doi: 10.1016/s1074-5521(98)90081-3. [DOI] [PubMed] [Google Scholar]
- 40.Steitz TA. DNA polymerases: structural diversity and common mechanisms. J. Biol. Chem. 1999;274:17395–17398. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 41.Kool ET, Sintim HO. The difluorotoluene debate—A decade later. Chem. Commun. (Cambridge) 2006:3665–3675. doi: 10.1039/b605414e. [DOI] [PubMed] [Google Scholar]
- 42.Kool ET. Active site tightness and substrate fit in DNA replication. Annu. Rev. Biochem. 2002;71:191–219. doi: 10.1146/annurev.biochem.71.110601.135453. [DOI] [PubMed] [Google Scholar]
- 43.Kunkel TA. DNA replication fidelity. J. Biol. Chem. 2004;279:16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- 44.Sawaya MR, Prasad R, Wilson SH, Kraut J, Pelletier H. Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry. 1997;36:11205–11215. doi: 10.1021/bi9703812. [DOI] [PubMed] [Google Scholar]
- 45.Friedberg EC, Lehmann AR, Fuchs RP. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol. Cell. 2005;18:499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 46.Pagès V, Fuchs RP. How DNA lesions are turned into mutations within cells? Oncogene. 2002;21:8957–8966. doi: 10.1038/sj.onc.1206006. [DOI] [PubMed] [Google Scholar]
- 47.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 48.Fuchs RP, Fujii S. Translesion synthesis in Escherichia coli: Lessons from the NarI mutation hot spot. DNA Rep. (Amsterdam) 2007;6:1032–1041. doi: 10.1016/j.dnarep.2007.02.021. [DOI] [PubMed] [Google Scholar]
- 49.Ling H, Boudsocq F, Woodgate R, Yang W. Snapshots of replication through an abasic lesion; structural basis for base substitutions and frameshifts. Mol. Cell. 2004;13:751–762. doi: 10.1016/s1097-2765(04)00101-7. [DOI] [PubMed] [Google Scholar]
- 50.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: A mechanism for error-prone and lesion-bypass replication. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 51.Ling H, Boudsocq F, Plosky BS, Woodgate R, Yang W. Replication of a cis-syn thymine dimer at atomic resolution. Nature. 2003;424:1083–1087. doi: 10.1038/nature01919. [DOI] [PubMed] [Google Scholar]
- 52.Vaisman A, Ling H, Woodgate R, Yang W. Fidelity of Dpo4: Effect of metal ions, nucleotide selection and pyrophosphorolysis. EMBO J. 2005;24:2957–2967. doi: 10.1038/sj.emboj.7600786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zang H, Goodenough AK, Choi JY, Irimia A, Loukachevitch LV, Kozekov ID, Angel KC, Rizzo CJ, Egli M, Guengerich FP. DNA adduct bypass polymerization by Sulfolobus solfataricus DNA polymerase Dpo4: Analysis and crystal structures of multiple base pair substitution and frameshift products with the adduct 1, N2-ethenoguanine. J. Biol. Chem. 2005;280:29750–29764. doi: 10.1074/jbc.M504756200. [DOI] [PubMed] [Google Scholar]
- 54.Trincao J, Johnson RE, Wolfle WT, Escalante CR, Prakash S, Prakash L, Aggarwal AK. Dpo4 is hindered in extending a G·T mismatch by a reverse wobble. Nat. Struct. Mol. Biol. 2004;11:457–462. doi: 10.1038/nsmb755. [DOI] [PubMed] [Google Scholar]
- 55.Boudsocq F, Kokoska RJ, Plosky BS, Vaisman A, Ling H, Kunkel TA, Yang W, Woodgate R. Investigating the role of the little finger domain of Y-family DNA polymerases in low fidelity synthesis and translesion replication. J. Biol. Chem. 2004;279:32932–32940. doi: 10.1074/jbc.M405249200. [DOI] [PubMed] [Google Scholar]
- 56.Zang H, Irimia A, Choi JY, Angel KC, Loukachevitch LV, Egli M, Guengerich FP. Efficient and high fidelity incorporation of dCTP opposite 7,8-dihydro-8-oxodeoxyguanosine by Sulfolobus solfataricus DNA polymerase Dpo4. J. Biol. Chem. 2006;281:2358–2372. doi: 10.1074/jbc.M510889200. [DOI] [PubMed] [Google Scholar]
- 57.Rechkoblit O, Malinina L, Cheng Y, Kuryavyi V, Broyde S, Geacintov NE, Patel DJ. Stepwise translocation of Dpo4 polymerase during error-free bypass of an oxoG lesion. PLoS Biol. 2006;4:e11. doi: 10.1371/journal.pbio.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eoff RL, Irimia A, Angel KC, Egli M, Guengerich FP. Hydrogen bonding of 7,8-dihydro-8-oxodeoxyguanosine with a charged residue in the little finger domain determines miscoding events in sulfolobus solfataricus DNA polymerase Dpo4. J. Biol. Chem. 2007;282:19831–19843. doi: 10.1074/jbc.M702290200. [DOI] [PubMed] [Google Scholar]
- 59.Eoff RL, Angel KC, Egli M, Guengerich FP. Molecular basis of selectivity of nucleoside triphosphate incorporation opposite O6-benzylguanine by Sulfolobus solfataricus DNA polymerase Dpo4: Steady-state and pre-steady-state kinetics and X-ray crystallography of correct and incorrect pairing. J. Biol. Chem. 2007;282:13573–13584. doi: 10.1074/jbc.M700656200. [DOI] [PubMed] [Google Scholar]
- 60.Eoff RL, Irimia A, Egli M, Guengerich FP. Sulfolobus solfataricus DNA polymerase Dpo4 is partially inhibited by “wobble” pairing between O6-methylguanine and cytosine, but accurate bypass is preferred. J. Biol. Chem. 2007;282:1456–1467. doi: 10.1074/jbc.M609661200. [DOI] [PubMed] [Google Scholar]
- 61.Ohashi E, Ogi T, Kusumoto R, Iwai S, Masutani C, Hanaoka F, Ohmori H. Error-prone bypass of certain DNA lesions by the human DNA polymerase κ. Genes Dev. 2000;14:1589–1594. [PMC free article] [PubMed] [Google Scholar]
- 62.Wang L, Yu X, Hu P, Broyde S, Zhang Y. A water-mediated and substrate-assisted catalytic mechanism for Sulfolobus solfataricus DNA polymerase IV. J. Am. Chem. Soc. 2007;129:4731–4737. doi: 10.1021/ja068821c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Human DNA polymerase ι incorporates dCTP opposite template G via a G.C+ Hoogsteen base pair. Structure (Cambridge) 2005;13:1569–1577. doi: 10.1016/j.str.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 64.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Hoogsteen base pair formation promotes synthesis opposite the 1,N6-ethenodeoxyadenosine lesion by human DNA polymerase ι. Nat. Struct. Mol. Biol. 2006;13:619–625. doi: 10.1038/nsmb1118. [DOI] [PubMed] [Google Scholar]
- 65.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. An incoming nucleotide imposes an anti to syn conformational change on the templating purine in the human DNA polymerase-iota active site. Structure. 2006;14:749–755. doi: 10.1016/j.str.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 66.Nair DT, Johnson RE, Prakash S, Prakash L, Aggarwal AK. Replication by human DNA polymerase ι occurs by Hoogsteen base-pairing. Nature. 2004;430:377–380. doi: 10.1038/nature02692. [DOI] [PubMed] [Google Scholar]
- 67.Lone S, Townson SA, Uljon SN, Johnson RE, Brahma A, Nair DT, Prakash S, Prakash L, Aggarwal AK. Human DNA polymerase kappa encircles DNA: Implications for mismatch extension and lesion bypass. Mol. Cell. 2007;25:601–614. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 68.Johnson SJ, Taylor JS, Beese LS. Processive DNA synthesis observed in a polymerase crystal suggests a mechanism for the prevention of frameshift mutations. Proc. Natl. Acad. Sci. U.S.A. 2003;100:3895–3900. doi: 10.1073/pnas.0630532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Feng B, Gorin A, Hingerty BE, Geacintov NE, Broyde S, Patel DJ. Structural alignment of the (+)-trans-anti-benzo[a]pyrene-dG adduct positioned opposite dC at a DNA template-primer junction. Biochemistry. 1997;36:13769–13779. doi: 10.1021/bi970069s. [DOI] [PubMed] [Google Scholar]
- 70.Xu P, Oum L, Beese LS, Geacintov NE, Broyde S. Following an environmental carcinogen N2-dG adduct through replication: Elucidating blockage and bypass in a high-fidelity DNA polymerase. Nucleic Acids. Res. 2007;35:4275–4288. doi: 10.1093/nar/gkm416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lin CH, Huang X, Kolbanovskii A, Hingerty BE, Amin S, Broyde S, Geacintov NE, Patel DJ. Molecular topology of polycyclic aromatic carcinogens determines DNA adduct conformation: a link to tumorigenic activity. J. Mol. Biol. 2001;306:1059–1080. doi: 10.1006/jmbi.2001.4425. [DOI] [PubMed] [Google Scholar]
- 72.Mao B, Gu Z, Gorin A, Chen J, Hingerty BE, Amin S, Broyde S, Geacintov NE, Patel DJ. Solution structure of the (+)-cis-anti-benzo[a]pyrene-dA ([BP]dA) adduct opposite dT in a DNA duplex. Biochemistry. 1999;38:10831–10842. doi: 10.1021/bi991212f. [DOI] [PubMed] [Google Scholar]
- 73.Tan J, Geacintov NE, Broyde S. Conformational determinants of structures in stereoisomeric cis-opened anti-benzo[a]pyrene diol epoxide adducts to adenine in DNA. Chem. Res. Toxicol. 2000;13:811–822. doi: 10.1021/tx000094q. [DOI] [PubMed] [Google Scholar]
- 74.Zhang L, Rechkoblit O, Wang L, Patel DJ, Shapiro R, Broyde S. Mutagenic nucleotide incorporation and hindered translocation by a food carcinogen C8-dG adduct in Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4): Modeling and dynamics studies. Nucleic Acids Res. 2006;34:3326–3337. doi: 10.1093/nar/gkl425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yasui M, Dong H, Bonala RR, Suzuki N, Ohmori H, Hanaoka F, Johnson F, Grollman AP, Shibutani S. Mutagenic properties of 3-(deoxyguanosin-N2-yl)-2-acetylaminofluorene, a persistent acetylaminofluorene-derived DNA adduct in mammalian cells. Biochemistry. 2004;43:15005–15013. doi: 10.1021/bi048279+. [DOI] [PubMed] [Google Scholar]
- 76.Perlow-Poehnelt RA, Likhterov I, Scicchitano DA, Geacintov NE, Broyde S. The spacious active site of a Y-family DNA polymerase facilitates promiscuous nucleotide incorporation opposite a bulky carcinogen-DNA adduct: elucidating the structure-function relationship through experimental and computational approaches. J. Biol. Chem. 2004;279:36951–36961. doi: 10.1074/jbc.M404332200. [DOI] [PubMed] [Google Scholar]
- 77.Bauer J, Xing G, Yagi H, Sayer JM, Jerina DM, Ling H. A structural gap in Dpo4 supports mutagenic bypass of a major benzo[a]pyrene dG adduct in DNA through template misalignment. Proc. Natl. Acad. Sci. U.S.A. 2007;104:14905–14910. doi: 10.1073/pnas.0700717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bauer J, Xing G, Yagi H, Sayer JM, Jerina DM, Ling H. Correction for “A structural gap in Dpo4 supports mutagenic bypass of a major benzo[a]pyrene dG adduct in DNA through template misalignment”. Proc. Natl. Acad. Sci. U.S.A. 2007;104:17240. doi: 10.1073/pnas.0700717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang L, Shapiro R, Broyde S. Molecular dynamics of a food carcinogen-DNA adduct in a replicative DNA polymerase suggest hindered nucleotide incorporation and extension. Chem. Res. Toxicol. 2005;18:1347–1363. doi: 10.1021/tx050132b. [DOI] [PubMed] [Google Scholar]