Abstract
The nuclease-hypersensitivity element III1 in the c-myc promoter is a good anticancer target since it largely controls transcriptional activation of the important c-myc oncogene. Recently, the guanine-rich strand of this element has been shown to form an equilibrium between G-quadruplex structures built from two different sets of G-stretches; two models of intramolecular fold-back antiparallel-stranded G-quadruplexes, called “basket” and “chair” forms, were proposed. Here, we show by NMR that two sequences containing these two sets of G-stretches form intramolecular propeller-type parallel-stranded G-quadruplexes in K+-containing solution. The two structures involve a core of three stacked G-tetrads formed by four parallel G-stretches with all anti guanines and three double-chain-reversal loops bridging three G-tetrad layers. The central loop contains two or six residues, while the two other loops contain only one residue.
Introduction
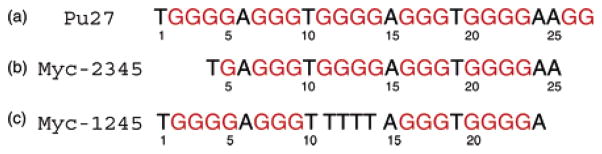
Human c-myc, a 65-kDa nuclear phosphoro-protein, is a central regulator of cellular proliferation and cell growth.1 The aberrant overexpression of the c-myc gene is associated with the progression of many cancers.2 There is an important element in the c-myc promoter region, termed nuclease-hypersensitivity element III1 (NHE), which controls about 90% of total c-myc transcription.3 The 27-nt purine-rich strand (Pu27) of this element contains six guanine stretches, five of which contain three or four guanines per stretch (Figure 1). Using chemical probing, Simonsson et al.4 proposed an intramolecular G-quadruplex structure for this G-rich strand. The model was a fold-back antiparallel-stranded G-quadruplex with a core of three G-tetrads formed by four G-tracks (namely, the first, second, fourth, and fifth), two lateral edgewise loops, and a central diagonal loop.
Figure 1.
DNA sequences of the NHE G-rich strand from the c-myc promoter and its derivatives: (a) Pu27, the 27-nt wild-type sequence; (b) myc-2345, a “chair” derivative5 containing G-tracks number 2, 3, 4, and 5; and (c) myc-1245, a “basket” derivative5 containing G-tracks number 1, 2, 4, and 5; the G-track number 3 is substituted by T4.
Recently, Siddiqui-Jain et al.5 have shown that the same sequence may form at least two different G-quadruplex structures in equilibrium in K+-containing solution. The authors proposed two models of intramolecular G-quadruplexes: while the first one (referred to as the “basket” form) was similar to Simonsson et al.’s structure (Figure 1C of ref 5), the second one (referred to as the “chair” form) was formed by another set of G-tracks (namely, the second, third, fourth, and fifth) with all three edgewise loops (Figure 1D of ref 5). Furthermore, the chair form has been shown to be kinetically favored and biologically relevant, since its destabilization results in a 3-fold increase in transcriptional activity of the c-myc promoter and its stabilization by a ligand decreases or suppresses c-myc transcriptional activation.5
We present here an NMR study on the structure of two different sequences derived from the G-rich strand of the NHE to favor either the basket or the chair form. Instead of these topologies, our results reveal novel distinct intramolecular propeller-type6 parallel-stranded G-quadruplexes for both sequences.
Materials and Methods
Sample Preparation
The unlabeled and the site-specific low-enrichment (2% 15N-labeled for myc-2345 or 2% 15N,13C-labeled for myc-1245) oligonucleotides were synthesized using solid-phase β-cya-noethyl phosphoramidite chemistry and purified by HPLC as previously described.6b,7 They were dialyzed successively against a 50 mM KCl solution and against water. Unless otherwise stated, the strand concentration of the NMR samples was typically 0.5–2 mM; the solutions contained 70 mM of KCl and 20 mM of potassium phosphate (pH 7).
Nuclear Magnetic Resonance
NMR experiments were performed on 600 MHz Varian and Bruker spectrometers. Experiments in H2O used the jump-and-return (JR) water suppression8,9 for detection. The recently developed strategy6b,10 was used for NMR characterization of G-quadruplex structures.
Resonances were assigned using site-specific low-enrichment labeling7 and through-bond correlations at natural abundance9 (imino–H8 by JRHMBC;11a H8–H2 by HMBC;11b H8/6–H1′ by HSQC and sHMBC;11c and H1′–H2′/2′′/3′/4′/5′/5″by COSY11d and TOCSY11e).
The establishment of equilibrium for stoichiometry and melting temperature measurements was monitored and ascertained by NMR. Measurements were performed on samples in H2O on the basis of signals of different imino and aromatic protons, using the JR pulse sequence with a repetition delay of 5 s. Stoichiometry determination was based on the titration of the concentration-dependent equilibrium between the structured and unfolded forms.9 The melting temperature was determined as the temperature where the equilibrium fractions of the structured and unfolded forms measured by NMR are equal.
Real-time imino proton-exchange experiments were performed at 25°C: samples were first dried and then quickly dissolved in D2O; NMR spectra were recorded at different times.
Results
Pu27 Forms Multiple G-Quadruplex Structures
The imino and aromatic proton spectrum of the 27-nt d(TG4AG3TG4AG3-TG4AAG2) c-myc sequence (Pu27) in K+ -containing solution is shown in Figure 2a. We observe a broad envelope including imino protons at 10–12 ppm with some fine structure, which indicates the presence of multiple G-quadruplex forms. Such NMR spectra provide minimal opportunities for further structural characterization, with line broadening, in part, reflecting the consequences of exchange between interconverting conformers.
Figure 2.
One-dimensional 600 MHz proton spectra of (a) Pu27, (b) myc-2345, and (c) myc-1245. Experimental conditions: strand concentration, 1 mM; temperature, 25 °C; 70 mM KCl; potassium phosphate, 20 mM; pH 7.
We therefore made systematic modifications in the Pu27 sequence (with over 50 sequences) in efforts to separately drive the equilibrium to either the basket or the chair form.12 Previously, chemical probing data5 indicated that the first (or third) G-track forms part of the quadruplex scaffold for only one fold, the basket fold (or the chair fold). Sequences myc-2345 and myc-1245 (Figure 1), having the first and third G-track modified, respectively, have been chosen for further structural analysis on the basis of their NMR spectral quality (Figure 2).
The myc-2345 sequence, in which the first G4 segment was replaced by a single G, can form the chair fold, but not the basket fold. The myc-1245 sequence, in which the third G4 segment was substituted by T4, can form the basket fold, but not the chair fold. In the latter sequence, the T4 substitution was chosen because it has been found previously to favor diagonal loops of several G-quadruplexes.13
Sequences myc-2345 and myc-1245 Form Intramolecular Propeller-Type Parallel-Stranded G-Quadruplexes
Proton spectra of myc-2345 and myc-1245 are plotted in Figure 2b and c, respectively. In both cases, the number and intensity of peaks indicated the presence of a major conformation. Sharp imino protons at 10–12 ppm correspond to guanine imino protons in G-tetrad formation.6b,10
The spectral line-widths are consistent with an intramolecular monomeric structure for both sequences, with the sharpest lines of 2–3 Hz at 25 °C (Figure S1). The stoichiometry was determined on the basis of NMR titration of the concentration-dependent equilibrium between the structured and unfolded forms.9 The latter was identified as the species which is predominant at high temperatures, showing sharp nonexchange-able protons but no imino proton signals (Figure S2). The results of the titrations (Figure 3) indicated the formation of monomeric structures, hence intramolecular monomeric G-quadruplexes. The concentration-independence of the melting temperature of these structures (see below) also supports the formation of monomers.
Figure 3.
Determination of stoichiometry by NMR titration of the equilibrium strand concentrations of the structured form and of the unfolded monomer. Squares and triangles represent myc-2345 and myc-1245, respectively. Lines with a slope of one are drawn through the data points. Experimental conditions: 7 mM KCl; potassium phosphate, 2 mM; pH 7; temperature, 60 °C for myc-2345 and 50 °C for myc-1245.
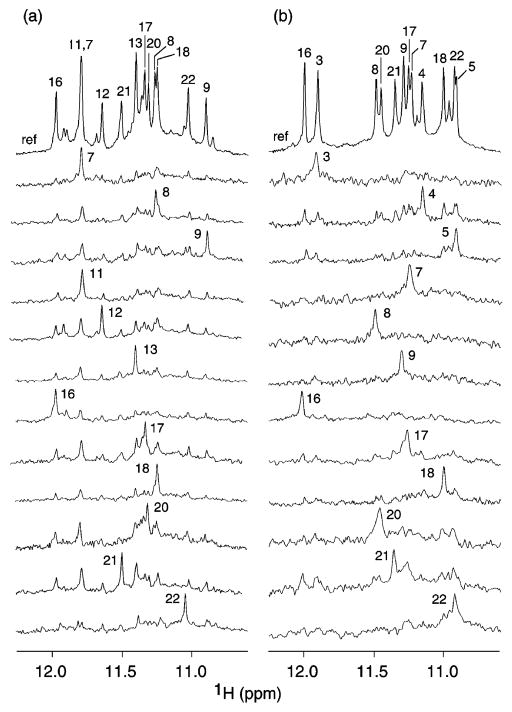
Guanine imino protons of myc-2345 and myc-1245 were assigned unambiguously to their positions in the sequence by the site-specific low-enrichment approach7 using 2% 15N-labeled or 2% 15N,13C-labeled samples. For both sequences, there are 12 major imino proton peaks, consistent with the formation of three G-tetrads. The guanine imino proton of each site-specific labeled residue was recognized on the basis of its intensity in the 15N-filtered spectrum (Figure 4).
Figure 4.
Imino proton NMR spectra of (a) myc-2345 and (b) myc-1245, with assignments listed over the reference spectrum (ref) at the top of the figure. Imino protons were assigned in 15N-filtered spectra of samples that were 2% (a) 15N-labeled and (b) 15N,13C-labeled at the indicated positions. The reference spectrum was recorded using the same pulse sequence but with a different phase cycle. Experimental conditions: 70 mM KCl; potassium phosphate, 20 mM; pH 7; temperature, 25 °C; strand concentration, 0.5–1 mM.
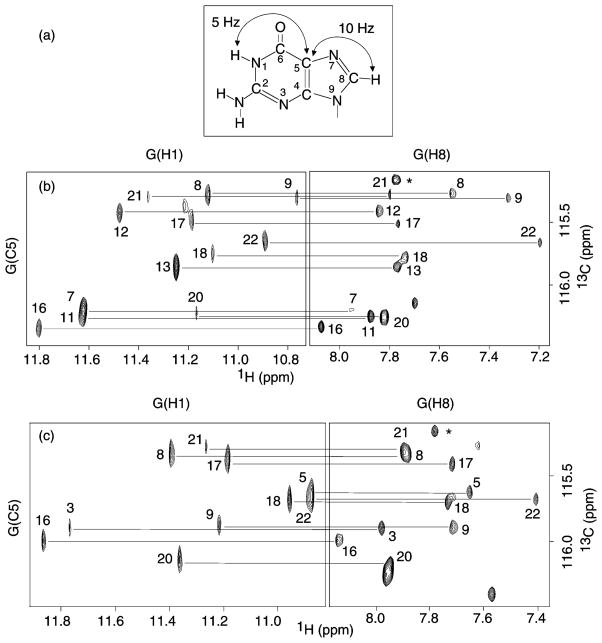
The assignments of guanine H8 protons were obtained by through-bond correlation with the already-assigned imino protons (Figure 5) via 13C5 at natural abundance.11a The assignments of several guanine H8 protons were confirmed independently using the site-specific labeled samples.7 Figure S3 plots some examples of guanine H8 assignments in myc-2345 and myc-1245 by [1H–15N] long-range and [1H–13C] one-bond correlation experiments, respectively.
Figure 5.
Through-bond correlations between imino and H8 protons via 13C5 at natural abundance for (b) myc-2345 and (c) myc-1245, using long-range J-couplings shown in (a). Assignments of guanine H8 protons, labeled with residue numbers, were obtained from the already-assigned imino protons. A peak from T(H6) is labeled with a star. Experimental conditions: 70 mM KCl; potassium phosphate, 20 mM; pH 7; temperature, 25 °C; strand concentration, 2 mM.
The assignments of guanine imino and H8 protons were supported by NOESY and other through-bond correlation experiments (see Materials and Methods).
The G-tetrad alignments were defined from NOESY spectra (Figure 6a, a′) on the basis of the specific imino–H8 connectivity pattern in a G-tetrad (Figure 6b). Such a connectivity pattern determines the alignment of G-tetrads and the hydrogen-bond directionality around each G-tetrad. Examination of NOESY imino–H8 connectivities between already-assigned imino and H8 protons (Figure 6a, a′) revealed the formation of three G-tetrads for each structure: (G9•G13•G18•G22), (G8•G12• G17•G21), and (G7•G11•G16•G20) for myc-2345; (G5•G9•G18• G22), (G4•G8•G17•G21), and (G3•G7•G16•G20) for myc-1245.
Figure 6.
NOESY spectra (mixing time, 200 ms) of (a) myc-2345 and (a′) myc-1245 (same conditions as in Figure 5). The imino–H8 cross-peaks are framed and labeled with the number of imino protons in the first position and that of H8 in the second position. (b) Specific imino–H8 connectivity pattern around a G-tetrad (Gα•Gβ•Gγ•Gδ) indicated with arrows (connectivity between Gδ and Gα implied). (c, c′) Schematic structures of myc-2345 and myc-1245 that satisfy NOE connectivities shown in parentheses.
The intramolecular G-quadruplex folding topologies of myc-2345 and myc-1245 were defined in two steps: first, the G-tetrad core was established from G-tetrad alignments; second, the loops were drawn by connecting sequential residues. This procedure revealed the formation of intramolecular propeller-type parallel-stranded G-quadruplexes for both myc-2345 and myc-1245 (Figure 6c, c′). The parallel-stranded G-quadruplex cores are consistent with all guanines adopting an anti conformation, as no strong H1′–H8 peaks were detected in NOESY spectra (data not shown). In each structure, there are three double-chain-reversal loops bridging three G-tetrad layers and connecting adjacent parallel strands: two of them are single-residue (A or T) loops; the third (central) ones are two-residue (GA) and six- residue (T5A) loops for myc-2345 and myc-1245, respectively (Figure 6c, c′). These topologies are also consistent with other NOESY data (e.g. detection of cross-peaks between protons of adjacent G-tetrads or absence of sequential H1′-H6/8 cross-peaks for single-residue loops).
Stability of Propeller-Type G-Quadruplexes
The propeller-type G-quadruplex structures of myc-2345 and myc-1245 are very stable in the presence of K+. The unfolded form was observed by NMR only at relatively high temperatures (Figure S2). The melting temperature (Tm) of a given G-quadruplex form was determined by NMR as the temperature where the equilibrium fractions of the G-quadruplex and unfolded forms are equal. The melting temperature was very slightly (or almost not) dependent on the DNA concentration, as expected for monomeric structures. By contrast, it depended strongly on the concentration of K+, with very high Tm observed for 90 mM K+. An example of Tm determination by NMR is shown in Figure S2. Table 1 lists the Tm values of the myc-2345 and myc-1245 G-quadruplexes at two different K+ concentrations (9 and 90 mM). In the presence of 9 mM K+, the melting temperature of the propeller-type G-quadruplex of myc-2345 is about 15 °C higher than that of myc-1245. In the presence of 90 mM K+, the melting temperature of the myc-2345 propeller-type G-quadruplex is higher than 80 °C and could not be measured by NMR.
Table 1.
Melting Temperatures (°C) of Intramolecular Propeller-type G-Quadruplexes of myc-2345 and myc-1245 at Different K+ Concentrations
| myc-2345 | myc-1245 | |
|---|---|---|
| 9 mM K+ | 63 | 47 |
| 90 mM K+ | >80a | 75 |
The melting temperature is higher than the limit of permitted temperature range for the NMR spectrometer; therefore, it could not be measured.
Imino Proton Exchange of the Central G-Tetrads
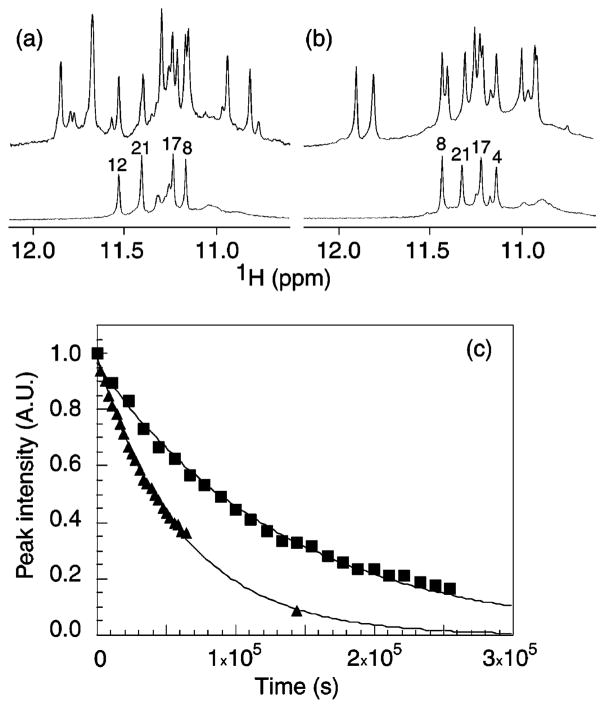
Imino proton spectra of myc-2345 and myc-1245 after 1 h in D2O are shown in Figure 7a and b. In each case, the most protected four peaks correspond to guanine imino protons of the central G-tetrad (G8, G12, G17, and G21 for myc-2345; G4, G8, G17, and G21 for myc-1245). This observation supports the folding topologies identified above (Figure 6c, c′).
Figure 7.
Imino proton spectra of (a) myc-2345 and (b) myc-1245 in H2O (upper) and after 1 h in D2O (lower) showing peaks from the central G-tetrad. (c) Intensities (in arbitrary units, A.U.) of G21 imino protons in myc-2345 and myc-1245 as a function of time. Squares and triangles represent myc-2345 and myc-1245, respectively. Experimental conditions: strand concentration, 1 mM; temperature, 25 °C; 70 mM KCl; potassium phosphate, 20 mM; pH 7.
The exchange times of the guanine imino protons in the central G-tetrad were measured in real time at 25 °C by NMR. The exchange times of the four guanine imino protons in the central G-tetrad of myc-1245 are similar, ranging from 14 to 18 h. In the central G-tetrad of myc-2345, the exchange time of the G12 imino proton is 12 h, similar to that of the corresponding protons in myc-1245. The exchange times of the three remaining imino protons of myc-2345 are much longer, ranging from 40 to 50 h. An example of the exchange time measurement for the G21 imino proton in myc-2345 and in myc-1245 is shown in Figure 7c. The G21 imino proton of myc-2345 exchanges about 2 times slower than the corresponding proton of myc-1245.
Discussion
Novel Propeller-Type G-Quadruplex Topologies
We have identified two derivative sequences of the guanine-rich strand Pu27 from the c-myc promoter, myc-2345 and myc-1245, which are suitable for structural characterization by NMR. These sequences contain two different sets of G-tracks that have been shown previously to participate in formation of G-quadruplexes of Pu27. We have shown here that both myc-2345 and myc-1245 form intramolecular propeller-type parallel-stranded G-quadruplexes in K+-containing solution: the core of three G-tetrads is formed by four G-stretches oriented in the same direction, with all anti guanines; all three loops are double-chain-reversal. A similar parallel-stranded G-quadruplex topology with three (three-residue) double-chain-reversal loops was recently observed for the human telomeric G-rich strand.6 The loop compositions of the structures presented here are different and represent some interesting features. In the structure of myc-2345, the central loop contains two nucleotides (GA), while the two flanking loops contain only one nucleotide (T). In the structure of myc-1245, each of the two flanking loops contains one residue (A or T), while the central loop contains six residues (T5A).
The double-chain-reversal loop, a structural element in G-quadruplexes that connects two adjacent parallel strands, has been observed previously on various occasions.6,14,15 A one-residue (A) loop has been found to bridge two G-tetrad layers and to participate in hydrogen-bond alignments with a G-tetrad edge.15 Two-residue (TT and CA) and three-residue (TTA) loops have been found to bridge three G-tetrad layers.6,14 In this study, besides a two-residue (GA) loop, we also observed systematically single-residue (A and T) double-chain-reversal loops bridging three layers of G-tetrads. On the other hand, the six-residue loop in the structure of myc-1245 is the longest double-chain-reversal loop that has ever been observed.
As mentioned above, although myc-2345 and myc-1245 contain two different sets of G-stretches from Pu27, their overall folding topologies are similar, with a parallel-stranded G-tetrad core and three double-chain-reversal loops, two of which are one-residue loops. The main difference between the two structures lies in the size of the central loop, which is a two-and six-residue loop, respectively. This might explain the difference in stability between the two structures, with myc-2345 having an approximately 15 °C higher melting temperature. This finding suggests that a two-residue double-chain-reversal loop is more stable than a six-residue loop for bridging three G-tetrad layers. In general, the imino proton-exchange time of the central tetrad of the myc-2345 G-quadruplex is also longer than that of the corresponding protons of the myc-1245 G-quadruplex, suggesting slower unfolding kinetics of the former structure.
Multiple Conformations in the Pu27 Sequence
The Pu27 sequence from the NHE element of the c-myc promoter contains six G-stretches, five of which contain three or four guanines per stretch (Figure 1a). One would envisage multiple possible ways of G-quadruplex formation using different G-stretches (including various possible stoichiometries). Indeed, the observed NMR spectra (Figure 2a) indicated the presence of multiple conformations. Usually, an intramolecular G-quadru-plex formation requires four G-tracks.16 For Pu27, there are five different possibilities of selecting four G-tracks (of at least three guanines each) out of five, hence at least five possible intramolecular G-quadruplex topologies. Siddiqui-Jain et al.’s data5 suggested that the most favorable topologies formed by two different sets (combinations) of four G-tracks, and the authors proposed two topologies, called “chair” and “basket”, respectively. (Structures using other sets of G-tracks may exist with lower proportions.) However, even for a given set of four G-tracks, there may be several possible intramolecular G-quaduplex topologies, which differ by strand orientations, syn/ anti distributions, and/or loop connections.6,16 The myc-2345 and myc-1245 sequences, derived from Pu27 by keeping two different sets of G-tracks reported by Siddiqui-Jain et al.,5 therefore may represent the two most favorable forms. In particular, myc-2345 is almost a natural fragment of Pu27, containing the four G-tracks used in the biologically relevant G-quadruplex structure(s).5 Interestingly, instead of chair and basket, we found that both sequences form propeller-type parallel-stranded G-quadruplexes in K+-containing solution. It should be noted that these topologies are also consistent with the previously reported chemical probing data.4,5
In a G-quadruplex structure involving three G-tetrads, there are four columns of three guanines, which are usually four three-guanine tracks. In each sequence, myc-2345 and myc-1245, there are two four-guanine stretches. There are multiple possibilities of selecting three guanines out of four, in particular, two possibilities of picking three consecutive guanines: three guanines at the 5′-end or at the 3′-end. Hence, when there are four guanines in a stretch, but only three are used for G-tetrad formation, there might be different possible G-quadruplex topologies or an equilibrium between them. However, we have observed only one major G-quadruplex form for each sequence. In myc-2345, G14 and G23 from the four-guanine G11–G14 and G20–G23 stretches, respectively, are excluded from the G-tetrad core (two 5′-end three-guanine tracks are in the G-tetrad core). In myc-1245, G2 and G23 from the four-guanine G2–G5 and G20–G23 stretches, respectively, are excluded from the G-tetrad core (a 3′-end and a 5′-end three-guanine tracks are in the G-tetrad core). In all cases, the selection of G-tracks participating in the G-tetrad core tends to favor the formation of one-residue loops. The observation of only one major form for each sequence is probably due to stable one-residue loop formation. This result suggests that the one-residue loop is the most stable double-chain-reversal loop bridging three G-tetrad layers. The systematic observation of single-residue double-chain-reversal loops here and in other structures studied in our laboratory (unpublished results) confirms the robustness of such looping topology.
In myc-2345, the shorter exchange time of the G12 imino proton (12 h at 25 °C) compared to the exchange time of other guanine imino protons in the central G-tetrad (40–50 h at 25 °C) may be due to the catalytic effect of G14 and/or A15 in the next GA loop (Figure 6c) or again may reflect an equilibrium between the G-quadruplex described above and other G-quadruplex conformations which do not adopt G12 in the central G-tetrad. For instance, myc-2345 may form another propeller-type G-quadruplex, where (within the four-guanine G11–G14 stretch) G11 would be in the two-residue (T10G11) double-chain-reversal loop and the G12–G14 track would participate in the G-tetrad core.
Biological Significance
In the presence of the complementary C-rich strand, the NHE element of the c-myc promoter can form different structures, including the i-motif of the C-rich strand,17 the G-quadruplex of the G-rich strand,4,5 and the Watson–Crick duplex of the two strands.18 For human telomeric DNA, the duplex is the predominant structure at physiological conditions.18b The high stability of the propeller-type G-quadruplex structure in the c-myc promoter (in particular, that of myc-2345) in K+-containing solution makes the existence of the G-quadruplex more likely. Folding/unfolding kinetics of these structures is another factor that determines their biological relevance.5,6,18,19 Proteins that bind specifically to either the C-rich strand, G-rich strand, or the duplex of the NHE have been identified.20 They may regulate the transcription level of c-myc by modulating the structures (or populations of structures) of the NHE. It would be interesting to study the interaction of the propeller-type G-quadruplex structures presented here with these proteins.
Our NMR data suggest that the NHE purine-rich strand (Pu27) of the c-myc promoter forms multiple, presumably interconverting, G-quadruplex structures. A G-quadruplex formed by G-tracks number 2, 3, 4, and 5 has been shown to be pharmacologically important, since its stabilization by a ligand decreases c-myc transcriptional activation.5 The present work identifies the major G-quadruplex conformation of a DNA fragment containing these four G-tracks. Such a structure might be a potential anticancer target, if one stipulates that it is part of the distribution of structures formed by the wild-type full-length sequence and the alternative structures are readily interconvertible to the target structure. With a drug targeting this structure, one expects to change the equilibrium between the G-quadruplex(es) of the G-rich strand, the i-motif of the C-rich strand, and the duplex association of the two strands.18
Supplementary Material
Acknowledgments
This research was supported by NIH grant GM34504. We thank Prof. Laurence Hurley of the University of Arizona for providing us with a preprint of his paper titled “The dynamic character of the G-quadruplex element in the c-MYC promoter and modification by TMPyP4” by Seenisamy, J. et al. & Hurley, L. H. and for mutually agreeing to publish his paper and ours back-to-back in this issue of the journal. D.J.P. is a member of the New York Structural Biology Center and acknowledges access to the NMR instrumentation purchased and maintained in part by NIH grant GM66354.
Footnotes
Supporting Information Available: Three figures S1–S3 (NMR spectra for stoichiometry, assignment, and stability characterization). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Gartel AL, Shchors K. Exp Cell Res. 2003;283:17–21. doi: 10.1016/s0014-4827(02)00020-4. [DOI] [PubMed] [Google Scholar]; (b) Pelengaris S, Khan M. Arch Biochem Biophys. 2003;416:129–136. doi: 10.1016/s0003-9861(03)00294-7. [DOI] [PubMed] [Google Scholar]
- 2.(a) Slamon DJ, deKernion JB, Verma IM, Cline MJ. Science. 1984;224:256–262. doi: 10.1126/science.6538699. [DOI] [PubMed] [Google Scholar]; (b) Marcu KB, Bossone SA, Patel AJ. Annu Rev Biochem. 1992;61:809–860. doi: 10.1146/annurev.bi.61.070192.004113. [DOI] [PubMed] [Google Scholar]; (c) Pelengaris S, Rudolph B, Littlewood T. Curr Opin Genet Dev. 2000;10:100–105. doi: 10.1016/s0959-437x(99)00046-5. [DOI] [PubMed] [Google Scholar]
- 3.(a) Siebenlist U, Hennighausen L, Battey J, Leder P. Cell. 1984;37:381–391. doi: 10.1016/0092-8674(84)90368-4. [DOI] [PubMed] [Google Scholar]; (b) Cooney M, Czernuszewicz G, Postel EH, Flint SJ, Hogan ME. Science. 1988;241:456–459. doi: 10.1126/science.3293213. [DOI] [PubMed] [Google Scholar]; (c) Davis TL, Firulli AB, Kinniburgh AJ. Proc Natl Acad Sci USA. 1989;86:9682–9686. doi: 10.1073/pnas.86.24.9682. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Berberich SJ, Postel EH. Oncogene. 1995;10:2343–2347. [PubMed] [Google Scholar]
- 4.Simonsson T, Pecinka P, Kubista M. Nucleic Acids Res. 1998;26:1167–1172. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Proc Natl Acad Sci USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Parkinson GN, Lee MPH, Neidle S. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]; (b) Phan AT, Patel DJ. J Am Chem Soc. 2003;125:15021–15027. doi: 10.1021/ja037616j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Phan AT, Patel DJ. J Am Chem Soc. 2002;124:1160–1161. doi: 10.1021/ja011977m. [DOI] [PubMed] [Google Scholar]; (b) Phan AT, Patel DJ. J Biomol NMR. 2002;23:257–262. doi: 10.1023/a:1020277223482. [DOI] [PubMed] [Google Scholar]
- 8.Plateau P, Guéron M. J Am Chem Soc. 1982;104:7310–7311. [Google Scholar]
- 9.Phan AT, Guéron M, Leroy JL. Methods Enzymol. 2001;338:341–371. doi: 10.1016/s0076-6879(02)38228-4. [DOI] [PubMed] [Google Scholar]
- 10.Phan AT, Modi YS, Patel DJ. J Mol Biol. 2004;338:93–102. doi: 10.1016/j.jmb.2004.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.(a) Phan AT. J Biomol NMR. 2000;16:175–178. doi: 10.1023/a:1008355231085. [DOI] [PubMed] [Google Scholar]; (b) van Dongen MJP, Wijmenga SS, Eritja R, Azorin F, Hilbers CW. J Biomol NMR. 1996;8:207–212. doi: 10.1007/BF00211166. [DOI] [PubMed] [Google Scholar]; (c) Phan AT. J Magn Reson. 2001;153:223–226. doi: 10.1006/jmre.2001.2445. [DOI] [PubMed] [Google Scholar]; (d) Piantini U, Sorensen OW, Ernst RR. J Am Chem Soc. 1982;104:6800–6801. [Google Scholar]; (e) Bax A, Davis DG. J Magn Reson. 1985;65:355–360. [Google Scholar]
- 12.NMR spectra of many modified sequences also indicated the presence of multiple G-quadruplex structures.
- 13.(a) Smith FW, Feigon J. Nature. 1992;356:164–168. doi: 10.1038/356164a0. [DOI] [PubMed] [Google Scholar]; (b) Schultze P, Hud NV, Smith FW, Feigon J. Nucleic Acids Res. 1999;27:3018–3028. doi: 10.1093/nar/27.15.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Smith FW, Lau FW, Feigon J. Proc Natl Acad Sci USA. 1994;91:10546–10550. doi: 10.1073/pnas.91.22.10546. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Wang Y, Patel DJ. J Mol Biol. 1995;251:76–94. doi: 10.1006/jmbi.1995.0417. [DOI] [PubMed] [Google Scholar]; (e) Horvath MP, Schultz SC. J Mol Biol. 2001;310:367–377. doi: 10.1006/jmbi.2001.4766. [DOI] [PubMed] [Google Scholar]; (f) Haider S, Parkinson GN, Neidle S. J Mol Biol. 2002;320:189–200. doi: 10.1016/S0022-2836(02)00428-X. [DOI] [PubMed] [Google Scholar]; (g) Crnugelj M, Hud NV, Plavec J. J Mol Biol. 2002;320:911–924. doi: 10.1016/s0022-2836(02)00569-7. [DOI] [PubMed] [Google Scholar]
- 14.(a) Wang Y, Patel DJ. Structure. 1994;2:1141–1156. doi: 10.1016/s0969-2126(94)00117-0. [DOI] [PubMed] [Google Scholar]; (b) Kuryavyi V, Majumdar A, Shallop A, Chernichenko N, Skripkin E, Jones R, Patel DJ. J Mol Biol. 2001;310:181–194. doi: 10.1006/jmbi.2001.4759. [DOI] [PubMed] [Google Scholar]
- 15.(a) Kettani A, Gorin A, Majumdar A, Hermann T, Skripkin E, Zhao H, Jones R, Patel DJ. J Mol Biol. 2000;297:627–644. doi: 10.1006/jmbi.2000.3524. [DOI] [PubMed] [Google Scholar]; (b) Zhang N, Gorin A, Majumdar A, Kettani A, Chernichenko N, Skripkin E, Patel DJ. J Mol Biol. 2001;311:1063–1079. doi: 10.1006/jmbi.2001.4916. [DOI] [PubMed] [Google Scholar]; (c) Matsugami A, Ouhashi K, Kanagawa M, Liu H, Kanagawa S, Uesugi S, Katahira M. J Mol Biol. 2001;313:255–269. doi: 10.1006/jmbi.2001.5047. [DOI] [PubMed] [Google Scholar]; (d) Matsugami A, Okuizumi T, Uesugi S, Katahira M. J Biol Chem. 2003;278:28147–28153. doi: 10.1074/jbc.M303694200. [DOI] [PubMed] [Google Scholar]
- 16.(a) Patel DJ, Bouaziz S, Kettani A, Wang Y. In: Oxford Handbook of Nucleic Acid Structures. Neidle S, editor. Oxford University Press; Oxford: 1999. pp. 389–453. [Google Scholar]; (b) Simonsson T. Biol Chem. 2001;382:621–628. doi: 10.1515/BC.2001.073. [DOI] [PubMed] [Google Scholar]
- 17.(a) Gehring K, Leroy JL, Guéron M. Nature. 1993;363:561–565. doi: 10.1038/363561a0. [DOI] [PubMed] [Google Scholar]; (b) Simonsson T, Pribylova M, Vorlickova M. Biochem Biophys Res Commun. 2000;278:158–166. doi: 10.1006/bbrc.2000.3783. [DOI] [PubMed] [Google Scholar]
- 18.(a) Rangan A, Fedoroff OY, Hurley LH. J Biol Chem. 2001;276:4640–4646. doi: 10.1074/jbc.M005962200. [DOI] [PubMed] [Google Scholar]; (b) Phan AT, Mergny JL. Nucleic Acids Res. 2002;30:4618–4625. doi: 10.1093/nar/gkf597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.(a) Fang G, Cech TR. Biochemistry. 1993;32:11646–11657. doi: 10.1021/bi00094a022. [DOI] [PubMed] [Google Scholar]; (b) Han H, Cliff CL, Hurley LH. Biochemistry. 1999;38:6981–6986. doi: 10.1021/bi9905922. [DOI] [PubMed] [Google Scholar]
- 20.(a) Postel EH, Berberich SJ, Flint SJ, Ferrone CA. Science. 1993;261:478–480. doi: 10.1126/science.8392752. [DOI] [PubMed] [Google Scholar]; (b) Takimoto M, Tomonaga T, Matunis M, Avigan M, Krutzsch H, Dreyfuss G, Levens D. J Biol Chem. 1993;268:18249–18258. [PubMed] [Google Scholar]; (c) Michelotti GA, Michelotti EF, Pullner A, Duncan RC, Eick D, Levens D. Mol Cell Biol. 1996;16:2656–2669. doi: 10.1128/mcb.16.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Michelotti EF, Tomonaga T, Krutzsch H, Levens D. J Biol Chem. 1995;270:9494–9499. doi: 10.1074/jbc.270.16.9494. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.