Abstract
We present the intramolecular G-quadruplex structure of human telomeric DNA in physiologically relevant K+ solution. This G-quadruplex, whose (3 + 1) topology differs from folds reported previously in Na+ solution and in a K+-containing crystal, involves the following: one anti•syn•syn•syn and two syn•anti•anti•anti G-tetrads; one double-chain reversal and two edgewise loops; three G-tracts oriented in one direction and the fourth in the opposite direction. The topological characteristics of this (3 + 1) G-quadruplex scaffold should provide a unique platform for structure-based anticancer drug design targeted to human telomeric DNA.
Introduction
The ends of eukaryotic chromosomes called telomeres are essential for genome integrity and play an important role in cellular aging and cancer.1 Telomeric DNA consists of tandem repeats of G-rich sequences, such as (GGGTTA)n in humans.2 In the presence of Na+ or K+, G-rich sequences can form G-quadruplex structures in vitro, built from stacking of multiple planar G•G•G•G tetrads3. The in vivo observation of G-quadruplex formation in telomeres, a process regulated by telomere end-binding proteins,4 highlights the biological importance of this DNA scaffold. Telomeres contain G-rich strand overhangs at their 3′-ends, about 200-nt long in humans,5 which can be elongated by the enzyme telomerase.6 Telomerase activity is not detected in normal human somatic cells, and telomeres progressively decrease in length after each round of cell division. In contrast, telomerase is activated in 80–85% cancer cells and helps to maintain the length of telomeres in these cells.7 It has been shown previously that formation of intramolecular G-quadruplexes by the telomeric G-rich strand inhibits the activity of telomerase.8–10 Therefore, ligand-induced stabilization of intramolecular telomeric G-quadruplexes has become an attractive strategy for the development of anticancer drugs.1
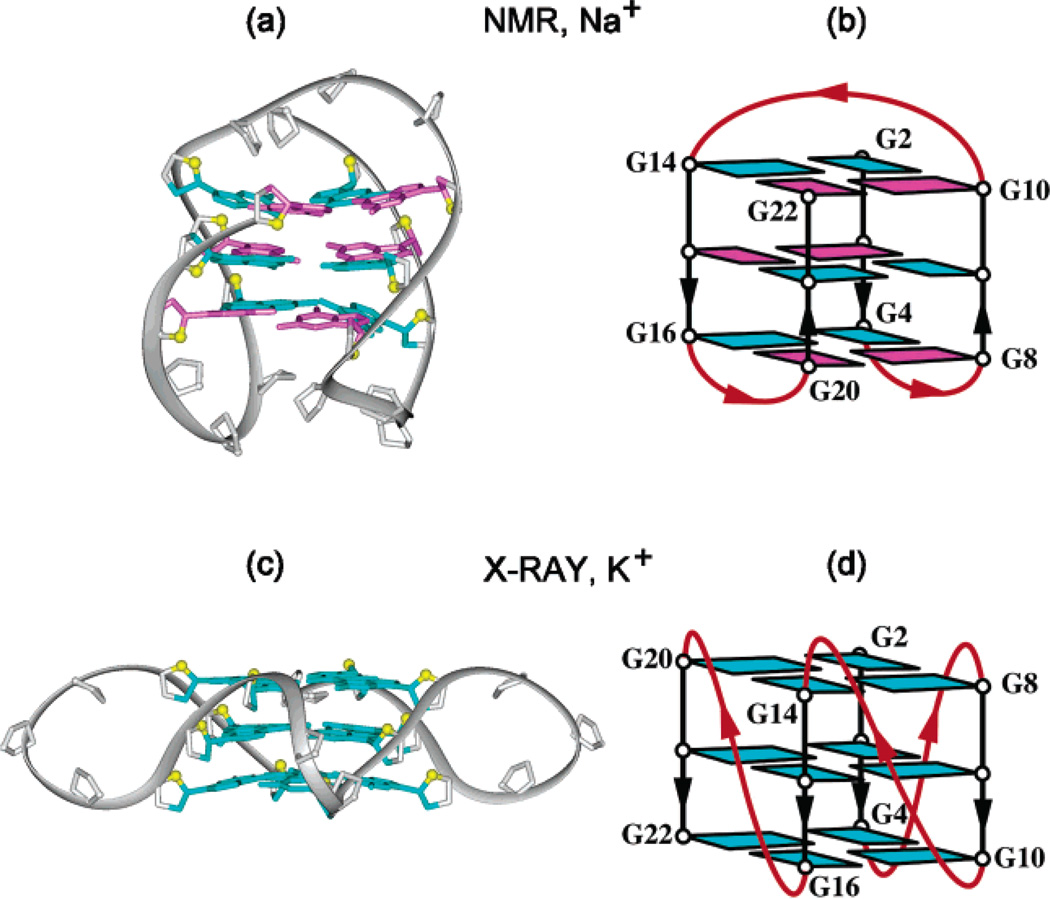
The structural elucidation of folding topologies adopted by oligonucleotides containing four human telomeric GGGTTA-tracts, the minimal length required for an intramolecular G-quadruplex formation, has posed a significant challenge in the field and has been intensively investigated using a range of physical and chemical techniques.11–21 Our laboratory previously showed that the four-repeat human telomeric d[AGGG(TTAGGG)3] sequence forms an intramolecular G-quadruplex in Na+ solution,11 where guanines around each tetrad are syn•syn• anti•anti, loops are successively edgewise-diagonal-edgewise, and each G-tract has both parallel and antiparallel adjacent strands (Figure 1a, b). By contrast, the crystal structure of the same sequence in the presence of K+ revealed a completely different intramolecular G-quadruplex,12 where all strands are parallel, guanines are anti and loops are double-chain reversal (Figure 1c, d). Our subsequent NMR studies have indicated that this sequence forms multiple G-quadruplexes in K+ solution.13 Studies from a number of laboratories14–21 suggest that the intramolecular G-quadruplex structure observed in the K+-containing crystal12 appears unlikely to be the major form in K+-containing solution. Because K+ is much more abundant than Na+ in cellular environments, knowledge of the structure of human telomeric G-quadruplexes in K+ solution is most important.
Figure 1.
Structure of intramolecular G-quadruplexes formed by the human telomeric sequence: (a, b) in Na+ solution;11 (c, d) in a K+-containing crystal.12 Loops are colored red; anti and syn guanines are colored cyan and magenta, respectively. O4′ atoms are colored yellow.
Here we report on the intramolecular G-quadruplex structure formed by a four-repeat human telomeric sequence in K+ solution, which is distinctly different from previously reported structures in Na+ solution11 (Figure 1a, b) and in a K+-containing crystal12 (Figure 1c, d). This structure of the human telomere under physiological conditions should turn out to be important for understanding telomere biology and for structure-based anticancer drug design.
Results and Discussion
Favoring a Major G-Quadruplex Conformation for Structural Study
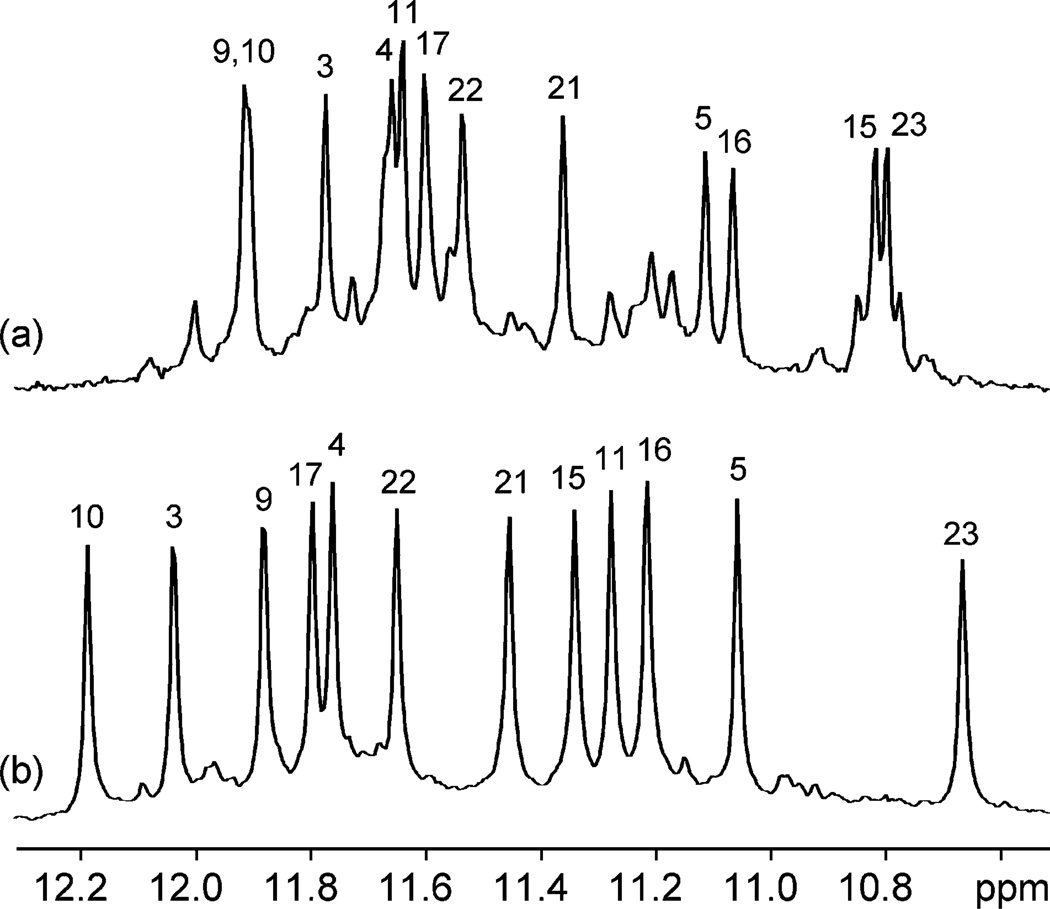
To determine the structure(s) of an intramolecular human telomeric G-quadruplex in K+ solution, we attempted to overcome the conformational heterogeneity associated with the presence of multiple conformations. We successfully achieved this goal through a strategy involving nucleotide substitution of residues flanking the four guanine tracts: the 24-nt d[TTGGG(TTAGGG)3A] sequence, which was derived from the natural 23-nt human telomeric d[TAGGG(TTAGGG)3] sequence by two modifications at terminal residues, favors a major G-quadruplex structure (about 95%) in K+ solution, and gives excellent NMR spectra suitable for structure determination (Figure 2b). The same major fold is observed for the natural 23-nt human telomeric sequence (see below), but only at 60–70% (Figure 2a).
Figure 2.
Imino proton spectra of (a) the natural 23-nt human telomeric d[TAGGG(TTAGGG)3] sequence and (b) the modified 24-nt d[TTGGG-(TTAGGG)3A] sequence, both in K+ solution. In each case, peaks for the major form are labeled with residue numbers obtained from unambiguous assignments.
Determination of G-Quadruplex Folding Topology
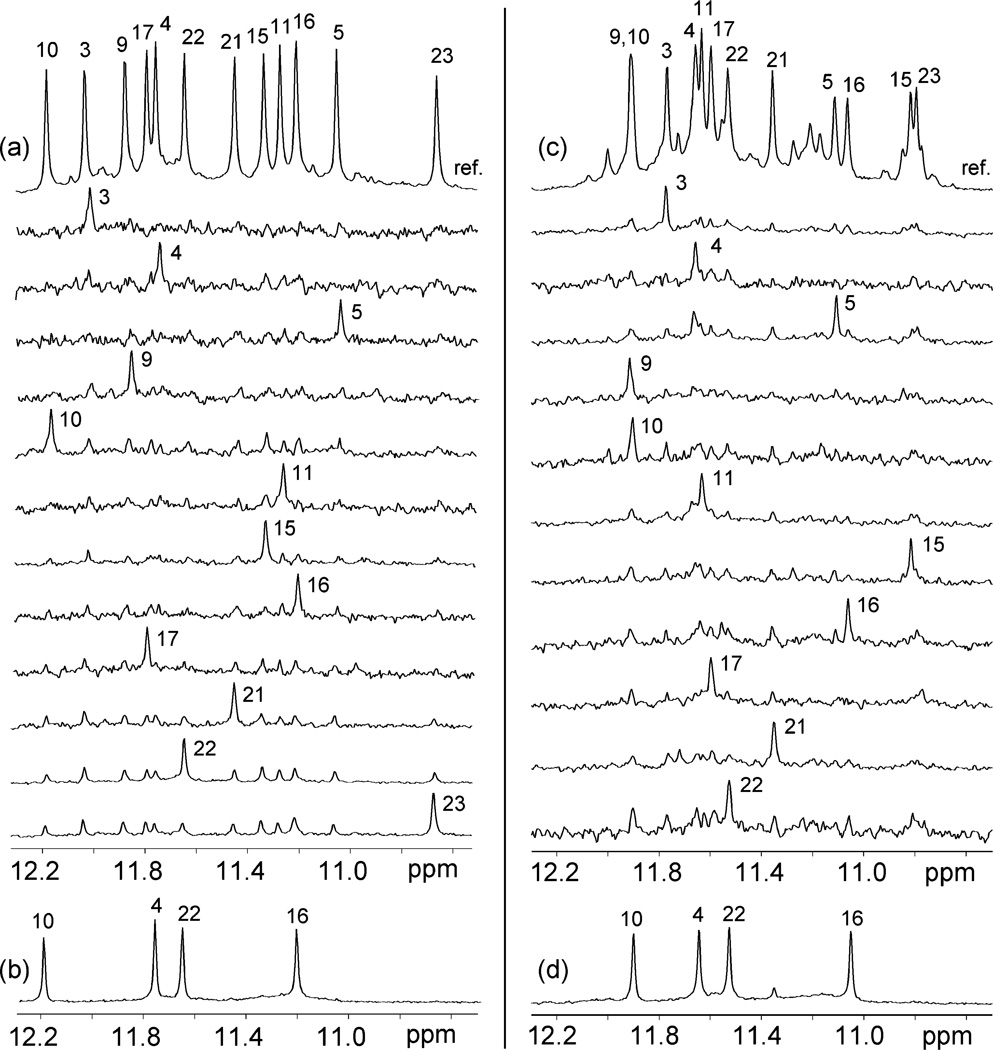
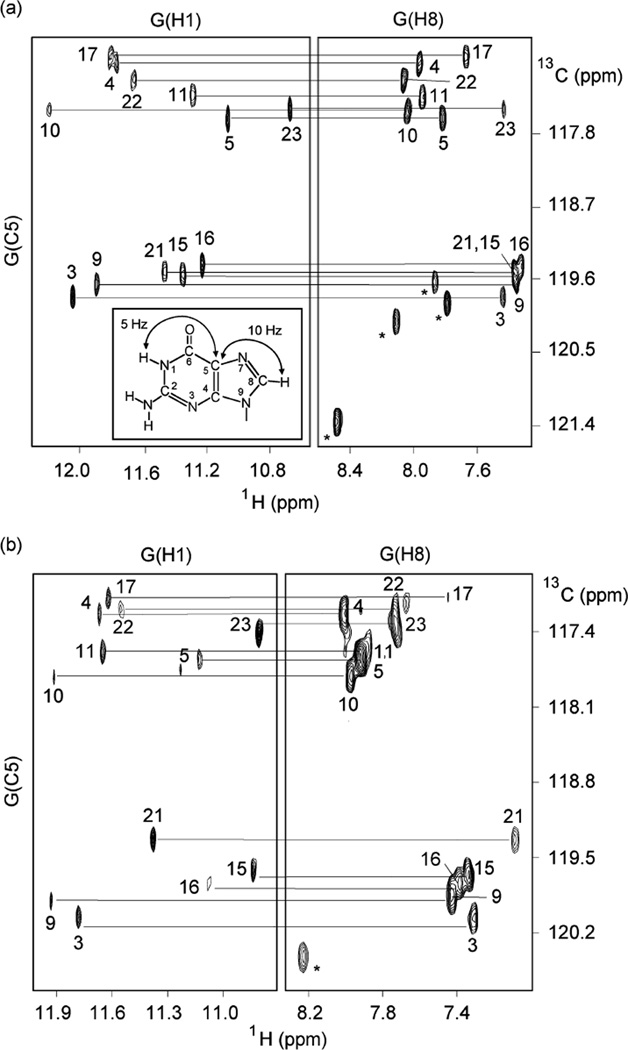
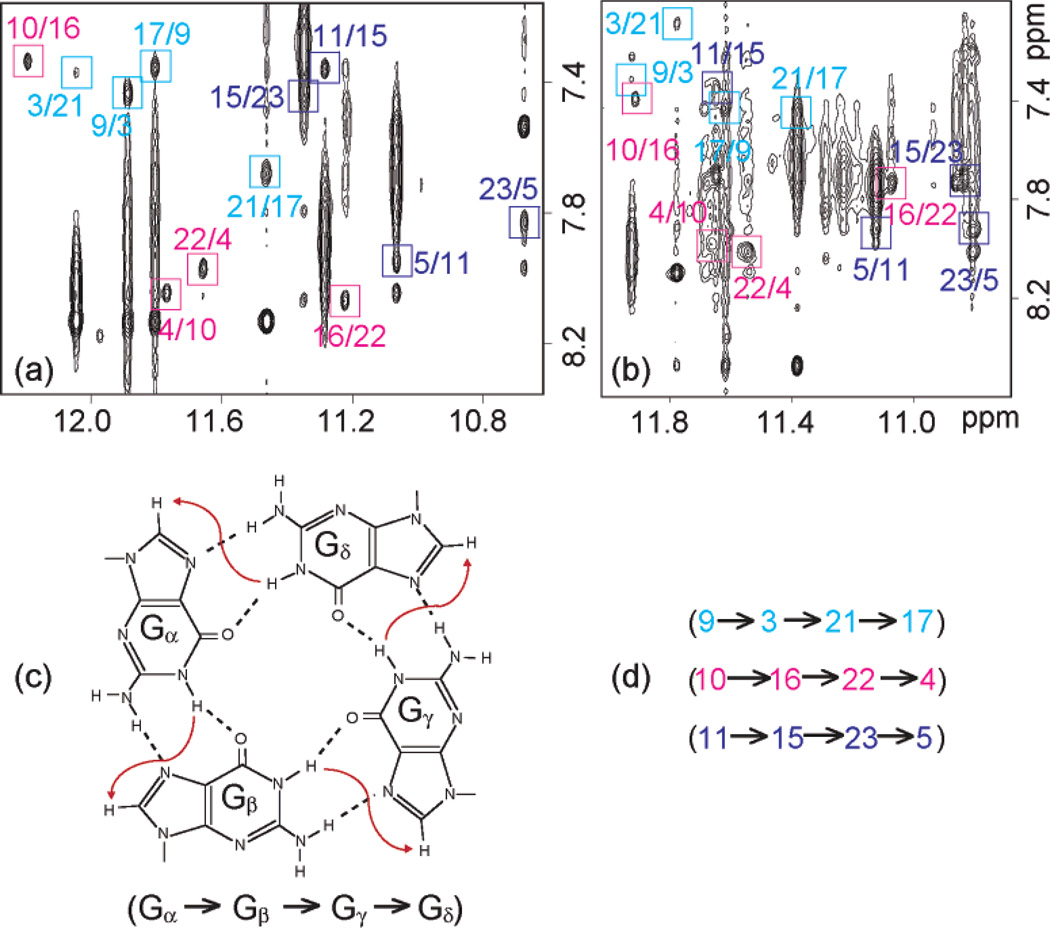
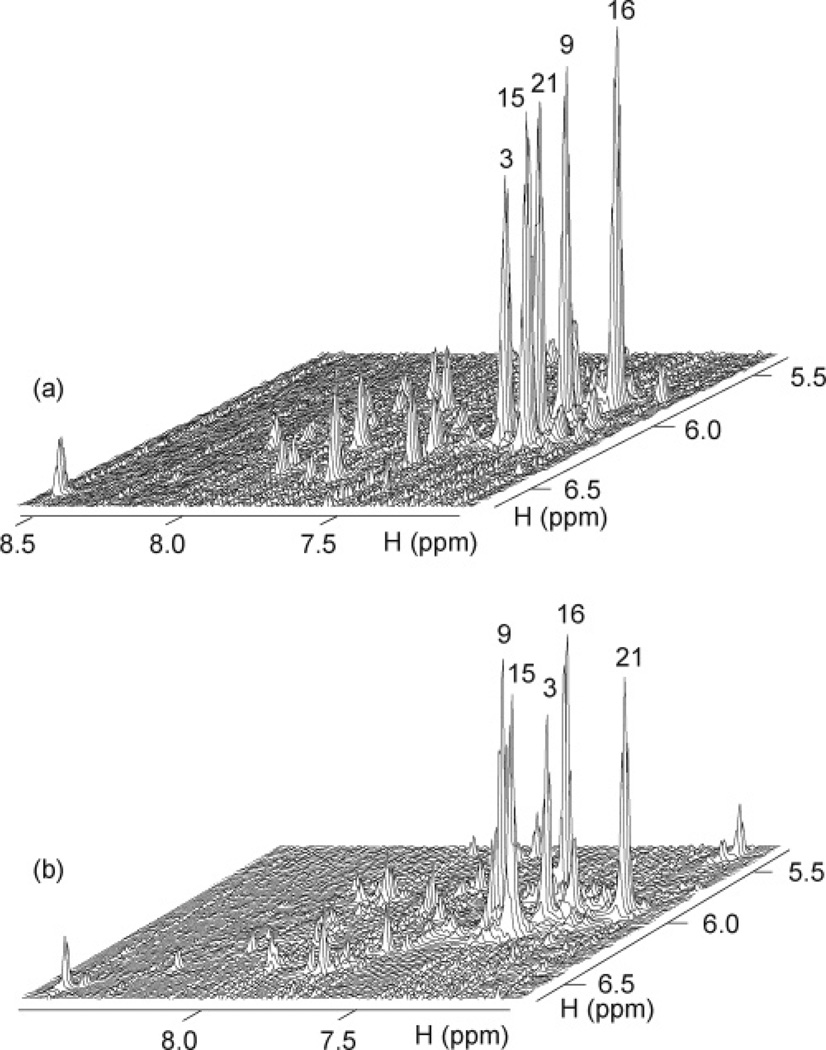
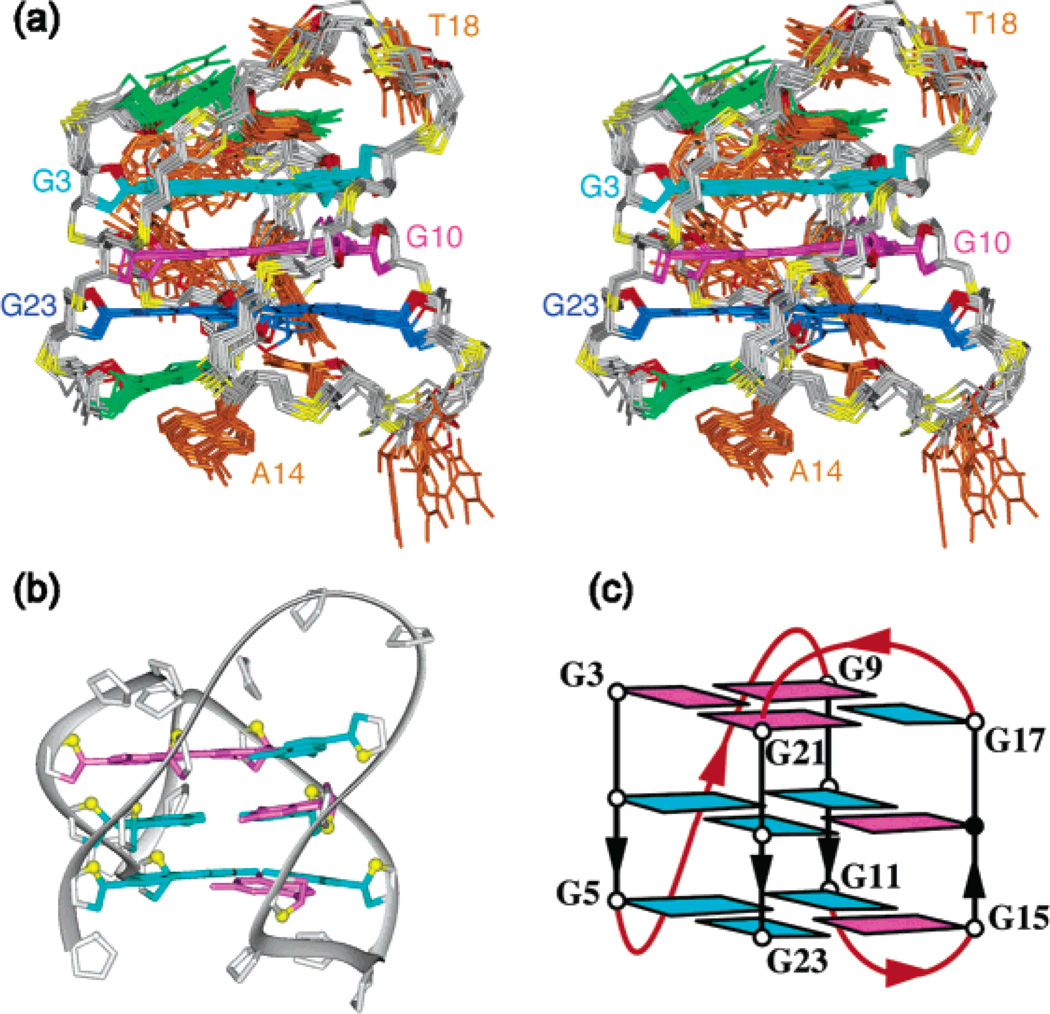
The spectral line widths of the modified 24-nt sequence (3–4 Hz for the sharpest peaks at 25 °C) are indicative of a monomeric intramolecular structure, and this was supported by the concentration-independence of the equilibrium between the structured and unfolded forms. We unambiguously assigned imino (Figure 3a) and H8 (Figure 4a) protons using the site-specific low-enrichment labeling22 and natural-abundance through-bond correlation23 strategies developed in our laboratory. Characteristic NOEs between imino and H8 protons (Figure 5a) established the fold of this G-quadruplex, involving three G-tetrads: G9•G3•G21•G17, G10•G16•G22•G4, and G11•G15•G23•G5. The glycosidic conformations of the first G-tetrad are anti•syn• syn•syn, those of the two other G-tetrads are syn•anti•anti•anti, as reflected by H1′–H8 NOE intensities observed for these residues (Figure 6a). These glycosidic conformations are consistent with the so-called (3 + 1) G-tetrad core24 containing three G-tracts (G9–G11, G3–G5, and G21–G23) oriented in one direction and the fourth (G15–G17) tract in the opposite direction (Figure 7c). The first linker (T6-T7-A8) forms a double-chain-reversal loop, while the two other linkers (T12-T13-A14 and T18-T19-A20) form edgewise loops. In this G-quadruplex fold G4, G10, G16 and G22 are in the central G-tetrad, consistent with imino protons of these residues being the most protected from exchange with water (Figure 3b).
Figure 3.
Imino proton spectra and assignments of (a, b) the modified 24-nt and (c, d) natural 23-nt human telomeric sequences in K+ solution. (a, c) Guanine imino proton spectra with assignments over the reference spectrum (ref). Imino protons were assigned in 15N-filtered spectra of samples, 2% 15N-labeled at the indicated positions. (b, d) Imino proton spectra after (b) 1 day and (d) 2 h in D2O at 25 °C.
Figure 4.
H8 proton assignments of (a) the modified 24-nt and (b) natural 23-nt human telomeric sequences by through-bond correlations between imino and H8 protons via 13C5 at natural abundance, using long-range J-couplings shown in the inset. Peaks from A(H8) are labeled with stars.
Figure 5.
Determination of G-quadruplex folding topology for the human telomeric sequence in K+ solution. (a, b) NOESY spectra (mixing time, 200 ms) of (a) the modified 24-nt and (b) natural 23-nt human telomeric sequences. Imino-H8 cross-peaks that identify three G-tetrads (colored cyan, magenta, and blue) are framed and labeled with the number of imino protons in the first position and that of H8 in the second position. (c) Characteristic guanine imino-H8 NOE connectivity patterns around a Gα•Gβ•Gγ•Gδ tetrad as indicated with arrows (connectivity between Gδ and Gα implied). (d) Characteristic guanine imino-H8 NOE connectivities observed for G9•G3•G21•G17 (cyan), G10•G16•G22•G4 (magenta), and G11•G15•G23•G5 (blue) tetrads.
Figure 6.
Stacked plot of NOESY spectra of (a) the modified 24-nt (mixing time, 75 ms) and (b) natural 23-nt (mixing time, 100 ms) human telomeric sequences that distinguish five strong intraresidue H8–H1′ cross-peaks (syn glycosidic bonds) from weak cross-peaks (anti glycosidic bonds).
Figure 7.
Solution structure of the d[TTGGG(TTAGGG)3A] quadruplex in K+ solution. (a) Stereoview of 12 superpositioned refined structures. Color code is as follows: G9•G3•G21•G17 tetrad, cyan; G10•G16•G22•G4 tetrad, magenta; G11•G15•G23•G5 tetrad, blue; terminal bases, green; loop bases, orange; backbone, gray; phosphorus atoms, yellow. (b) Ribbon and (c) schematic views of a representative refined structure. Color code is the same as that in Figure 1.
Similarly, data for the natural 23-nt human telomeric sequence, including unambiguous spectral assignments for imino (Figure 3c) and H8 protons (Figure 4b), NOE connectivities (Figure 5b) and intensities (Figure 6b), and hydrogen exchange (Figure 3d), independently established the same G-quadruplex fold for its major form.
Structure of Human Telomere G-Quadruplex in K+ Solution
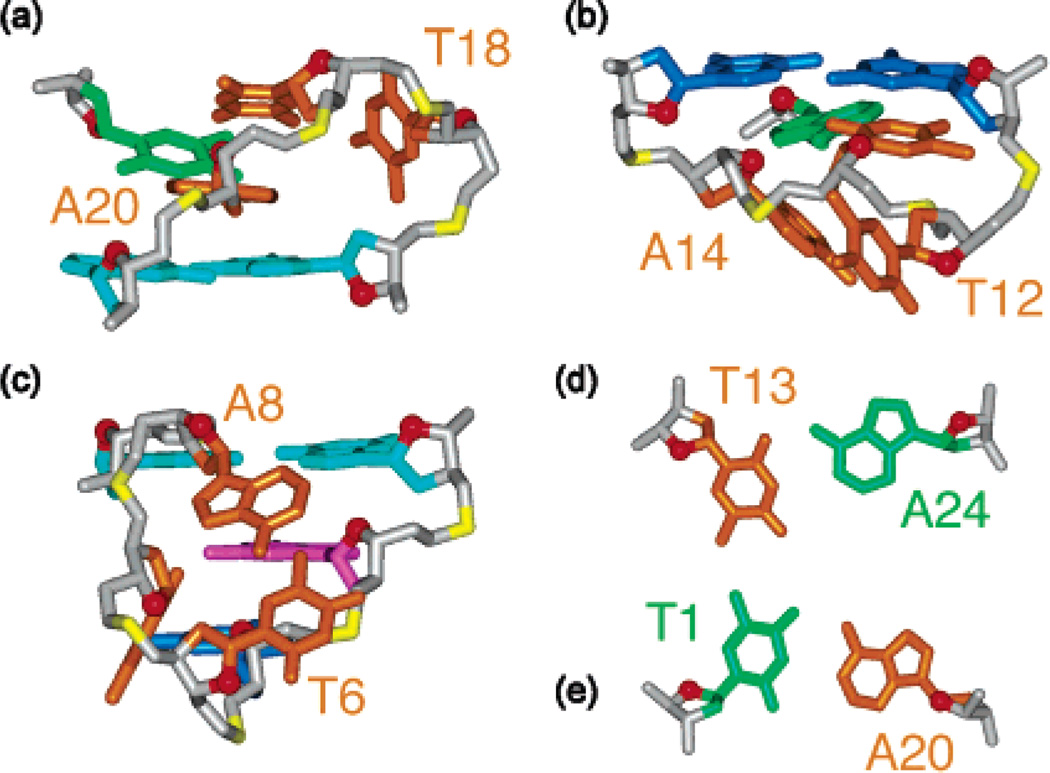
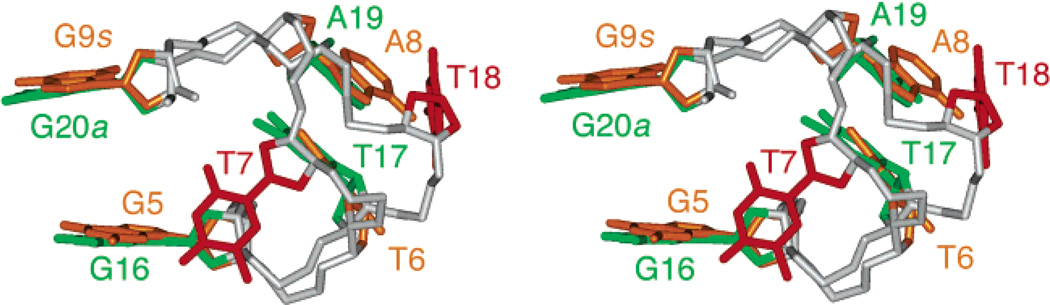
The G-quadruplex structure of the modified 24-nt sequence with a core of three G-tetrad layers (Figure 7) was calculated on the basis of NMR restraints (Table 1). The edgewise T18-T19-A20 and T12-T13-A24 loops (Figure 8a, b) are on the top and bottom of the G-tetrad core, respectively. In the ensemble of refined structures, we have observed Watson-Crick T1•A20 (Figure 8e) and reversed Watson-Crick T13•A24 (Figure 8d) pairs, stacked on either side of the G-tetrad core. These findings are supported by the detection of two thymine imino protons in the 12–13 ppm range at low temperatures (data not shown), thereby partly explaining the role of A24 in helping to favor this G-quadruplex fold. The double-chain-reversal T6-T7-A8 loop is less well defined (Figure 7a). This might reflect the existence of multiple conformations for this loop. Comparison between the structures of this TTA loop and the double-chain-reversal TTA loop observed in the crystal structure12 shows similar positions of the first T and third A but different positions of the middle T (Figure 9).
Table 1.
Statistics of the Computed Structures of the d[TTGGG(TTAGGG)3A] Quadruplex in K+ Solution
| A. NMR restraints | ||
| distance restraints | nonexchangeable | exchangeable |
| intraresidue distance restraints | 164 | 0 |
| sequential (i, i + 1) distance restraints | 92 | 17 |
| long-range (i, ≥ i + 2) distance restraints | 29 | 63 |
| other restraints | ||
| hydrogen bonding restraints | 48 | |
| torsion angle restraints | 55 | |
| intensity restraints | ||
| nonexchangeable protons | 255 | |
| B. Statistics for 12 structures following intensity refinement | ||
| NOE violations | ||
| number (>0.2 Å) | 0.300 ± 0.060 | |
| maximum violation (Å) | 0.218 ± 0.018 | |
| rmsd of violations (Å) | 0.022 ± 0.002 | |
| deviations from the ideal covalent geometry | ||
| bond lengths (Å) | 0.004 ± 0.000 | |
| bond angles (deg) | 0.956 ± 0.012 | |
| impropers (deg) | 0.302 ± 0.015 | |
| NMR R-factor ((R1/6) | 0.021 ± 0.006 | |
| Pairwise all heavy atom rmsd values (Å) | ||
| all heavy atoms except T6, T7, A8, T12 | 0.83 ± 0.12 | |
| all heavy atoms | 1.34 ± 0.32 | |
Figure 8.
Loop connectivities and base pairing in the d[TTGGG-(TTAGGG)3A] quadruplex in K+ solution. (a) Edgewise T18-T19-A20 loop. (b) Edgewise T12-T13-A14 loop. (c) Double-chain-reversal T6-T7-A8 loop. (d) Reversed Watson-Crick T13•A24 pair. (e) Watson-Crick T1•A20 pair.
Figure 9.
Comparison between TTA double-chain-reversal loops bridging three G-tetrads: Stereoview of the T6-T7-A8 loop in the d[TTGGG(TTAGGG)3A] quadruplex in K+ solution (this work) and the T17-T18-A19 loop in the d[AGGG(TTAGGG)3] quadruplex in a K+-containing crystal structure.12 Notice that the first T and last A are better superpositioned than the central T; G9 is syn, while G20 is anti.
Comparison with Earlier Structures of Human Telomere G-Quadruplex
The intramolecular (3+1) G-quadruplex fold of human telomeric DNA observed in K+ solution here (Figure 7b, c) is very different from the structures observed in Na+ solution (Figure 1a, b) or in a K+-containing crystal (Figure 1c, d), in terms of strand orientations, loop topologies, and glycosidic conformations of guanines. In the present fold, 5′- and 3′- ends of the sequence are located at opposite ends of the G-tetrad core (similar to the K+ crystal structure12 and different from the Na+ solution structure11), thereby potentially allowing continuous stacking of successive blocks within a longer sequence context.12
(3+1) Core Architecture Represents a Robust Scaffold
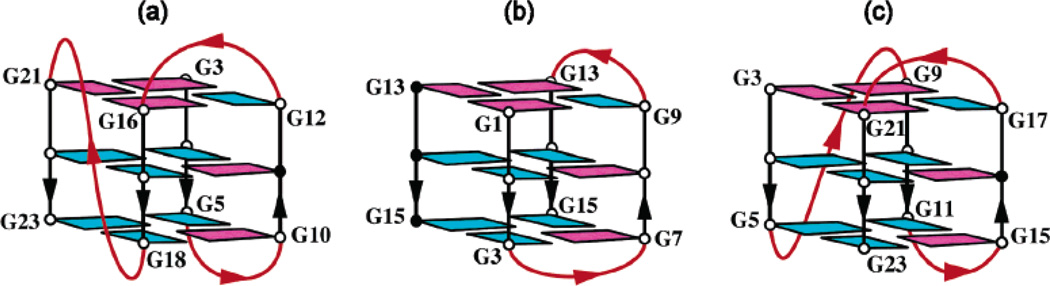
This G-quadruplex topology can also be compared with the intramolecular G-quadruplex formed by the four-repeat Tetrahymena TTGGGG telomeric sequence25 (Figure 10a) and with the dimeric G-quadruplex formed by the three-repeat human telomeric sequence24 (Figure 10b). All these structures contain the (3 + 1) core G-quadruplex topology, in which three strands are oriented in one direction and the fourth is in the opposite direction and G-tetrads are anti•syn•syn•syn or syn•anti•anti• anti. However, there are differences in the order of G-tracts in these three structures: when their G-tetrad cores are aligned, the 5′-end in each structure starts from a distinct corner of the core (Figure 10). In intramolecular (3 + 1) G-quadruplexes (Figure 10a, c), either the first loop (Figure 10c) or the third loop (Figure 10a) can be double-chain-reversal. Recently, a variant sequence of the four G-tract human bcl-2 promoter26 was shown to adopt the G-quadruplex fold with the same (3 + 1) core topology as the G-quadruplex fold formed by the four-repeat Tetrahymena telomeric sequence.25 Thus, the (3 + 1) core topology of the G-quadruplex, initially identified in 1994 (ref 25) and thought to be an anomaly at that time, now appears to be a robust G-quadruplex scaffold, adopted by both telomeric and oncogenic promoter sequences.
Figure 10.
Schematic structures of the (3 + 1) core-containing G-quadruplexes formed by (a) the four-repeat Tetrahymena telomeric d[(TTGGGG)4] sequence in Na+ solution,25 (b) the three-repeat human telomeric d[GGG(TTAGGG)2T] sequence in Na+ solution,24 and (c) the four-repeat human telomeric d[TAGGG(TTAGGG)3] sequence in K+ solution (this work). Color code is the same as that in Figure 1.
Ligand Recognition of the (3+1) Core G-Quadruplex Scaffold
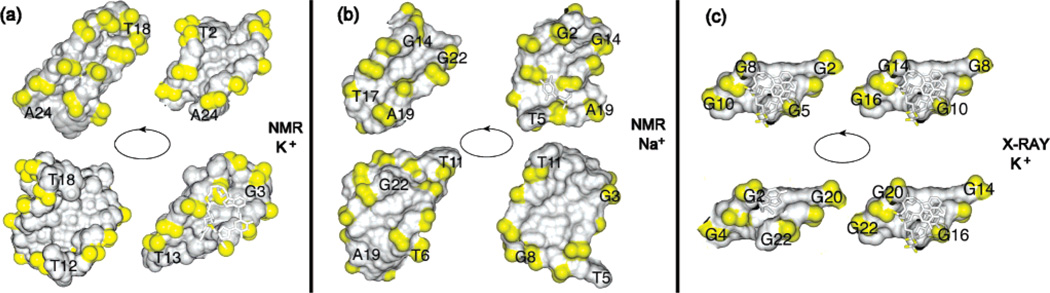
The present (3 + 1) G-tetrad core is identified by one narrow, one wide, and two medium grooves (Figure 11a), thereby providing a unique target for small-molecule ligands. One of the grooves is occupied by the double-chain-reversal loop, but the other three grooves are accessible for recognition. It should be noted that while planar ligands have the potential to stack over the ends of the quadruplex,27–29 either planar30 or nonplanar ligands can be positioned in the grooves of the quadruplex. The groove dimensions of the (3 + 1) core topology (Figure 11a) are quite different from those reported previously for the human telomeric G-quadruplex structures in Na+ solution11 (Figure 11b) and in the K+-containing crystalline state12 (Figure 11c).
Figure 11.
Surface view of four grooves in intramolecular G-quadruplexes formed by the human telomeric sequence: (a) in K+ solution (this work); (b) in Na+ solution;11 (c) in a K+-containing crystal.12 Phosphorus atoms and backbone oxygens are colored yellow.
Conclusion
We have achieved the long-time goal of solving the structure of an intramolecular human telomeric G-quadruplex in physiological K+ solution conditions. Our study revealed an intramolecular G-quadruplex with a (3 + 1) core topology (Figure 7b, c), which is distinctly different from previously reported structures in Na+ solution11 (Figure 1a, b) and in a K+-containing crystal12 (Figure 1c, d). The robustness of the (3 + 1) core G-quadruplex topology, as demonstrated here for the four-repeat human telomere sequence in K+ solution and observed elsewhere within other sequence contexts24–26 (Figure 10), makes this scaffold a unique platform for structure-based anticancer drug design.1 Our future efforts will be directed toward deciphering the folding topologies of other G-quadruplex conformations that are formed by human telomere sequences in K+ solution.13
Methods
Sample Preparation
The unlabeled and the site-specific low-enrichment (2% 15N-labeled) oligonucleotides were synthesized and purified as described previously.31,22 Unless otherwise stated, the strand concentration of the NMR samples was typically 0.5–5 mM; the solutions contained 70 mM KCl and 20 mM potassium phosphate (pH 7).
NMR Spectroscopy
Experiments were performed on 600 MHz Varian and 800 MHz Bruker spectrometers at 25 °C, unless otherwise specified. Resonances were assigned unambiguously by using site-specific low-enrichment labeling22 and through-bond correlations at natural abundance.23 The resonances for T residues were assigned following systematic T-to-U replacements. Spectral assignments were also assisted and supported by NOESY spectra. Interproton distances were measured by using NOESY experiments at different mixing times.
Structure Calculation
The structures of the d[TTGGG(TTAGGG)3A] quadruplex were calculated using the X-PLOR program.32 NMR-restrained molecular dynamics (torsion dynamics33,34 and relaxation-matrix refinement) computations were performed as described previously.31
Data Deposition
The coordinates for the d[TTGGG(TTAGGG)3A] quadruplex have been deposited in the Protein Data Bank (accession code 2GKU).
Acknowledgments
This research was supported by National Institutes of Health Grant GM34504. D.J.P. is a member of the New York Structural Biology Center supported by National Institutes of Health Grant GM66354.
Footnotes
Note Added after ASAP Publication. There was a typographical error in the sequence notation in the caption for Figure 10a when this paper was published ASAP on July 7, 2006, that was corrected on the same day.
Contributor Information
Anh Tuân Phan, Email: phantuan@mskcc.org.
Dinshaw J. Patel, Email: pateld@mskcc.org.
References
- 1.(a) Neidle S, Parkinson G. Nat. Rev. Drug Discovery. 2002;1:383–393. doi: 10.1038/nrd793. [DOI] [PubMed] [Google Scholar]; (b) Hurley LH. Nat. Rev. Cancer. 2002;2:188–200. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]; (c) Mergny JL, Riou JF, Mailliet P, Teulade-Fichou MP, Gilson E. Nucleic Acids Res. 2002;30:839–865. doi: 10.1093/nar/30.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR. Proc. Natl. Acad. Sci. U.S.A. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Patel DJ, Bouaziz S, Kettani A, Wang Y. In: Oxford Handbook of Nucleic Acid Structures. Neidle S, editor. Oxford: Oxford University Press; 1999. pp. 389–453. [Google Scholar]; (b) Davis JT. Angew. Chem., Int. Ed. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]; (c) Phan AT, Kuryavyi V, Patel DJ. Curr. Opin. Struct. Biol. 2006;16:288–298. doi: 10.1016/j.sbi.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Nat. Struct.. Mol. Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 5.(a) Makarov VL, Hirose Y, Langmore JP. Cell. 1997;88:657–666. doi: 10.1016/s0092-8674(00)81908-x. [DOI] [PubMed] [Google Scholar]; (b) Wright WE, Tesmer VM, Huffman KE, Levene SD, Shay JW. Genes Dev. 1997;11:2801–2809. doi: 10.1101/gad.11.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) McElligott R, Wellinger RJ. EMBO J. 1997;16:3705–3714. doi: 10.1093/emboj/16.12.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greider CW, Blackburn EH. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 7.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 8.Zahler AM, Williamson JR, Cech TR, Prescott DM. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 9.Zaug AJ, Podell ER, Cech TR. Proc. Natl. Acad. Sci. U.S.A. 2005;102:10864–10869. doi: 10.1073/pnas.0504744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oganesian L, Moon IK, Bryan TM, Jarstfer MB. EMBO J. 2006;25:1148–1159. doi: 10.1038/sj.emboj.7601006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Patel DJ. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 12.Parkinson GN, Lee MPH, Neidle S. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 13.Phan AT, Patel DJ. J. Am. Chem. Soc. 2003;125:15021–15027. doi: 10.1021/ja037616j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ying L, Green JJ, Li H, Klenerman D, Balasubramanian S. Proc. Natl. Acad. Sci. U.S.A. 2003;100:14629–14634. doi: 10.1073/pnas.2433350100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He Y, Neumann RD, Panyutin IG. Nucleic Acids Res. 2004;32:5359–5367. doi: 10.1093/nar/gkh875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rujan IN, Meleney JC, Bolton PH. Nucleic Acids Res. 2005;33:2022–2031. doi: 10.1093/nar/gki345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qi J, Shafer RH. Nucleic Acids Res. 2005;33:3185–3192. doi: 10.1093/nar/gki632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vorlickova M, Chladkova J, Kejnovska I, Fialova M, Kypr J. Nucleic Acids. Res. 2005;33:5851–5860. doi: 10.1093/nar/gki898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.(a) Ourliac-Garnier I, Elizondo-Riojas MA, Redon S, Farrell NP, Bombard S. Biochemistry. 2005;44:10620–10634. doi: 10.1021/bi050144w. [DOI] [PubMed] [Google Scholar]; (b) Xu Y, Sugiyama H. J. Am. Chem. Soc. 2004;126:6274–6279. doi: 10.1021/ja031942h. [DOI] [PubMed] [Google Scholar]
- 20.Li J, Correia JJ, Wang L, Trent JO, Chaires JB. Nucleic Acids. Res. 2005;33:4649–4659. doi: 10.1093/nar/gki782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee JY, Okumus B, Kim DS, Ha T. Proc. Natl. Acad. Sci. U.S.A. 2005;102:18938–18943. doi: 10.1073/pnas.0506144102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phan AT, Patel DJ. J. Am. Chem. Soc. 2002;124:1160–1161. doi: 10.1021/ja011977m. [DOI] [PubMed] [Google Scholar]
- 23.Phan AT, Guéron M, Leroy JL. Methods Enzymol. 2001;338:341–371. doi: 10.1016/s0076-6879(02)38228-4. [DOI] [PubMed] [Google Scholar]
- 24.Zhang N, Phan AT, Patel DJ. J. Am. Chem. Soc. 2005;127:17277–17285. doi: 10.1021/ja0543090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Patel DJ. Structure. 1994;2:1141–1156. doi: 10.1016/s0969-2126(94)00117-0. [DOI] [PubMed] [Google Scholar]
- 26.Dai J, Dexheimer TS, Chen D, Carver M, Ambrus A, Jones RA, Yang D. J. Am. Chem. Soc. 2006;128:1096–1098. doi: 10.1021/ja055636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han H, Langley DR, Rangan A, Hurley LH. J. Am. Chem. Soc. 2001;123:8902–8913. doi: 10.1021/ja002179j. [DOI] [PubMed] [Google Scholar]
- 28.Haider SM, Parkinson GN, Neidle S. J. Mol. Biol. 2003;326:117–125. doi: 10.1016/s0022-2836(02)01354-2. [DOI] [PubMed] [Google Scholar]
- 29.Phan AT, Kuryavyi V, Gaw HY, Patel DJ. Nat. Chem. Biol. 2005;1:167–173. doi: 10.1038/nchembio723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kettani A, Gorin A, Majumdar A, Hermann T, Skripkin E, Zhao H, Jones R, Patel DJ. J. Mol. Biol. 2000;297:627–644. doi: 10.1006/jmbi.2000.3524. [DOI] [PubMed] [Google Scholar]
- 31.Phan AT, Kuryavyi V, Ma JB, Faure A, Andréola ML, Patel DJ. Proc. Natl. Acad. Sci. U.S.A. 2005;102:634–639. doi: 10.1073/pnas.0406278102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brünger AT. X-PLOR: A system for X-ray crystallography and NMR. New Haven, CT: Yale University Press; 1992. [Google Scholar]
- 33.Rice LM, Brünger AT. Proteins. 1994;19:277–290. doi: 10.1002/prot.340190403. [DOI] [PubMed] [Google Scholar]
- 34.Stein EG, Rice LM, Brünger AT. J. Magn. Reson. 1997;124:154–164. doi: 10.1006/jmre.1996.1027. [DOI] [PubMed] [Google Scholar]