Abstract
Silencing of the tumor suppressor protein BRCA2 and its detection by conventional biochemical analyses represent a great technical challenge owing to the large size of the human BRCA2 protein (approximately 390 kDa). We report modifications of standard siRNA transfection and immunoblotting protocols to silence human BRCA2 and detect endogenous BRCA2 protein, respectively, in human epithelial cell lines. Key steps include a high siRNA to transfection reagent ratio and two subsequent rounds of siRNA transfection within the same experiment. Using these and other modifications to the standard protocol we consistently achieve more than 70% silencing of the human BRCA2 gene as judged by immunoblotting analysis with anti-BRCA2 antibodies. In addition, denaturation of the cell lysates at 55 °C instead of the conventional 70-100 °C and other technical optimizations of the immunoblotting procedure allow detection of intact BRCA2 protein even when very low amounts of starting material are available or when BRCA2 protein expression levels are very low. Efficient silencing of BRCA2 in human cells offers a valuable strategy to disrupt BRCA2 function in cells with intact BRCA2, including tumor cells, to examine new molecular pathways and cellular functions that may be affected by pathogenic BRCA2 mutations in tumors. Adaptation of this protocol for efficient silencing and analysis of other 'large' proteins like BRCA2 should be readily achievable.
Keywords: Molecular Biology, Issue 102, BRCA2, siRNA, human cell lines, gene silencing, transfection, immunoblotting
Introduction
The BRCA2 (BReast CAncer susceptibility gene-2) gene encodes for a tumor suppressor protein that plays a crucial role in repairing DNA double-strand breaks by regulating the function of the recombinase enzyme Rad51 1. BRCA2 has also been implicated in the modulation of transcription and in cell cycle control 2. Germline mutations in the BRCA2 gene induce an autosomal dominant susceptibility to breast and ovarian cancer in women and prostate cancer in men, as well as predisposition to other cancer types 3,4. However, despite the increased risk in developing cancer, BRCA2 mutations that suppress or reduce BRCA2 function may render cancer cells more vulnerable to chemotherapeutic agents that cause DNA damage 5-7. Sporadic cancers exhibit a low rate of BRCA2 mutations (< 3%) though reduced levels of BRCA2 protein have been detected in some cancer types, suggesting that BRCA2 protein may be lost during tumorigenesis in sporadic cancers through non-mutational-dependent mechanisms 7,8. Thus, it is important to fully understand BRCA2 functions in the context of cancer biology as well as in other biological settings.
A powerful tool used to identify novel functions of a gene in mammalian cells is to silence its expression. As an example, silencing BRCA2 expression in a variety of normal epithelial cells has recently led to the identification of a novel function of BRCA2 as regulator of anoikis resistance, an important step during acquisition of cancer cell invasive and metastatic ability 9. Gene silencing can be achieved by introducing in the cells either small interfering (si) RNA or short hairpin (sh) RNA molecules targeting the specific gene. The high potency of siRNA and its ease of use make it the preferential tool for silencing experiments aimed at gaining new insights into critical biological processes and to identify novel therapeutic targets. However, efficient knockdown of gene expression may not be easily achievable for all genes and may be highly variable depending on the cell type. Because a decrease in intracellular protein levels is the most relevant phenotype under investigation, it is crucial to quantify gene silencing by immunoblotting analysis of the gene product. With this respect, the BRCA2 protein presents a further challenge: being a large protein (approximately 390 kDa), technical difficulties do exist for conventional biochemical analysis, including immunoblotting.
We report here a protocol optimized for efficient silencing of BRCA2 in human epithelial cell lines and for rapid and successful detection of BRCA2 protein knockdown by immunoblotting analysis.
Protocol
1. Prepare siRNA solutions
Resuspend 5 nmol of scrambled (Ctrl) and 5 nmol of BRCA2 siRNA dried pellets in 100 µl of RNase-free water (final siRNA concentration: 50 µM). This is the siRNA stock and must be stored at -20 °C in 10 µl aliquots.
Take an aliquot of 10 µl from the siRNA stock and add 40 µl of RNase-free water to obtain the 10 µM siRNA working solution. This dilution must be stored at -20 °C until use. NOTE: The siRNA working solution should not undergo freezing/thawing more than 3 times.
2. Seeding of Human Epithelial Cell Lines
Remove the growth medium from human epithelial cell lines growing as cell monolayers in tissue culture plates in a humidified incubator at 37 °C, 5% CO2.
Wash the cells in the plate once with RT PBS (10 ml of PBS for a 100 mm plate).
To detach the cells from the plate, add 2 ml of 0.25% trypsin/0.53 mM EDTA solution and incubate 3-5 min depending on the cell type.
Before collecting the detached cells, neutralize trypsin by adding 2 ml of complete growth medium containing 10% fetal bovine serum.
Transfer the cells to a 15 ml tube.
Centrifuge the cells at 320-330 x g for 3 min at 4 °C in a swinging bucket rotor.
Resuspend the cell pellets in 5 ml of antibiotics-free medium.
Count the cells under the microscope in a Burker chamber or by using an automated counting system according to the manufacturer’s instructions.
Seed the cells in 6-well plates to be about 80% confluent after 24 hr (0.5-1 x106 cells in 2 ml of medium for each well). Optimal cell number to be seeded may vary depending on the specific cell lines and the growth rate. NOTE: Do not add antibiotics in the medium at this point because antibiotics will negatively interfere with the efficiency of transfection. It is very important that the cells are at low culture passages (< 10) to achieve high efficiency of BRCA2 siRNA transfection.
3. Transfect cells with siRNA
- Twenty-four hours after plating the cells, proceed with the first round of transfection.
- Dilute 6 µl of lipid-based 10 siRNA specific transfection reagent in 130 µl of reduced serum medium.
- Dilute 3 µl of 10 µM siRNA (30 pmol) in 130 µl of reduced serum medium.
- Combine the diluted siRNA with the diluted transfection reagent and incubate for 5-10 min at RT.
- During the incubation, remove the medium from the cells and add 2 ml of fresh medium pre-warmed at 37 °C (without antibiotics).
- Add 250 µl of the siRNA-transfection reagent complexes to the cells (this amount is for a single well of a 6-well plate).
- Incubate the cells in a humidified incubator at 37 °C with 5% CO2.
- After 24 hr, proceed with the second round of transfection by repeating steps 3.1.1-3.1.5 with the following modification for step 3.1.2: dilute 5 µl of 10 µM siRNA (50 pmol) in 130 µl of reduced serum medium.
- Incubate the cells for 1-2 days at 37 °C before analysis. NOTE: Two rounds of transfections and use of a high siRNA/transfection reagent ratio in the second round are essential to get high efficiency of BRCA2 silencing.
4. Lyse cells for immunoblotting analysis
Prepare fresh lysis buffer with PBS (pH 7.4) containing 1% of the nonionic, non-denaturing detergent octylphenoxypolyethoxyethanol, 5 mM sodium pyrophosphate, 1 mM sodium orthovanadate, 10 mM sodium fluoride, 2 mM phenylmethylsulfonyl fluoride, 10 µg/ml leupeptin, 10 µg/ml aprotinin. Keep it on ice.
Remove the medium from the cells and wash them once with cold PBS (2 ml/well) in the plate. To prevent any PBS carryover, completely remove it from the plate.
Add 200 µl of lysis buffer to each well. NOTE: If the cells have to be used also for other assays, trypsinize the cells as described in step 2.1-2.6 and resuspend the cells in appropriate buffer/medium according to the application to be used.
Gently rotate the plate to uniformly distribute the lysis buffer and incubate for 15 min at 4 °C on a rotator.
Collect the cell lysate in a microcentrifuge tube on ice using a cell scraper.
Centrifuge at 17,900 x g for 25 min at 4 °C and collect the supernatant in a new tube.
Measure the protein concentration using a commercially available protein assay according to the manufacturer's instructions.
5. BRCA2 immunoblotting analysis
Prepare the running buffer by diluting 50 ml of 20x Tris-Acetate SDS running buffer (50 mM Tricine, 50 mM Tris Base, 0.1% SDS, pH 8.24) with 950 ml of distilled water.
Prepare the sample as follows: add 15 µg of total cell lysate, 5 µl of 4x sample buffer containing lithium dodecyl sulfate at pH 8.4, 2 µl of 10x sample reducing agent (0.5 M DTT), and lysis buffer to a final volume of 20 µl. NOTE: This protocol allows detection of BRCA2 in normal cell lines also when using a lower amount of total proteins (down to 5 µg).
Denature samples at 55 °C for 10 min. NOTE: It is crucial to use 55 °C. Since BRCA2 protein is thermosensitive 11, higher denaturing temperatures result in consistent loss of intact full-length (390 kDa) BRCA2 protein.
Set up the electrophoretic chamber according to the manufacturer's instructions.
Load the samples and 10 µl of high-molecular weight pre-stained protein standard on a precasted gel (Tris-Acetate 3-8%) and run at 120 V for about 2 hr 12. Stop the electrophoretic run before the 55 kDa blue marker leaves the precasted gel.
Prepare the transfer buffer with 50 ml of 20x transfer buffer, 100 ml of 100% methanol and 850 ml water.
Activate PVDF membrane (0.45 µm pore size) by soaking in 100% methanol for 5 min, rinse with distilled water and equilibrate in transfer buffer at least for 5 min. Soak the sponges of the transfer apparatus in Transfer buffer at 4 °C until use.
When the electrophoretic run is over, assemble the sandwich in the transfer stack in the following order (bottom-up): three sponges saturated in transfer buffer, one 3MM grade paper sheet saturated in transfer buffer, the gel, activated PVDF membrane, one 3MM grade paper sheet saturated in transfer buffer, three sponges saturated in transfer buffer 13,14. Place the stack into the apparatus and transfer proteins at 350 mA for 4 hr at 4 °C or at 180 mA overnight at 4 °C. NOTE: Being BRCA2 a very large protein, it is crucial to extend the transfer time from 1 hr (as suggested by the manufacturer and most transfer protocols) to 4 hr or overnight to ensure complete transfer of high-molecular weight proteins. In addition, due to the long transfer time, it is essential to perform the blotting in a cold room at 4 °C to minimize overheating and facilitate sandwich disassembling. Semi-dry transfer systems are not good for transferring large proteins.
After transfer, wash the membrane with TBS, block it for 1 hr at RT in TBS-Tween 0.25% (TBS-T) containing 5% nonfat dry milk and probe overnight at 4 °C with 30 μl anti-BRCA2 rabbit polyclonal antibody (200 μg/ml), diluted in 9 ml TBS-T + 5% nonfat dry milk [1:300 (v/v) dilution].
Wash the membrane three times (10 min each) with TBS-T, then incubate the filter for 1 hr with 1 μl of horseradish peroxidase-conjugated anti-rabbit secondary antibody diluted in 10 ml of TBS-T + 5% nonfat dry milk. Thereafter, wash the membrane three times (10 min each) with TBS-T.
Perform immunodetection by ECL to reveal BRCA2 protein using enhanced chemiluminescence (ECL) Western Blotting Detection Reagent, following the manufacturer's instructions. Visualize immunochemiluminescent bands on high resolution ECL films.
Perform immunodetection with an antibody for a housekeeping gene, like β-tubulin (55 kDa), on the same filter as loading control by using a monoclonal antibody to β-tubulin diluted at 1:1,000 (10 µl of anti-tubulin antibody in 10 ml TBS-T+5% non-fat dry milk) for 1 hr at RT. Wash and incubate secondary antibody as described for BRCA2 (5.10-5.11), except for the use of horseradish peroxidase-conjugated anti-mouse instead of anti-rabbit secondary antibody.
Acquire a digital image of the films impressed with immunochemiluminescent bands and analyze the band intensity by dedicated image analysis software (e.g., ImageJ).
Representative Results
Before proceeding with a biological/biochemical assay to examine the effect of BRCA2 silencing on cell functions, the first step is to confirm the specificity of the silencing by using a scrambled siRNA (non-targeting siRNA) side by side with BRCA2 siRNA (Figure 1A). It is important to perform a second round of siRNA transfection with a high siRNA/transfection ratio to get a high efficiency of BRCA2 silencing. Indeed, performing only one round of siRNA transfection or using a lower siRNA to transfection ratio in the second round (25 pmol of siRNA and 6 µl of transfection reagent) would result in lower knockdown efficiency (Figure 1B). The second step is to assess the optimal minimum time to get the highest level of gene silencing. Depending on the cell type, this may vary between 24 and 48 hr after the second round of siRNA transfection. For example, 24 hr are sufficient for Nthy thyroid cells but up to 48 hr are needed for efficient BRCA2 knockdown in PNT1A prostate cells (Figure 2A). BRCA2 silencing is quite stable until day 7 after the second transfection cycle (Figure 2B). Of note, denaturing the cell lysates at a temperature above 55 °C (100 °C for 5 min), results in up to 50% loss of BRCA2 protein (Figure 2C), due to the thermosensitivity of BRCA2 11.
Once optimal silencing time is established, the effects of loss of BRCA2 protein may be exploited in several assays. Since in vivo mutations that cause loss of function of BRCA2 promote increased sensitivity to PARP inhibitors as well as to other chemotherapeutic drugs that cause DNA damage 15-18, a common assay is to determine the sensitivity of BRCA2-silenced cells to the PARP inhibitor rucaparib or oloparib. In the experiment reported in Figure 3, PNT1A cells depleted of BRCA2 protein through siRNA have been treated with 10 µM rucaparib for 24 hr, after which cell proliferation has been assessed by an MTT-based cell proliferation assay. Depletion of BRCA2 protein results in a time-dependent decrease in cell proliferation after rucaparib treatment (Figure 3).
 Figure 1. Two rounds of transfection and high siRNA to transfection reagent ratio improve BRCA2 silencing. (A) Specificity of BRCA2 silencingwas confirmed in Nthy cells by immunoblotting analysis 24 hr after the second round of transfection, using scrambled siRNA (Ctrl) as control for comparison of BRCA2 protein levels. Molecular weight markers are reported on the left. (B) PNT1A cells were subjected to 1 or 2 cycles of BRCA2 siRNA transfection as described in the protocol [Cycles (#) 1 and 2] and BRCA2 protein depletion was quantified by immunoblotting. Two cycles of transfection with scrambled siRNA were used as control (Ctrl). In 2b, PNT1A cells were subjected to two cycles of BRCA2 siRNA transfection using a lower siRNA to transfection reagent ratio in the second cycle as a modification (25 pmol siRNA/6 µl transfection reagent). In all cases, BRCA2 protein levels were analyzed 48 hr after the second cycle. At the bottom, quantification of BRCA2 protein levels is reported as BRCA2/Tubulin ratio. Please click here to view a larger version of this figure.
Figure 1. Two rounds of transfection and high siRNA to transfection reagent ratio improve BRCA2 silencing. (A) Specificity of BRCA2 silencingwas confirmed in Nthy cells by immunoblotting analysis 24 hr after the second round of transfection, using scrambled siRNA (Ctrl) as control for comparison of BRCA2 protein levels. Molecular weight markers are reported on the left. (B) PNT1A cells were subjected to 1 or 2 cycles of BRCA2 siRNA transfection as described in the protocol [Cycles (#) 1 and 2] and BRCA2 protein depletion was quantified by immunoblotting. Two cycles of transfection with scrambled siRNA were used as control (Ctrl). In 2b, PNT1A cells were subjected to two cycles of BRCA2 siRNA transfection using a lower siRNA to transfection reagent ratio in the second cycle as a modification (25 pmol siRNA/6 µl transfection reagent). In all cases, BRCA2 protein levels were analyzed 48 hr after the second cycle. At the bottom, quantification of BRCA2 protein levels is reported as BRCA2/Tubulin ratio. Please click here to view a larger version of this figure.
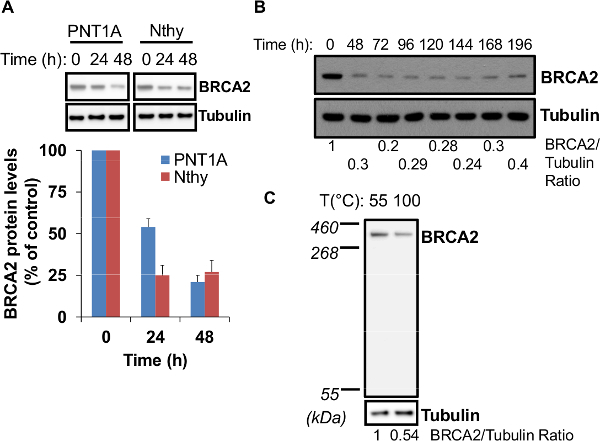 Figure 2. Optimization of BRCA2 knockdown. (A) Optimal BRCA2 knockdown may require 24 to 48 hr after the second siRNA transfection cycle. Two different cell lines (PNT1A and Nthy) were assessed for BRCA2 knockdown 24 hr and 48 hr after the second siRNA transfection cycle by immunoblotting analysis. At the bottom, BRCA2 protein levels are reported as percentage of the levels at time 0 hr, set at 100. Data represent the mean ± SE of three independent experiments. (B) BRCA2 knockdown is stable for 7 days. PNT1A cells were assessed for BRCA2 knockdown until 196 hr after the second siRNA transfection cycle by immunoblotting. The ratio of BRCA2 to tubulin signal is reported at the bottom. (C) BRCA2 protein is thermosensitive. Non-transfected PNT1A cells were collected 24 hr after plating (about 70% confluency) and lysed according to the protocol. Two aliquots of the same cell lysate (15 µg total proteins) were denatured either at 55 °C for 10 min or at 100 °C for 5 min and BRCA2 protein levels were assessed by immunoblotting. At the bottom, the ratio of BRCA2 to tubulin signal is reported. Please click here to view a larger version of this figure.
Figure 2. Optimization of BRCA2 knockdown. (A) Optimal BRCA2 knockdown may require 24 to 48 hr after the second siRNA transfection cycle. Two different cell lines (PNT1A and Nthy) were assessed for BRCA2 knockdown 24 hr and 48 hr after the second siRNA transfection cycle by immunoblotting analysis. At the bottom, BRCA2 protein levels are reported as percentage of the levels at time 0 hr, set at 100. Data represent the mean ± SE of three independent experiments. (B) BRCA2 knockdown is stable for 7 days. PNT1A cells were assessed for BRCA2 knockdown until 196 hr after the second siRNA transfection cycle by immunoblotting. The ratio of BRCA2 to tubulin signal is reported at the bottom. (C) BRCA2 protein is thermosensitive. Non-transfected PNT1A cells were collected 24 hr after plating (about 70% confluency) and lysed according to the protocol. Two aliquots of the same cell lysate (15 µg total proteins) were denatured either at 55 °C for 10 min or at 100 °C for 5 min and BRCA2 protein levels were assessed by immunoblotting. At the bottom, the ratio of BRCA2 to tubulin signal is reported. Please click here to view a larger version of this figure.
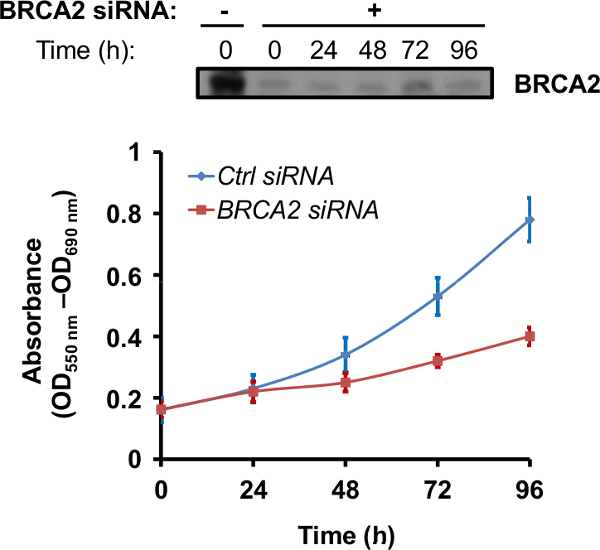 Figure 3. BRCA2 silencing increases the cell sensitivity to the PARP inhibitor rucaparib. PNT1A cells either transfected with control (Ctrl) or BRCA2 siRNA were collected 48 hr after transfection and plated in 96-well plate (2000 cells/well). After 24 hr, 10 µM rucaparib was added to the cells for 24 hr, after which the cells were kept in absence of the drug up to 96 hr. Cell proliferation was assessed at the indicated time using the MTT assay by measuring the absorbance at 550 - 690 nm. Each time point represents the mean ± S.D. of three wells from a representative experiment. Ctrl siRNA, blue line; BRCA2 siRNA, red line. At the top, BRCA2 protein profile in BRCA2 siRNA-transfected cells is reported. Please click here to view a larger version of this figure.
Figure 3. BRCA2 silencing increases the cell sensitivity to the PARP inhibitor rucaparib. PNT1A cells either transfected with control (Ctrl) or BRCA2 siRNA were collected 48 hr after transfection and plated in 96-well plate (2000 cells/well). After 24 hr, 10 µM rucaparib was added to the cells for 24 hr, after which the cells were kept in absence of the drug up to 96 hr. Cell proliferation was assessed at the indicated time using the MTT assay by measuring the absorbance at 550 - 690 nm. Each time point represents the mean ± S.D. of three wells from a representative experiment. Ctrl siRNA, blue line; BRCA2 siRNA, red line. At the top, BRCA2 protein profile in BRCA2 siRNA-transfected cells is reported. Please click here to view a larger version of this figure.
Discussion
Because germline mutations of the BRCA2 gene lead to increased risk of several cancer types, including female and male breast cancer, ovarian cancer, prostate cancer, pancreatic cancer, and melanoma 3,4, a number of studies have been undertaken to understand the biological function of the BRCA2 protein. Most of these studies are genetic-based mainly due to technical difficulties in analyzing a giant protein like BRCA2. The method described here for silencing and analyzing BRCA2 protein expression provides an efficient tool to investigate the functions of the BRCA2 tumor suppressor gene in a broad spectrum of biological assays. The silencing protocol is based on standard siRNA transfection procedures yet contains several crucial modifications as well as tips for successful and consistent knockdown of BRCA2. The siRNA transfection protocol is safe, rapid and efficient and, differently from shRNA, which usually makes use of lentiviral vectors, does not need specific biosafety procedures and can be easily performed in any laboratory equipped with a cell culture facility. Similarly, the immunoblotting protocol has been optimized for efficient detection of a large protein like BRCA2. The immunoblotting protocol has been also validated for detection of recombinant human BRCA2 protein heterologously expressed in yeast cells 9.
Several important factors for efficient delivery of siRNA are reported here. They include two cycles of transfections and a high siRNA to transfection reagent ratio, especially in the second cycle. Absence of antibiotics prior and during the transfection procedure is another critical factor. However, this procedure exposes cells in culture to increased risk of bacterial contamination; thus extra precautions should be taken to guarantee a high standard of sterility at all steps. It should be noted that, depending on the cell type, efficient gene knockdown will typically require 24-48 hr after the second cycle. The current method has the advantage of keeping gene silencing quite stable at least until day 7 after the second transfection cycle. This allows the study of the effects of BRCA2 knockdown in a variety of biological assays, including the cellular response to different chemotherapeutic drugs. However, a limitation of the present technique is represented by the fact that exogenous siRNA molecules are not integrated in the genome, thus stable knockdown of gene expression cannot be achieved. Alternatively, to get stable knockdown, shRNA lentiviral infection may be performed, providing that the lab is prepared for additional biosafety requirements (BSL-2 standards according to the NIH guidelines). In addition, when transfection is not an applicable approach to study the gene function in a specific cell type (e.g., T lymphocytes), use of cell-specific conditional Brca2 knockout mice remains the only alternative 19.
This method reports also several tips for successful detection of a high-molecular weight protein by immunoblotting and it is appropriate for both high and low abundance proteins. In particular, due to the thermosensitivity of the BRCA2 protein, a lower denaturing temperature is critical for its optimal detection, in agreement with a previous study 11. This technical 'trick' may also be useful for other proteins for which detection by standard immunoblotting protocols has been proven unsuccessful. We anticipate that this method contains tips of general validity and thus may facilitate silencing and detection of many other challenging proteins.
Disclosures
The authors have nothing to disclose.
Acknowledgments
This work was supported by FIRB-Merit grants RBNE08YFN3_005 and RBNE08HWLZ_012, and the Italian Ministry of Economy and Finance to the CNR for the Project “FaReBio di Qualità”.
References
- Thorslund T, West SC. BRCA2: a universal recombinase regulator. Oncogene. 2007;26:7720–7730. doi: 10.1038/sj.onc.1210870. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Miki Y. Role of BRCA1 and BRCA2 as regulators of DNA repair, transcription, and cell cycle in response to DNA damage. Cancer Sci. 2004;95:866–871. doi: 10.1111/j.1349-7006.2004.tb02195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AM, et al. Germline mutations in BRCA1 and BRCA2 in breast-ovarian families from a breast cancer risk evaluation clinic. J Clin Oncol. 2001;19:2247–2253. doi: 10.1200/JCO.2001.19.8.2247. [DOI] [PubMed] [Google Scholar]
- Castro E, Eeles R. The role of BRCA1 and BRCA2 in prostate cancer. Asian J Androl. 2012;14:409–414. doi: 10.1038/aja.2011.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vencken PM, et al. Chemosensitivity and outcome of BRCA1- and BRCA2-associated ovarian cancer patients after first-line chemotherapy compared with sporadic ovarian cancer patients. Ann Oncol. 2011;22:1346–1352. doi: 10.1093/annonc/mdq628. [DOI] [PubMed] [Google Scholar]
- Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- Arbini AA, et al. Mitochondrial DNA depletion sensitizes cancer cells to PARP inhibitors by translational and post-translational repression of BRCA2. Oncogenesis. 2013;2:e82. doi: 10.1038/oncsis.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Guo X, Yang G, Rosen DG, Liu J. AURKA and BRCA2 expression highly correlate with prognosis of endometrioid ovarian carcinoma. Mod Pathol. 2011;24:836–845. doi: 10.1038/modpathol.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guaragnella N, Marra E, Galli A, Moro L, Giannattasio S. Silencing of BRCA2 decreases anoikis and its heterologous expression sensitizes yeast cells to acetic acid-induced programmed cell death. Apoptosis. 2014;19:1330–1341. doi: 10.1007/s10495-014-1006-z. [DOI] [PubMed] [Google Scholar]
- Mahato RI, Rolland A, Tomlinson E. Cationic lipid-based gene delivery systems: pharmaceutical perspectives. Pharm Res. 1997;14:853–859. doi: 10.1023/a:1012187414126. [DOI] [PubMed] [Google Scholar]
- Su LK, et al. Characterization of BRCA2: temperature sensitivity of detection and cell-cycle regulated expression. Oncogene. 1998;17:2377–2381. doi: 10.1038/sj.onc.1202162. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette WN'. 'Western blotting': electrophoretic transfer of proteins from sodium dodecyl sulfate--polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981;112:195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- Hunter FW, et al. Dual targeting of hypoxia and homologous recombination repair dysfunction in triple-negative breast cancer. Mol Cancer Ther. 2014. [DOI] [PubMed]
- Drew Y, et al. Therapeutic potential of poly(ADP-ribose) polymerase inhibitor AG014699 in human cancers with mutated or methylated BRCA1 or BRCA2. J Natl Cancer Inst. 2011;103:334–346. doi: 10.1093/jnci/djq509. [DOI] [PubMed] [Google Scholar]
- Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- Ihnen M, et al. Therapeutic potential of the poly(ADP-ribose) polymerase inhibitor rucaparib for the treatment of sporadic human ovarian cancer. Mol Cancer Ther. 2013;12:1002–1015. doi: 10.1158/1535-7163.MCT-12-0813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong JH, Jo A, Park P, Lee H, Lee HO. Brca2 Deficiency Leads to T Cell Loss and Immune Dysfunction. Mol Cells. 2015. [DOI] [PMC free article] [PubMed]


