Abstract
Aggregation of amyloid-β (Aβ) peptides is a characteristic pathological feature of Alzheimer's disease. We have exploited the relationship between solvent exposure and intrinsic fluorescence of a single tyrosine residue, Tyr10, in the Aβ sequence to probe structural features of the monomeric, oligomeric and fibrillar forms of the 42-residue Aβ1-42. By monitoring the quenching of Tyr10 fluorescence upon addition of water-soluble acrylamide, we show that in Aβ1-42 oligomers this residue is solvent-exposed to a similar extent to that found in the unfolded monomer. By contrast, Tyr10 is significantly shielded from acrylamide quenching in Aβ1-42 fibrils, consistent with its proximity to the fibrillar cross-β core. Furthermore, circular dichroism measurements reveal that Aβ1-42 oligomers have a considerably lower β-sheet content than the Aβ1-42 fibrils, indicative of a less ordered molecular arrangement in the former. Taken together these findings suggest significant differences in the structural assembly of oligomers and fibrils that are consistent with differences in their biological effects.
Keywords: Amyloid-β, Aβ oligomer, Amyloid fibril, tyrosine fluorescence, Acrylamide quenching
Highlights
-
•
Acrylamide quenching of Tyr10 provides insights into Aβ1-42 aggregate structure.
-
•
Tyr10 is solvent-exposed in Aβ1-42 oligomers, but protected in Aβ1-42 fibrils.
-
•
Aβ1-42 oligomers exhibit less β-sheet content than fibrils.
-
•
Monomer arrangement in oligomers and fibrils may explain their different toxicities.
1. Introduction
Alzheimer's disease (AD) and a range of related disorders are associated with the self-assembly, aggregation, and fibril formation of disease-specific peptides and proteins [1]; in the case of AD such processes are involved with aggregation of the amyloid-β (Aβ) peptide [2], [3]. In its fibrillar form, this peptide is the main proteinaceous component of the extracellular plaque deposits that are characteristic of AD pathology [4], [5], but in the brain Aβ also exists in a variety of monomeric and oligomeric forms [6]. It has been reported that soluble Aβ concentrations correlate more closely with dementia than the amount of amyloid plaques [7], [8], and indeed soluble Aβ oligomers are now thought to be the main culprits in the pathogenesis of AD and related conditions [9], particularly since they have been associated with impaired cognitive function [10], [11], [12] and have been shown to induce cellular toxicity [13], [14], [15], [16], [17]. Unfortunately, the small size and low abundance of Aβ oligomers, in combination with their considerable heterogeneity and high sensitivity to environmental changes, has rendered them challenging to characterise. Although a detailed molecular model of a stabilised Aβ1-42 protofibril has been recently reported [18], little is known in detail how monomers arrange to build up the smaller, globular oligomers that are populated during Aβ aggregation reactions and which can form in the AD brain [19].
In the present paper we have used fluorescence and circular dichroism spectroscopies to examine structural differences, particularly related to the microenvironment around the N-terminal region, to compare soluble, globular Aβ1-42 oligomers, prepared in vitro, with monomers and fibrils formed by both Aβ1-40 and Aβ1-42. The oligomers were prepared using established methods for producing stable oligomers of a type that is often referred to as amyloid-derived diffusible ligands (ADDLs) [19]. These ADDLs have been reported to be neurotoxic [20] and it has been found that antibodies raised against in vitro prepared oligomers of this type also recognise oligomeric species that are elevated in AD brains [21], suggesting their resemblance to naturally occurring forms. Although it has been proposed that oligomers of this type could be of a fibrillar nature [22], [23], it is not known how monomers are present within these oligomer assemblies or to what extent the microscopic architecture resembles that found in mature fibrils. To address this issue, we have taken advantage of the fact that the Aβ peptide contains only a single intrinsically fluorescent residue, tyrosine at position 10 (Tyr10), which is located in between the β-core and the flexible N-terminus of fibrillar Aβ1-40 and Aβ1-42 [24], [25], [26], [27], [28] (Fig. 1A). This residue, therefore, has the potential to report on the participation of this region of the Aβ peptide in the different aggregated states. In this study, we have exploited the intrinsic fluorescent properties of Tyr10 and also its susceptibility to fluorescence quenching by water-soluble acrylamide to examine the characteristics of the monomeric, oligomeric and fibrillar forms of Aβ. Our results indicate that Aβ1-42 oligomers show the presence of a degree of β-sheet structure, but that they are distinctly less ordered than fibrils, and this is confirmed by a substantially higher degree of solvent exposure around Tyr10. Taken together these results highlight significant differences in the molecular architecture of the oligomeric versus the fibrillar forms of the Aβ peptides which may offer new insights into the differential biological activities.
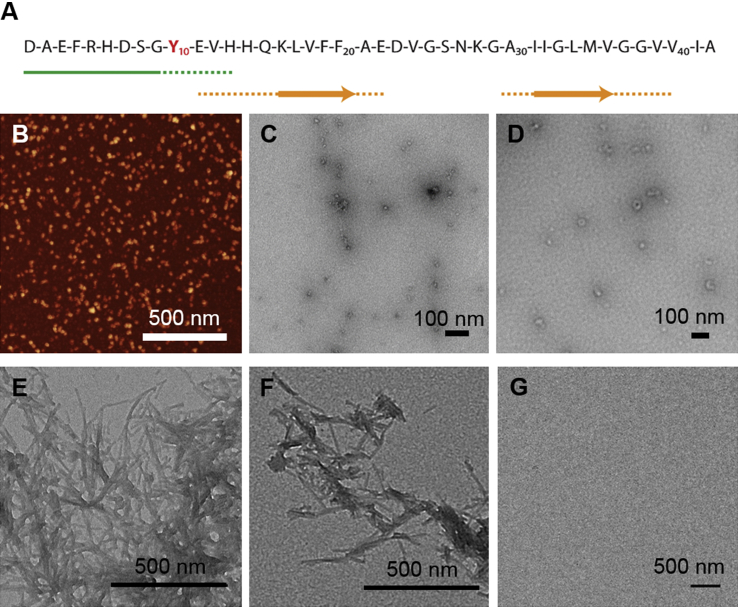
Fig. 1.
(A) Schematic representation of the Aβ1-42 peptide sequence. The arrows show the reported engagement of different parts of the sequence in the mature amyloid fibrils [24], [25], [26], [27], [28], with the unstructured N-terminus marked in green and the β-sheet regions participating in the core marked in orange. Tyr10 is highlighted in red. (B) Atomic force microscopy image of oAβ1-42. (C–D) TEM image of oAβ1-42 at higher magnification. (E) TEM image of fAβ1-40. (F) TEM image of fAβ1-42. (G) TEM image of mAβ1-40 showing the absence of aggregates in the monomeric preparation. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2. Materials and methods
2.1. Materials
Synthetic Aβ1-40 and Aβ1-42 peptides were acquired as lyophilised powders from Anaspec EGT (Fremont, USA) and were prepared for use as described below. All other reagents were purchased from Sigma–Aldrich (Dorset, UK).
2.2. Sample preparation
The Aβ peptide powders were dissolved in ice-cold trifluoroacetic acid, sonicated (30 s, on ice), flash frozen and again lyophilised. The samples were redissolved in ice-cold hexafluoroisopropanol (1 mL) and the solutions were kept on ice (10 min) then divided into aliquots (50 μL) whilst working at 4 °C, and dried by rotary evaporation. The peptide concentration was determined by amino acid analysis and all experiments were performed in 50 mM sodium phosphate buffer (pH 7.4). The monomeric form of the Aβ peptides was obtained by dissolving a fresh peptide aliquot to a final concentration of 5 μM directly in buffer. The solution was analysed immediately to minimise the formation of aggregates. Fibrillar Aβ samples were prepared by incubation at room temperature for 48 h under shaking conditions (1400 rpm in a Titramax 100 shaker, Heidolph Instruments GmbH, Schwabach, Germany). Oligomers of Aβ1-42 were prepared by resuspending peptide aliquots in DMSO (2 μL), followed by dilution in ice-cold buffer (10 mM NaCl, 10 mM sodium phosphate, pH 7.4), to a final peptide concentration of 100 μM followed by incubation (overnight, 4 °C under quiescent conditions) [20]. The solutions were diluted in 50 mM sodium phosphate buffer (pH 7.4) (5 μM based on monomer concentration) prior to experimental analysis. The oligomer yield was determined by amino acid analysis following separation of the oligomers from residual monomer by ultracentrifugation (1 h, 90,000 g) using an Optima TLX ultracentrifuge (Beckman Coulter Inc, Brea, USA).
2.3. Fluorescence spectroscopy
Fluorescence spectra were recorded on a Cary Eclipse fluorimeter (Agilent Technologies, Stockport, UK) using a reduced pathlength quartz cuvette (4 mm excitation/10 mm emission). The excitation wavelength was 275 nm and emission spectra were recorded at 1 nm increments between 290 and 350 nm, with excitation and emission slit widths of 5 nm and 10 nm, respectively, and a scan rate of 60 nm/min. Samples were subjected to an acrylamide gradient following additions of a 0.2 M stock solution (10 μL aliquots), the fluorescence spectra being recorded immediately after the addition of the acrylamide aliquots. The recorded spectra were corrected for the background contributions and the fluorescence intensity in each case was taken as the sum of the intensities in a 6 nm range centred around the emission maxima, to increase signal-to-noise. Data were analysed using the Stern-Volmer equation [29].
| (1) |
where F0 and F are the fluorescence intensities in the absence and presence of quencher, [Q] is the concentration of quencher, and KSV the Stern–Volmer constant. All experiments were performed in triplicate and are reported as mean ± SD. As acrylamide absorbs significantly at the excitation wavelength of tyrosine and therefore acts as an inner filter, corrections were made using separate experiments in which a non-quenching molecule (here DNA) was titrated into solutions of tyrosine at the same absorbance increments in order to determine correction factors for primary inner filter effects.
2.4. Circular dichroism (CD) spectroscopy
CD spectra were recorded on a JASCO J-810 spectropolarimeter (JASCO Inc. Tokyo, Japan) between 190 and 250 nm using a 1 mm quartz cuvette. 10 scans were recorded and averaged using a bandwidth of 2 nm and a scan speed of 50 nm/min. The peptide concentration was 30 μM and all spectra were corrected for background contributions by subtracting buffer blanks.
2.5. Transmission electron microscopy (TEM)
Aβ samples were adsorbed (2 min) onto carbon-coated copper grids (Taab Laboratories Equipment Ltd, Berks, UK). The grids were blotted, washed with milliQ water (2X) and negatively stained with 2% (w/v) uranyl acetate. Samples were imaged on a FEI Tecnai G2 transmission electron microscope (Eindhoven, Netherlands) and images were analysed using the SIS Megaview II Image Capture system (EMSIS GmbH, Muenster, Germany).
2.6. Atomic force microscopy (AFM)
AFM measurements were performed using a NanoWizard AFM system (JPK Instruments AG, Berlin, Germany). Samples were diluted using dH2O and deposited onto freshly cleaved mica surfaces and slowly dried before imaging. AFM imaging was carried out in the intermittent (air) contact mode using a silicon nitride cantilever (μmasch, NSC36/No Al, 65–130 kHz, 0.6–2 N/m). AFM images were analysed using Gwyddion software package (http://gwyddion.net/).
3. Results and discussion
The intrinsic fluorescence of Tyr10 in combination with circular dichroism spectroscopy has been used to examine five different preparations of Aβ1-40 and Aβ1-42; monomers (mAβ1-40 and mAβ1-42), fibrils (fAβ1-40 fAβ1-42) and Aβ1-42 oligomers (oAβ1-42) in order to assess the conformational differences between the various forms of Aβ1-42, and also to explore potential differences between Aβ1-40 and Aβ1-42 fibrils.
Prior to spectroscopic characterisation the morphology of the different Aβ species was examined (Fig. 1B–G). Analysis of the oligomeric oAβ1-42 samples by AFM (Fig. 1B) and TEM (Fig. 1C–D) showed that they contain a relatively homogeneous population of small aggregates with approximately spherical morphology; analysis of the TEM images indicates that their approximate diameters were 10–20 nm. The fibrillar fAβ1-40 and fAβ1-42 samples contained typical amyloid fibrils; importantly no such fibrillar structures were detected in the oligomeric samples used in this study and no discernible aggregates were observed in the monomeric samples (Fig. 1G,).
Next, the secondary structure content in the different Aβ preparations was assessed (Fig. 2); the monomeric samples displayed CD spectra typical of random coils with negative peaks centred at 195 nm, consistent with their intrinsically disordered nature and confirming the absence of amyloid aggregates in the monomer preparations. The oligomeric and fibrillar samples, by contrast, exhibited characteristic β-sheet features with CD spectra displaying a negative peak at ∼218 nm and a positive peak at ∼196 nm. The CD signal from the oligomers, however, was considerably weaker than that of the fibrils indicating that they have significantly less β-sheet content.
Fig. 2.
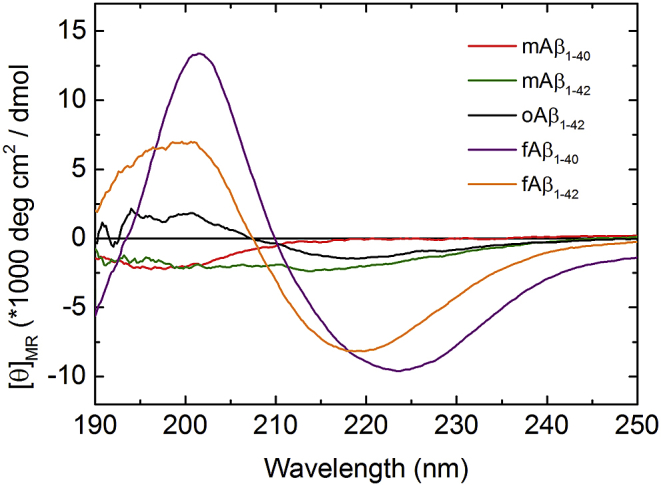
Circular dichroism spectra of different Aβ species; mAβ1-40 (red), mAβ1-42 (green), oAβ1-42 (black), fAβ1-40 (purple) and fAβ1-42 (orange). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The fluorescence spectra of the various Aβ samples were recorded to monitor the intrinsic emission from Tyr10. Fig. 3A shows the spectra of the five different Aβ preparations along with corresponding spectra for free tyrosine and for a tyrosine-containing tri-peptide (VYV), the latter was used to determine the generic effects on tyrosine emission due to its incorporation into a polypeptide sequence. It was observed that the tyrosine emission intensity is decreased by as much as a factor of 2 upon incorporation into a peptide sequence, consistent with previous observations where the quenching of tyrosine in proteins has been attributed to photoinduced electron transfer to the peptide bond in presence of electron withdrawing groups [30]. All the Aβ conformers examined, except fAβ1-42, were observed to exhibit even lower intrinsic tyrosine fluorescence intensities than the VYV control peptide, indicating the existence of additional quenching interactions within the Aβ sequence, attributable to interactions with neighbouring side-chains. Spectral broadening of the tyrosine emission peak was observed in all the Aβ samples (Fig. 3B); the broadening on the blue edge of the emission peak is particularly apparent for fAβ1-40, fAβ1-42 and oAβ1-42 and can be explained by light scattering by the aggregates present in these solutions, whereas the red edge broadening, which is mainly apparent for the monomeric Aβ peptides, may be indicative of tyrosinate formation in the excited state [31], [32].
Fig. 3.
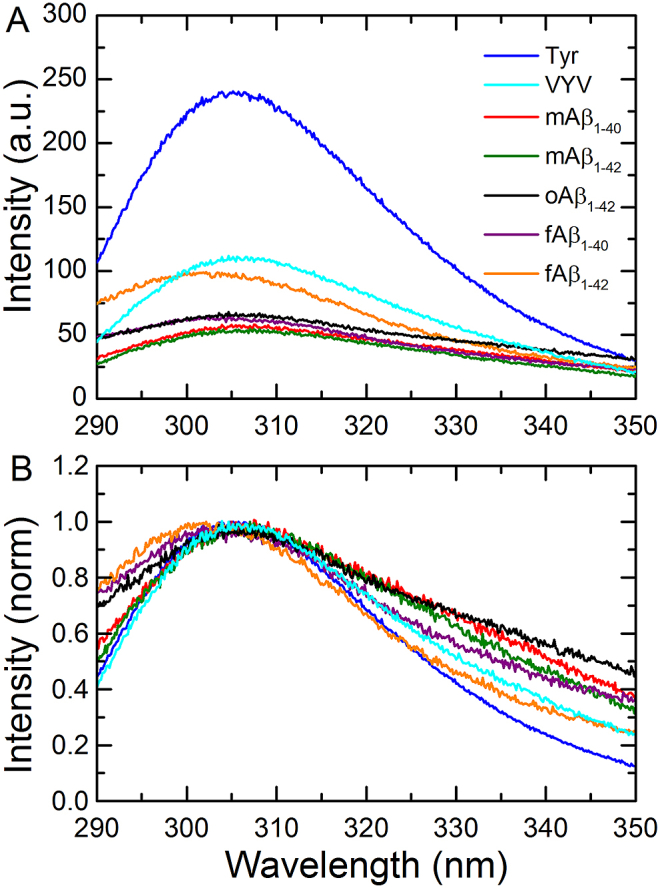
Intrinsic tyrosine fluorescence of different Aβ species. (A) Emission spectra of free tyrosine (blue), VYV (cyan), mAβ1-40 (red), mAβ1-42 (green), oAβ1-42 (black), fAβ1-40 (purple), fAβ1-42 (orange). (B) Normalised emission intensity, corresponding to the spectra shown in (A). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
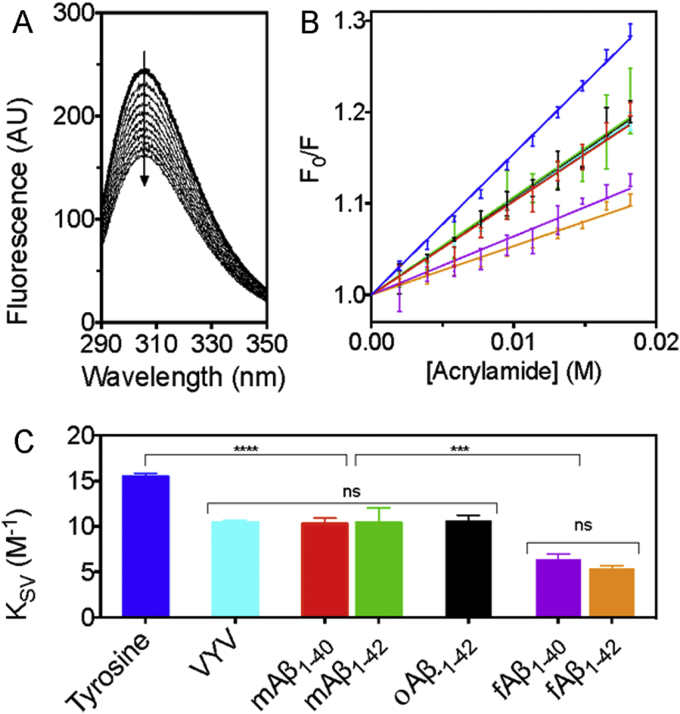
Acrylamide quenching experiments were conducted to explore the extent to which tyrosine is exposed to solvent in its free form (Fig. 4A) compared to its exposure when it is incorporated as Tyr10 in the different monomeric and aggregated forms of Aβ. The resulting Stern-Volmer plots (Fig. 4B) were linear in all cases, an observation consistent with acrylamide being a predominately collisional quencher [33]. Fig. 4C summarises the calculated Stern–Volmer quenching constants (KSV) and shows that incorporation of tyrosine into a peptide sequence, even into the short VYV model peptide which displays a ∼30% reduction of the KSV value, results in shielding from the polar but non-charged water soluble acrylamide quencher. This effect is likely to be due to steric hindrance imparted by the neighbouring residues, which limits the number of possible quenching interactions but does not reflect shielding caused by secondary structure constraints. No appreciable differences in the solvent exposure of tyrosine in the monomeric mAβ1-40 and mAβ1-42 forms compared to the VYV peptide were observed. This finding is consistent with other observations showing that the Aβ chain is highly unfolded in solution [34], [35], at least on timescales in the order of the excited state lifetime of tyrosine (≤3.4 ns [36]). Analysis of the Stern-Volmer plots showed, however, that Tyr10 in fAβ1-40 and fAβ1-42 is considerably less exposed to acrylamide quenching than in mAβ1-40 and mAβ1-42, suggesting that the incorporation of the Aβ molecules into a fibrillar structure protects this residue from solvent even though existing structural models [24], [25], [26], [27], [28] suggest that Tyr10 does not participate directly in the cross-β core (see summary in Fig. 1). This effect can, however, be explained by the close packing of monomer units in the fibril protofilaments [37], which sterically hinders the acrylamide-Tyr10 collisions, or it may also be a consequence of a fraction of the Aβ N-termini becoming buried within the mature fibril; indeed, limited proteolysis data for fAβ1-40 suggests that at least 20% of Aβ monomers have an N-terminal segment that is protected within the fibril structure [38]. The considerable similarity observed for fAβ1-40 and fAβ1-42 is interesting in relation to hydrogen/deuterium-exchange rates measured by NMR, which indicate significant differences between the two fibril types with respect to the amide solvent protection of the N-terminus, whereby the amide group of Tyr10 is more solvent accessible in fAβ1-42 than in fAβ1-40 [26].
Fig. 4.
Acrylamide quenching of the intrinsic tyrosine fluorescence in the different species of Aβ. (A) Fluorescence emission spectra for free tyrosine (5 μM) titrated with acrylamide (added in 2 mM increments). The bold line represents the fluorescence of unquenched tyrosine and the arrow indicates the decrease in fluorescence with increasing concentration of the quencher. (B) Stern-Volmer plots of the acrylamide quenching of Tyr10 in the different Aβ species showing free tyrosine (blue), VYV (cyan), mAβ1-40 (red), mAβ1-42 (green), oAβ1-42 (black) fAβ1-40 (purple), fAβ1-42 (orange). The error bars represent the standard deviations (n = 3). (C) Stern–Volmer quenching constants (KSV) ±SD calculated from the linear-fit of the data in (B). The statistical significance of the differences between the various KSV values was tested by one-way ANOVA with Tukey's post hoc test; ns denotes not significantly different (p > 0.05), *** (p < 0.001), **** (p < 0.0001). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Interestingly, the oligomeric oAβ1-42 samples exhibit KSV values that are significantly higher than those observed for the fibrils (Fig. 4C). This observation suggests that Tyr10 is considerably less shielded in these globular oligomers than in Aβ fibrils and is consistent with our finding that oAβ1-42 species have a lower β-sheet content than fAβ1-42 (Fig. 2A). More surprisingly, we also find that the KSV values of the oligomers are similar, within experimental error, to those of monomeric Aβ implying that Tyr10 is exposed to a similar extent as in a random coil monomer. To ensure that the high KSV value truly reflects the structure of the oAβ1-42 species and is not attributable to sample heterogeneity, and as it has been suggested previously that oligomers of the type we examine here can exist in binary mixtures with monomers, a further control experiment was performed [39]. Quantitative amino acid analysis following ultracentrifugation to separate the oAβ1-42 species from residual monomers showed that, in our samples, nearly 90% of the Aβ monomers were incorporated into oligomers (Supplementary Information). This result is comparable to our previous report of the yield in fibril forming reactions with Aβ1-40 and Aβ1-42 [40]. Furthermore, these control experiments confirm that the difference in β-sheet content indicated in the oAβ1-42 and fAβ1-42 CD spectra, is indeed related to the oAβ1-42 species exhibiting some random coil nature, rather than due to the presence of significant quantities of unstructured monomers in the samples. Therefore, this study shows that the Aβ1-42 oligomers differ from fAβ1-42, not only in size and shape, but also in secondary structure content and internal architecture. Our finding that Tyr10 in the oligomers is significantly solvent exposed, and thus likely to be present on the surface of the oligomers is in agreement with a previous study of a disc shaped Aβ1-42 pentamer [41]. These observations are also consistent with the conclusion that the Aβ1-42 peptides in these oligomers do not adopt the same β-hairpin arrangement as that formed in the mature Aβ fibrils. This suggestion supports the idea that oligomers of this type may accumulate because their structural properties are such that they are not able to convert into fibrils [42], [43].
In conclusion, this study has set out to examine the fluorescent properties and the degree of solvent exposure of Tyr10 in Aβ to gain insights into structural similarities and dissimilarities between different Aβ species. We report significant differences between monomers and fibrils, suggesting that even though Tyr10 is not directly part of the cross-β core, the side-chain is, on average, well shielded within the fibrils. In addition, we have shown that Aβ1-42 oligomers, due to their lower β-sheet content and extensive exposure of Tyr10 to solvent, have significantly different structural properties from those of Aβ fibrils.
Acknowledgements
This work was funded by grants to E.K.E from the Wenner-Gren Foundations, the Hasselblad Foundation, and the Swedish Innovation Agency (Vinnova): 2011-03488 and to C.M.D from the Wellcome Trust. The TEM imaging was carried out in the Multi-Imaging Unit in the Department of Physiology, Development and Neuroscience, University of Cambridge, UK and quantitative amino acid analysis was carried out at the Protein and Nucleic Acid Chemistry Facility, Department of Biochemistry, University of Cambridge, UK.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.bbrc.2015.11.018.
Transparency document related to this article can be found online at http://dx.doi.org/10.1016/j.bbrc.2015.11.018.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
Transparency document
References
- 1.Chiti F., Dobson C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 2.Hardy J., Allsop D. Amyloid deposition as the central event in the aetiology of Alzheimer's disease. Trends Pharmacol. Sci. 1991;12:383–388. doi: 10.1016/0165-6147(91)90609-v. [DOI] [PubMed] [Google Scholar]
- 3.Hardy J.A., Higgins G.A. Alzheimer's disease: the amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 4.Glenner G.G., Wong C.W. Alzheimer's disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem. Biophys. Res. Commun. 1984;120:885–890. doi: 10.1016/s0006-291x(84)80190-4. [DOI] [PubMed] [Google Scholar]
- 5.Masters C.L., Simms G., Weinman N.A., Multhaup G., McDonald B.L., Beyreuther K. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc. Natl. Acad. Sci. U. S. A. 1985;82:4245–4249. doi: 10.1073/pnas.82.12.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glabe C.G. Structural classification of toxic amyloid oligomers. J. Biol. Chem. 2008;283:29639–29643. doi: 10.1074/jbc.R800016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lue L.F., Kuo Y.M., Roher A.E., Brachova L., Shen Y., Sue L., Beach T., Kurth J.H., Rydel R.E., Rogers J. Soluble amyloid beta peptide concentration as a predictor of synaptic change in Alzheimer's disease. Am. J. Pathol. 1999;155:853–862. doi: 10.1016/s0002-9440(10)65184-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McLean C.A., Cherny R.A., Fraser F.W., Fuller S.J., Smith M.J., Beyreuther K., Bush A.I., Masters C.L. Soluble pool of Abeta amyloid as a determinant of severity of neurodegeneration in Alzheimer's disease. Ann. Neurol. 1999;46:860–866. doi: 10.1002/1531-8249(199912)46:6<860::aid-ana8>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 9.Haass C., Selkoe D.J. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer's amyloid beta-peptide. Nat. Rev. Mol. Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 10.Cleary J.P., Walsh D.M., Hofmeister J.J., Shankar G.M., Kuskowski M.A., Selkoe D.J., Ashe K.H. Natural oligomers of the amyloid-beta protein specifically disrupt cognitive function. Nat. Neurosci. 2005;8:79–84. doi: 10.1038/nn1372. [DOI] [PubMed] [Google Scholar]
- 11.Lesne S., Koh M.T., Kotilinek L., Kayed R., Glabe C.G., Yang A., Gallagher M., Ashe K.H. A specific amyloid-beta protein assembly in the brain impairs memory. Nature. 2006;440:352–357. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- 12.Selkoe D.J. Soluble oligomers of the amyloid beta-protein impair synaptic plasticity and behavior. Behav. Brain Res. 2008;192:106–113. doi: 10.1016/j.bbr.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campioni S., Mannini B., Zampagni M., Pensalfini A., Parrini C., Evangelisti E., Relini A., Stefani M., Dobson C.M., Cecchi C., Chiti F. A causative link between the structure of aberrant protein oligomers and their toxicity. Nat. Chem. Biol. 2010;6:140–147. doi: 10.1038/nchembio.283. [DOI] [PubMed] [Google Scholar]
- 14.Bolognesi B., Kumita J.R., Barros T.P., Esbjörner E.K., Luheshi L.M., Crowther D.C., Wilson M.R., Dobson C.M., Favrin G., Yerbury J.J. ANS binding reveals common features of cytotoxic amyloid species. ACS Chem. Biol. 2010;5:735–740. doi: 10.1021/cb1001203. [DOI] [PubMed] [Google Scholar]
- 15.Dahlgren K.N., Manelli A.M., Stine W.B., Jr., Baker L.K., Krafft G.A., LaDu M.J. Oligomeric and fibrillar species of amyloid-beta peptides differentially affect neuronal viability. J. Biol. Chem. 2002;277:32046–32053. doi: 10.1074/jbc.M201750200. [DOI] [PubMed] [Google Scholar]
- 16.Sandberg A., Luheshi L.M., Söllvander S., Barros T.P., Macao B., Knowles T.P.J., Biverståhl H., Lendel C., Ekholm-Petterson F., Dubnovitsky A., Lannfelt L., Dobson C.M., Härd T. Stabilization of neurotoxic Alzheimer amyloid-β oligomers by protein engineering. Proc. Natl. Acad. Sci. U. S. A. 2010;107:15595–15600. doi: 10.1073/pnas.1001740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walsh D.M., Klyubin I., Fadeeva J.V., Cullen W.K., Anwyl R., Wolfe M.S., Rowan M.J., Selkoe D.J. Naturally secreted oligomers of amyloid beta protein potently inhibit hippocampal long-term potentiation in vivo. Nature. 2002;416:535–539. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- 18.Lendel C., Bjerring M., Dubnovitsky A., Kelly R.T., Filippov A., Antzutkin O.N., Nielsen N.C., Hard T. A hexameric peptide barrel as building block of amyloid-beta protofibrils. Angew. Chem. Int. Ed. 2014;53:12756–12760. doi: 10.1002/anie.201406357. [DOI] [PubMed] [Google Scholar]
- 19.Narayan P., Orte A., Clarke R.W., Bolognesi B., Hook S., Ganzinger K.A., Meehan S., Wilson M.R., Dobson C.M., Klenerman D. The extracellular chaperone clusterin sequesters oligomeric forms of the amyloid-beta(1-40) peptide. Nat. Struct. Mol. Biol. 2012;19:79–83. doi: 10.1038/nsmb.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lambert M.P., Barlow A.K., Chromy B.A., Edwards C., Freed R., Liosatos M., Morgan T.E., Rozovsky I., Trommer B., Viola K.L., Wals P., Zhang C., Finch C.E., Krafft G.A., Klein W.L. Diffusible, nonfibrillar ligands derived from Abeta1-42 are potent central nervous system neurotoxins. Proc. Natl. Acad. Sci. U. S. A. 1998;95:6448–6453. doi: 10.1073/pnas.95.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gong Y., Chang L., Viola K.L., Lacor P.N., Lambert M.P., Finch C.E., Krafft G.A., Klein W.L. Alzheimer's disease-affected brain : presence of oligomeric Abeta ligands ( ADDLs ) suggests a molecular basis for reversible memory loss. Proc. Natl. Acad. Sci. U. S. A. 2003;100:10417–10422. doi: 10.1073/pnas.1834302100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lacor P.N., Buniel M.C., Chang L., Fernandez S.J., Gong Y., Viola K.L., Lambert M.P., Velasco P.T., Bigio E.H., Finch C.E., Krafft G.A., Klein W.L. Synaptic targeting by Alzheimer's-related amyloid beta oligomers. J. Neurosci. 2004;24:10191–10200. doi: 10.1523/JNEUROSCI.3432-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lambert M.P., Velasco P.T., Chang L., Viola K.L., Fernandez S., Lacor P.N., Khuon D., Gong Y., Bigio E.H., Shaw P., De Felice F.G., Krafft G.A., Klein W.L. Monoclonal antibodies that target pathological assemblies of Abeta. J. Neurochem. 2007;100:23–35. doi: 10.1111/j.1471-4159.2006.04157.x. [DOI] [PubMed] [Google Scholar]
- 24.Fandrich M., Schmidt M., Grigorieff N. Recent progress in understanding Alzheimer's beta-amyloid structures. Trends Biochem. Sci. 2011;36:338–345. doi: 10.1016/j.tibs.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luhrs T., Ritter C., Adrian M., Riek-Loher D., Bohrmann B., Dobeli H., Schubert D., Riek R. 3D structure of Alzheimer's amyloid-beta(1-42) fibrils. Proc. Natl. Acad. Sci. U. S. A. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olofsson A., Lindhagen-Persson M., Sauer-Eriksson A.E., Ohman A. Amide solvent protection analysis demonstrates that amyloid-beta(1-40) and amyloid-beta(1-42) form different fibrillar structures under identical conditions. Biochem. J. 2007;404:63–70. doi: 10.1042/BJ20061561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olofsson A., Sauer-Eriksson A.E., Ohman A. The solvent protection of Alzheimer amyloid-beta-(1-42) fibrils as determined by solution NMR spectroscopy. J. Biol. Chem. 2006;281:477–483. doi: 10.1074/jbc.M508962200. [DOI] [PubMed] [Google Scholar]
- 28.Petkova A.T., Ishii Y., Balbach J.J., Antzutkin O.N., Leapman R.D., Delaglio F., Tycko R. A structural model for Alzheimer's beta-amyloid fibrils based on experimental constraints from solid state NMR. Proc. Natl. Acad. Sci. U. S. A. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stern O., Volmer M. Uber die Abklingzeit der Fluoreszenz. Z Phys. 1919;20:183–188. [Google Scholar]
- 30.Seidel C., Orth A., Greulich K.O. Electronic effects on the fluorescence of tyrosine in small peptides. Photochem. Photobiol. 1993;58:178–184. doi: 10.1111/j.1751-1097.1993.tb09546.x. [DOI] [PubMed] [Google Scholar]
- 31.Pundak S., Roche R.S. Tyrosine and tyrosinate fluorescence of bovine testes calmodulin: calcium and pH dependence. Biochemistry. 1984;23:1549–1555. doi: 10.1021/bi00302a032. [DOI] [PubMed] [Google Scholar]
- 32.Szabo A.G., Lynn K.R., Krajcarski D.T., Rayner D.M. Tyrosinate fluorescence maxima at 345 nm in proteins lacking tryptophan at pH 7. FEBS Lett. 1978;94:249–252. doi: 10.1016/0014-5793(78)80948-x. [DOI] [PubMed] [Google Scholar]
- 33.Eftink M.R., Ghiron C.A. Exposure of tryptophanyl residues in proteins. Quantitative determination by fluorescence quenching studies. Biochemistry. 1976;15:672–680. doi: 10.1021/bi00648a035. [DOI] [PubMed] [Google Scholar]
- 34.Danielsson J., Andersson A., Jarvet J., Gräslund A. 15N relaxation study of the amyloid beta-peptide: structural propensities and persistence length. Magn. Reson. Chem. 2006;44:S114–S121. doi: 10.1002/mrc.1814. [DOI] [PubMed] [Google Scholar]
- 35.Riek R., Guntert P., Dobeli H., Wipf B., Wuthrich K. NMR studies in aqueous solution fail to identify significant conformational differences between the monomeric forms of two Alzheimer peptides with widely different plaque-competence, A beta(1-40)(ox) and A beta(1-42)(ox) Eur. J. Biochem. 2001;268:5930–5936. doi: 10.1046/j.0014-2956.2001.02537.x. [DOI] [PubMed] [Google Scholar]
- 36.Ross J.B.A., Laws W.R., Rousslang K.W., Wyssbrod H.R. Tyrosine Fluorescence and Phosphorescence from Proteins and Polypeptides. In: Lakowicz J., editor. Topics in Fluorescence Spectroscopy. vol. 3. Plenum Press; New York: 1992. pp. 1–63. (Biochemical Applications). [Google Scholar]
- 37.Serpell L.C. Alzheimer's amyloid fibrils: structure and assembly. BBA Mol. Bas. Dis. 2000;1502:16–30. doi: 10.1016/s0925-4439(00)00029-6. [DOI] [PubMed] [Google Scholar]
- 38.Kheterpal I., Williams A., Murphy C., Bledsoe B., Wetzel R. Structural features of the Abeta amyloid fibril elucidated by limited proteolysis. Biochemistry. 2001;40:11757–11767. doi: 10.1021/bi010805z. [DOI] [PubMed] [Google Scholar]
- 39.Hepler R.W., Grimm K.M., Nahas D.D., Breese R., Dodson E.C., Acton P., Keller P.M., Yeager M., Wang H., Shughrue P., Kinney G., Joyce J.G. Solution state characterization of amyloid beta-derived diffusible ligands. Biochemistry. 2006;45:15157–15167. doi: 10.1021/bi061850f. [DOI] [PubMed] [Google Scholar]
- 40.Lindberg D.J., Wranne M.S., Gilbert Gatty M., Westerlund F., Esbjörner E.K. Steady-state and time-resolved Thioflavin-T fluorescence can report on morphological differences in amyloid fibrils formed by Abeta(1-40) and Abeta(1-42) Biochem. Biophys. Res. Commun. 2015;458:418–423. doi: 10.1016/j.bbrc.2015.01.132. [DOI] [PubMed] [Google Scholar]
- 41.Ahmed M., Davis J., Aucoin D., Sato T., Ahuja S., Aimoto S., Elliott J.I., Van Nostrand W.E., Smith S.O. Structural conversion of neurotoxic amyloid-beta(1-42) oligomers to fibrils. Nat. Struct. Mol. Biol. 2010;17:561–567. doi: 10.1038/nsmb.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen S.W., Drakulic S., Deas E., Ouberai M., Aprile F.A., Arranz R., Ness S., Roodveldt C., Guilliams T., De-Genst E.J., Klenerman D., Wood N.W., Knowles T.P., Alfonso C., Rivas G., Abramov A.Y., Valpuesta J.M., Dobson C.M., Cremades N. Structural characterization of toxic oligomers that are kinetically trapped during alpha-synuclein fibril formation. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E1994–E2003. doi: 10.1073/pnas.1421204112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Necula M., Kayed R., Milton S., Glabe C.G. Small molecule inhibitors of aggregation indicate that amyloid beta oligomerization and fibrillization pathways are independent and distinct. J. Biol. Chem. 2007;282:10311–10324. doi: 10.1074/jbc.M608207200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.