Abstract
Bacteriocins are antimicrobial peptides of bacterial origin that are considered as a promising alternative to the use of conventional antibiotics. Recently, our laboratory reported the purification and characterization of two lantibiotics, suicin 90–1330 and suicin 3908, produced by the swine pathogen and zoonotic agent Streptococcus suis (serotype 2). In this study, a novel bacteriocin produced by S. suis has been identified and characterized. The producing strain S. suis 65 (serotype 2) was found to belong to the sequence type 28, that includes strains known to be weakly or avirulent in a mouse model. The bacteriocin, whose production was only possible following growth on solid culture medium, was purified to homogeneity by cationic exchange and reversed-phase high-pressure liquid chromatography. The bacteriocin, named suicin 65, was heat, pH and protease resistant. Suicin 65 was active against all S. suis isolates tested, including antibiotic resistant strains. Amino acid sequencing of the purified bacteriocin by Edman degradation revealed the presence of modified amino acids suggesting a lantibiotic. Using the partial sequence obtained, a blast was performed against published genomes of S. suis and allowed to identify a putative lantibiotic locus in the genome of S. suis 89–1591. From this genome, primers were designed and the gene cluster involved in the production of suicin 65 by S. suis 65 was amplified by PCR. Sequence analysis revealed the presence of ten open reading frames, including a duplicate of the structural gene. The structural genes (sssA and sssA’) of suicin 65 encodes a 25-amino acid residue leader peptide and a 26-amino acid residue mature peptide yielding an active bacteriocin with a deducted molecular mass of 3,005 Da. Mature suicin 65 showed a high degree of identity with class I type B lantibiotics (globular structure) produced by Streptococcus pyogenes (streptococcin FF22; 84.6%), Streptococcus macedonicus (macedocin ACA-DC 198; 84.6%), and Lactococcus lactis subsp. lactis (lacticin 481; 74.1%). Further studies will evaluate the ability of suicin 65 or the producing strain to prevent experimental S. suis infections in pigs.
Introduction
Streptococcus suis is a major swine pathogen worldwide that is transmitted via the respiratory route and colonizes the palatine tonsils of pigs [1]. Given its ability to produce a large array of virulence factors, it can cause meningitis, septicemia, arthritis, and endocarditis [2, 3]. Although S. suis infections in humans remain sporadic and affect mainly individuals in close contact with sick or pig-derived products, important outbreaks that occurred in Asia modified the world viewpoint regarding the threat of S. suis for humans [4, 5]. While 35 serotypes (1 to 34 and 1/2) have been identified, serotype 2 is the most frequently associated with pathology [1, 2]. However, in the last decade, other serotypes with a particular geographical distribution have also been identified as the source of many infections [6]. For instance, in North America, serotypes 2 and 3 show a prevalence of 24.3% and 21%, respectively, followed by serotypes 1/2, 8, and 7 [6].
Due to the wide diversity among strains, current vaccines for S. suis infections confer only a limited and serotype-specific protection [7]. For this reason, studies are still needed to develop an effective and broadly protective vaccine against the most prevalent if not all pathogenic serotypes. Antibiotics administrated as prophylactic or methaphylactic treatment are usually effective for controlling disease progression in the herds [8]. However, antibiotics should be used carefully and adequately to reduce the selection of resistant S. suis isolates. While most strains of S. suis are sensitive to penicillin and amoxicillin, resistance to macrolides, lincosamides, sulphonamides, and tetracyclines has been reported with up to 85% of strains showing resistance [9, 10].
With the rising concern on resistance of pathogens to conventional antibiotics, bacteriocins, which are ribosomally synthesized antimicrobial peptides of bacterial origin, have been proposed as a promising alternative antimicrobial strategy in animal agriculture [11, 12]. Bacteriocins are cationic bactericidal peptides that kill bacteria by disrupting the cell membrane integrity [13]. More specifically, lantibiotics are an important family of heat stable low molecular weight bacteriocins that possess unusual post-translationally modified amino acids, such as lanthionine, methyllanthionine, dehydroalanine, and dehydrobutyrine with thioether linkages [14]. Nisin A represents the most studied lantibiotic, being used commercially as a food preservative especially in dairy products in more than 50 countries [15]. Recently, our laboratory purified and characterized two lantibiotics produced by S. suis serotype 2 [16, 17]. Suicin 90–1330 is a type A (linear) lantibiotic secreted by a non-virulent strain of S. suis that exhibits high homology with nisin U produced by Streptococcus uberis [16]. Suicin 3908 is a type B (N-terminal linear and C-terminal globular moieties) lantibiotic produced by a strain of S. suis isolated from an healthy carrier pig and that shows a high homology with bovicin HJ50 (Streptococcus bovis) and thermophilin 1277 (Streptococcus thermophilus) [17]. As a continuation of our ongoing work aimed to investigate S. suis bacteriocins, in this study, we identified and characterized a novel class I type B lantibiotic produced by S. suis.
Materials and Methods
Bacterial strains and growth conditions
Strains of S. suis used in this study are listed in Table 1. Bacteria were routinely grown aerobically under static conditions at 37°C in Todd Hewitt broth (THB; BD-Canada, Mississauga, ON, Canada).
Table 1. Strains of S. suis used in this study, their characteristics and susceptibility to S. suis 65.
| Strain | Country | Origin | ST | Resistance to antibiotics 1 | Inhibitory zone (mm) 2 produced by S. suis 65 | |
|---|---|---|---|---|---|---|
| Erythromycin | Tetracycline | |||||
| 31533 | France | Meningitis | 1 | Sensitive | Sensitive | 4.7 ± 0.6 |
| MNCM01 | Thailand | Endocarditis (human) | 1 | Resistant | Resistant | 2.0 ± 1.0 |
| MNCM06 | Thailand | Meningitis (human) | 1 | Resistant | Resistant | 3.7 ± 0.6 |
| P1/7 | United Kingdom | Meningitis | 1 | Sensitive | Sensitive | 4.3 ± 0.6 |
| MGGUS3 | United States | Meningitis | 1 | Unknown | Unknown | 5.3 ± 0.5 |
| 89–1591 | Canada | Septicemia/meningitis | 25 | Unknown | Unknown | 1.2 ± 0.3 |
| 1043248 | Canada | Meningitis | 25 | Unknown | Unknown | 4.3 ± 0.6 |
| MNCM51 | Thailand | Septicemia (human) | 25 | Unknown | Unknown | 6.0 ± 1.0 |
| LPH4 | Thailand | Septicemia (human) | 25 | Resistant | Resistant | 5.0 ± 1.0 |
| MGGUS4 | United States | Septicemia | 25 | Unknown | Unknown | 3.3 ± 0.6 |
| 1054471 | Canada | Meningitis | 28 | Sensitive | Resistant | 3.3 ± 0.6 |
| 1057906 | Canada | Meningitis | 28 | Unknown | Unknown | 3.7 ± 0.6 |
| 1088563 | Canada | Meningitis | 28 | Resistant | Resistant | 3.3 ± 0.6 |
| 65 | France | Tonsils, healthy pig carrier | 28 | Unknown | Unknown | 0 |
| DAT245 | Japan | Meningitis | 28 | Unknown | Unknown | 4.0 ± 1.0 |
| MGGUS10 | United States | Pneumonia | 28 | Unknown | Unknown | 4.5 ± 0.7 |
1 A strain was considered resistant if the MIC was ≥ 400 μg/ml
2 Means ± standard deviations of triplicate assays
Plate diffusion assay for bacteriocin production
Overnight cultures of S. suis were spotted (2 μl) on Todd Hewitt agar (THA; BD-Canada) plates (100 mm) supplemented with 0.01% Tween 80 (sorbitan polyoxyethylene monooleate; Sigma-Aldrich Canada Co., Oakville, ON, Canada), which were incubated at 37°C for 24 h. Plates were then overlaid with THB soft-agar (0.75%, w/v) that had been inoculated (750 μl in 7.5 ml) with a 24-h culture of the indicator strain, and were further incubated at 37°C for 24 h. The zones of inhibition (in mm) were measured from the edge of the growth of S. suis to the margin of the inhibitory zone.
Detection of suiA and sslA structural genes
The presence in the S. suis 65 genome of the structural genes suiA and sslA, coding respectively for suicin 3908 and suicin 90–1330 previously characterized in our laboratory [16, 17], was investigated by PCR. PCR reactions consisted of 41.5 μl of PCR grade water, 5 μl of 10× Taq reaction buffer, 1 μl of nucleotide mix, 0.6 μl each of the appropriate forward and reverse primers for suiA (A284: 5’-CAAACTGCAACTGATCAAGAAATTA-3’ and A398R: 5’-AATTTTTGCACCCAGAGATGAATCC-3’, respectively), sslA (G48: 5’-AAACAACTCAGGAGCTTCAC-3’ and G130R: 5’-CACAGGTCATCAAAATACCC-3’, respectively), or gdh (used as a positive control; GDH645: 5’-TTTGGTTTACTTCACTGATAACATG-3’ and GDH794R: 5’-GAGTCTGAAACAGAAATAACTTTTG-3’, respectively), 1 μl of EconoTaq DNA polymerase (5 U/μl), and 1 μl of genomic DNA as a template. The PCR was performed with a DNA Thermal Cycler 480 (Perkin-Elmer, Waltham, MA, USA) according to the EconoTaq reaction protocol of Lucigen Corporation (Middleton, WI, USA). The reaction was carried out for 30 cycles with the following temperature-time profile: 95°C for 1 min, 50°C for 1 min, and 72°C for 1 min. At the end of the amplification protocol, the samples were incubated at 72°C for 3 min. A 1% agarose gel was used to analyze the PCR products.
Purification of bacteriocin
One hundred μl of an overnight culture of S. suis 65 was spread onto twelve THA plates supplemented with 0.01% Tween 80. Following incubation at 37°C under aerobic conditions for 24 h, bacterial cells were removed by scraping the surface of plates, and the solid medium of each plate was chopped into small pieces and transferred into 50-ml polypropylene tubes containing 2 ml of 20 mM 2-(N-morpholino)ethanesulfonic acid (MES) buffer pH 5.5 and 0.01% Tween 80 prior to freezing at -80°C for a minimum of 2 h. Thereafter, the culture medium was thawed at 55°C and the tubes were subjected to centrifugation (10,000 x g for 10 min). The liquid phases were recovered and pooled. The above procedure was repeated and all fractions were pooled. Residual bacteria and agar particles in the final bacteriocin-containing fraction were removed by two centrifugations at 10,000 x g for 10 min. The sample was then subjected to cationic exchange high-pressure liquid chromatography (HPLC; MonoS 5/50 GL column; GE Healthcare, Baie d’Urfé, QC, Canada) using an ÄKTA Purifier system (GE Healthcare). Elution was performed at a flow rate of 1 ml/min using a linear gradient of KCl from 0 to 0.65 M in the above MES buffer. The active fractions were detected by spotting 5 μl onto the surface of THA plates inoculated with a lawn of S. suis MGGUS3 used as the indicator strain (spot test plate assay). Following growth (37°C/24 h), positive fractions showing an inhibitory zone were pooled and dialyzed (1,000 Da cut-off) overnight against 0.01% trifluoroacetic acid (TFA) + 10% acetonitrile. The resulting fraction was then subjected to reversed-phase HPLC (SOURCE 15RPC column; GE Healthcare). Elution was performed at a flow rate of 1 ml/min using a linear gradient of acetonitrile from 10 to 90%. The acetonitrile and TFA were removed by a rotary evaporator prior to analyze the fractions for bacteriocin activity using the spot test plate assay as above. The active fractions were pooled and glycerol was added to a final concentration of 15%, prior to aliquot (100 μl) and store at -80°C. Total purified bacteriocin was quantified as arbitrary units, which correspond to the reciprocal of the highest two-fold serial dilution giving a clear inhibitory zone following application of 5 μl of the bacteriocin solution on a lawn of S. suis MGGUS3.
SDS-PAGE analysis
The purified bacteriocin was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 16.5% Tris-Tricine gel (Bio-Rad Laboratories, Mississauga, ON, Canada), fixed (30 min) in 40% methanol-10% acetic acid, and stained with Coomassie brilliant blue G-250. A gel was also fixed in 10% acetic acid—20% propanol (30 min), washed thoroughly in sterile distilled water (5 x 30 min), and bacteriocin activity was detected using an overlay of THB-soft agar inoculated with the indicator strain S. suis MGGUS3. Nisin A (Sigma-Aldrich Canada Co.) was used as a positive control.
Characterization of bacteriocin
The susceptibility of the purified bacteriocin to temperature, pH, and enzymatic treatments was investigated using the spot test plate assay described above and S. suis MGGUS3 as the indicator bacteria. To evaluate temperature stability, the purified bacteriocin was incubated at 45, 70, 100 or 121°C for 15 min prior to determine the antibacterial activity. To investigate the susceptibility to extreme pHs, the bacteriocin solution was adjusted to pH 2 or 11 by using 0.125 N HCl or 0.125 N NaOH, respectively. After 15 min at room temperature, the bacteriocin solution was diluted 1:2 in PBS to neutralize pH, and the antibacterial activity was then determined. Lastly, the proteolytic enzymes trypsin, chymotrypsin, and proteinase K (Sigma-Aldrich Canada Co.), each at a final concentration of 500 μg/ml, were used to evaluate the susceptibility of the bacteriocin to proteolytic cleavage. Following incubation at 37°C for 30 min, samples were treated for 5 min at 68°C to inactivate the enzymes and the bacteriocin activity was determined.
Amino acid sequencing
The purified bacteriocin was subjected to SDS-PAGE as above and then electroblotted onto a polyvinylidene diofluoride (PVDF) membrane. The bacteriocin band, localized based on the migration of pre-stained molecular weight markers, was excised and transferred into a microtube. Ethanethiol derivatization of post-translationally modified amino acids of the PVDF-blotted bacteriocin was carried out as previously described by Meyer et al. [18] with slight modifications. In an anaerobic chamber, 200 μl of a reaction mixture containing 280 μl methanol, 200 μl of H2O, 65 μl 5 M of NaOH and 60 μl of ethanethiol was added. After incubation at 50°C for 1 h, the solution was acidified by adding 66 μl of 70% (v/v) formic acid and the bacteriocin-blotted PVDF membrane was vacuum-dried. The treated bacteriocin was then sent to the SPARC BioCentre (The Hospital for Sick Children, Toronto, ON, Canada) and subjected to Edman degradation using an Applied Biosystems ABI 492 Procise cLC sequencer (Life Technologies Inc., Burlington, ON, Canada).
Identification of the putative gene cluster encoding suicin 65
Published S. suis genomes were blasted for the presence of proteins related to the partial amino acid sequence determined above using the NCBI Protein blast web-based platform (blast.ncbi.nlm.nih.gov) [19]. The putative bacteriocin gene cluster that was identified was used to design primers for PCR amplification using genomic DNA extracted from S. suis 65. The sequences of the forward and reverse primers were: ext84170: 5’- CTCCTGTAAGATGAACTTGT-3’ and ext84641R: 5’-CTTTATATGAGTCTGTCTGT-3’, respectively. The reaction mixture was prepared as above and the reaction was carried out for 25 cycles with the following temperature-time profile: 95°C for 1 min, 45°C for 1 min, and 72°C for 40 sec. At the end of the amplification protocol, the samples were incubated at 72°C for 3 min. The putative suicin 65 locus was then sequenced.
Multilocus sequence typing of S. suis 65
Multilocus sequence typing of S. suis 65 was performed by PCR amplification and DNA sequencing of seven housekeeping genes (cpn60, dpr, recA, aroA, thrA, gki, mutS), as previously described [20].
Antibiotic susceptibility of S. suis 65
The susceptibility of S. suis 65 to antibiotics (amoxicillin, ceftiofur, and penicillin G) was determined as follows. Briefly, a 24-h bacterial culture in THB was diluted in fresh broth medium to obtain an optical density at 660 nm (OD660) of 0.2. Equal volumes (100 μl) of bacteria and serial dilutions of antibiotics in THB were mixed in the wells of 96-well plates. Wells with no bacteria or no antibiotics were used as controls. Following a 24-h incubation at 37°C, bacterial growth was recorded visually. Minimal inhibitory concentration (MIC) values (μg/ml) were expressed as the lowest concentration at which no growth occurred. The MIC values were determined in three independent experiments.
Results
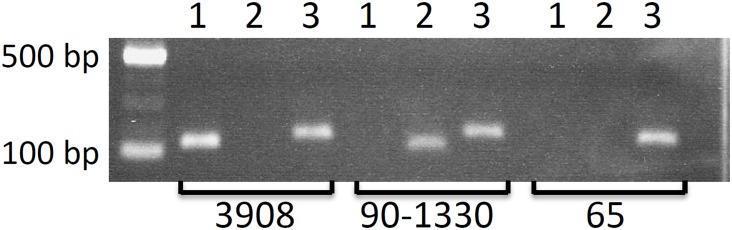
In a previous study [21], S. suis 65 has been found to exert antagonism against other strains of S. suis suggesting that it may produce a bacteriocin-like inhibitory substance. We first confirmed the antibacterial activity of S. suis 65 using the indicator strain S. suis MGGUS3 in a plate diffusion assay (inhibitory zone = 5.3 ± 0.5 mm). To exclude the possibility that this antibacterial activity resulted from the production of the two previously characterized S. suis lantibiotics, the presence of the structural genes suiA and sslA, coding respectively for suicin 3908 and suicin 90–1330, was investigated by PCR. As shown in Fig 1, both genes were absent in S. suis 65, while suiA was identified in S. suis 3908, and sslA in S. suis 90–1330, thus suggesting that S. suis 65 produces a novel bacteriocin. The control gene gdh was identified in all three S. suis isolates.
Fig 1. PCR detection of the lantibiotic structural genes suiA and sslA, and the control gene gdh in S. suis 3908, 90–1330, and 65.
Lane 1: suiA. Lane 2: sslA. Lane 3: gdh.
The antibacterial activity of S. suis 65 was investigated with the diffusion plate assay on additional strains of S. suis. As reported in Table 1, all strains tested belonging to different STs, isolated in different countries, and showing resistance to antibiotics, including erythromycin and tetracycline, were inhibited to various extents.
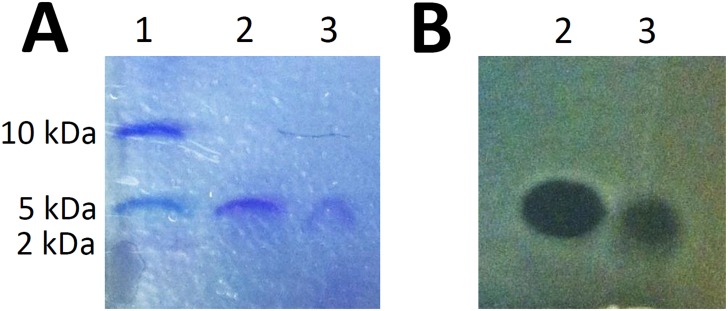
A preliminary analysis revealed that a 24-h culture supernatant of S. suis 65 does not possess antibacterial activity thus indicating that the bacteriocin was not secreted following growth in a liquid medium. Consequently, a protocol was developed to prepare a bacteriocin-rich fraction following growth of S. suis 65 on THA plates. This fraction was then used to purify the bacteriocin to homogeneity by cationic exchange and reversed-phase HPLC. Bacteriocin activity recovered from the reversed-phase chromatography eluted in a single peak and Tricine-SDS-PAGE analysis of the purified bacteriocin yielded a single band stained by Coomassie blue and migrating slightly lower than the commercial lantibiotic nisin A, which has a molecular mass of 3,354 Da (Fig 2A). An overlay with the indicator strain (S. suis MGGUS3) correlated the bacteriocin activity with the protein band (Fig 2B). From the twelve THA plates of S. suis 65, the purification protocol allowed the recovery of 8,000 arbitrary units of bacteriocin, as defined in Materials and Methods.
Fig 2. Tris-Tricine SDS-PAGE analysis of the purified bacteriocin produced by S. suis 65.
Panel A. Coomassie blue staining. Panel B. Antibacterial activity detected by an overlay with S. suis MGGUS3 as the indicator strain. Lane 1: molecular weight markers; Lane 2: commercial nisin A; Lane 3: purified suicin 65.
The purified bacteriocin was subjected to various treatments to determine its stability to heat, pH, and proteolytic enzymes (Table 2). Using the spot test assay, the bacteriocin was highly heat stable as the antibacterial activity against the indicator strain (MGGUS3) was still detected even after treatment at 121°C for 15 min. The bacteriocin was also found to be stable under a wide range of pH since antibacterial activity was still observed following exposure at pH 2 and 11. Moreover, the antibacterial activity remained active following a treatment of the purified bacteriocin with proteolytic enzymes (trypsin, chymotrypsin, proteinase K). Lastly, using the spot test assay, the purified bacteriocin was found to be active against all strains that have been tested in the plate diffusion assay with the producing strain.
Table 2. Stability of the purified suicin 65 determined using the spot test assay and S. suis MGGUS3 as the indicator strain.
| Treatment | Inhibitory activity |
|---|---|
| 45°C / 15 min | + |
| 70°C / 15 min | + |
| 100°C / 15 min | + |
| 121°C / 15 min | + |
| pH 2 / 15 min | + |
| pH 11 / 15 min | + |
| Trypsin / 30 min | + |
| Chymotrypsin / 30 min | + |
| Proteinase K / 30 min | + |
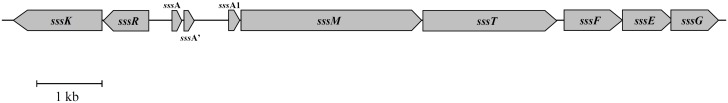
Peptide sequencing of the purified bacteriocin was carried out by Edman degradation. The first nine amino acids yielded the following sequence: Gly1-Lys2-Asn3-Gly4-Val5-Phe6-Lys7-X(Ser or Cys or Thr)8-Ile9. Residue 8 yielded a peak close to the Phe peak, accompanied by a relatively strong Leu signal, which is characteristic of Thr residues modified to either dehydrobutyrine (Dhb) or a methyllanthionine (MeLan) moiety [18]. Using the NCBI Protein blast web-based platform, published genomes of S. suis were analyzed for the presence of a protein showing similarity with the above 9-amino acid sequence. This analysis led to the identification of two genes (ssuisDraft_3894 and ssuisDraft_3893), annotated as type A lantibiotic, in the genome of strain S. suis 89–1591 (serotype 2, ST25) that were contiguous and shared 92% identity of residues (nucleotides or amino acid). Further analysis of the 89–1591 genome revealed the presence of a complete lantibiotic biosynthesis locus. No additional locus that may correspond to suicin 90–1330 or suicin 3908 were found. Based on this gene locus identified in strain 89–1591, primers were designed for PCR amplification in S. suis 65. Following sequencing, the complete sss (streptococcus suis suicin) gene cluster of S. suis 65 was identified and was found to contain ten open reading frames. As reported in Fig 3, the locus encodes the suicin 65 precursors (sssA, sssA’), a synthetase involved in lantibiotic modification (sssM), an ABC transporter (sssT), a response regulator (sssR), a sensor histidine kinase (sssK), and three immunity proteins (sssF, sssE, sssG). A small open reading frame (sssA1), for which no functions has been assigned yet, was located upstream the sssM gene.
Fig 3. Genetic organization of the putative suicin 65 gene cluster.
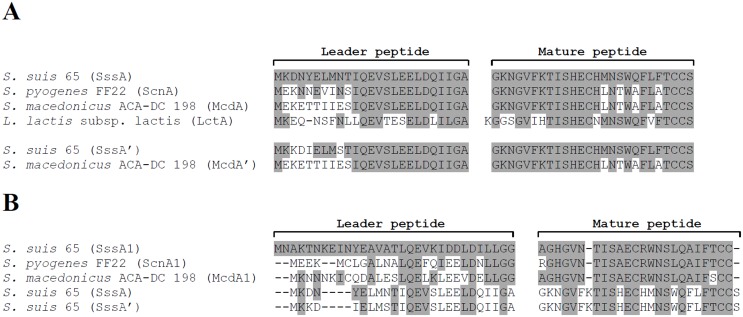
From the structural genes sssA and sssA’, the inferred amino acid sequence of the leader peptide contains 25 amino acids while that of mature unmodified peptide consists of 26 amino acids (Fig 4). The deduced molecular mass of the unmodified mature bacteriocin is 3,005 Da. Comparison of the SssA amino acid sequence with previously characterized lantibiotics highlighted high levels of identity with streptococcin FF22 (80%) produced by S. pyogenes FF22, macedocin ACA-DC 198 (75%) produced by S. macedonicus ACA-DC 198, and lacticin 481 (62%) produced by Lactococcus lactis subsp. lactis 481 (Table 3). Fig 4 indicates that the mature peptides of S. pyogenes FF22 and S. macedonicus ACA-DC 198 lantibiotics differ from that of suicin 65 in only four amino acids. In regard to the duplicate structural gene sssA’, the inferred amino acid sequence of the leader peptide and the mature peptide showed respectively 92% and 100% of identity with the corresponding peptides encoded by the sssA gene. Regarding nucleic acid residues, sssA and sssA’ genes related to mature peptide showed 86.4% identity. The macedocin ACA-DC 198 locus in S. macedonicus also contains a duplicate structural gene mcdA’ that showed 76% of identity (amino acid sequence) with sssA’ (Fig 4). Lastly, the gene sssA1 whose function in the sss locus is not identified yet showed poor homology (65%) in amino acid sequence with its homologues scnA1 and mcdA1 found in S. pyogenes and S. macedonicus, respectively (Fig 4).
Fig 4. Comparison of the amino acid sequence of suicin 65 with other lantibiotics produced by S. macedonicus ACA-DC 198 (macedocin), S. pyogenes FF22 (streptococcin), and L. lactis subsp. lactis 481 (lacticin).
The left and right blocks of the sequence refer to the leader and mature peptides, respectively. Panel A: SuiA and SuiA’ and their homologues; Panel B: SuiA1 and its homologues.
Table 3. Percentage identity in deduced amino acid sequences between the products of the S. suis suicin 65 gene cluster and the gene cluster bacteriocins of S. macedonicus ACA-DC 198, S. pyogenes FF22, L. lactis subsp. lactis 481 and S. suis 89–1591.
| Genes | % identity | |||
|---|---|---|---|---|
| S. macedonicus ACA-DC 198 | S. pyogenes FF22 | L. lactis subsp. lactis 481 | S. suis 89–1591 | |
| K | 73 | 78 | - | 100 |
| R | 82 | 86 | - | 100 |
| A | 75 | 80 | 62 | 100 |
| A’ | 76 | - | - | 100 |
| A1 | 65 | 65 | - | - 1 |
| M | 62 | 62 | 30 | - 1 |
| T | 72 | 74 | 39 | 100 1 |
| F | 82 | 81 | 44 | 99 |
| E | 34 | 33 | 22 | 100 |
| G | 49 | 44 | 26 | 100 |
1The complete genes A1 and M, as well as half of the gene T are deleted
Table 3 reports the percentage identity of S. suis 65 genes of the lantibiotic biosynthesis locus with the corresponding genes found in S. macedonicus ACA-DC 198, S. pyogenes FF22, L. lactis subsp. lactis 481, and S. suis 89–1591. The genes E and G coding for immunity proteins showed the lowest percentage identity. As in the lantibiotic locus of S. suis 65, the lantibiotic locus of S. macedonicus also contains a duplicate of the structural gene (A and A’) as well as a non-related gene (A1). The lantibiotic locus of S. suis 89–1591 has a 4,473 bp deletion that results in the lack of the gene A1, the gene M, and half of the gene T. These observations suggest that active bacteriocin may not be produced by this strain. Using the plate diffusion assay, it was confirmed that S. suis 89–1591 does not possess antibacterial activity against the indicator strain S. suis MGGUS3 (data not shown). Moreover, S. suis 89–1591 did not possess immunity against the bacteriocin produced by S. suis 65 (Table 1).
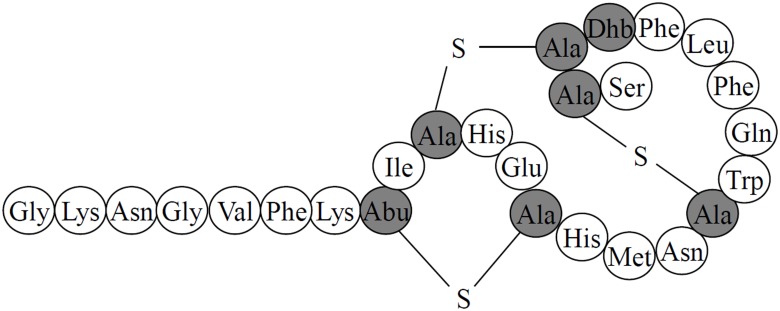
Fig 5 presents the proposed structure of suicin 65 based on the deducted amino acid sequence of the structural genes sssA and sssA’ and previously reported structures of lantibiotics with high identity with suicin 65. It has a linear part in N-terminal and a globular part in C-terminal. Moreover, it shows the presence of two lanthionine groups (Ala-S-Ala) between residues 17 and 25 and 10 and 24, and one methyllanthionine group (Aminobutyrate-S-Ala) between residue 8 and 13, and a dehydrobutyrine (Dhb) in residue 23.
Fig 5. Proposed structure of suicin 65.
Mature peptide where Ser and Thr residues which are posttranslationally dehydrated to dehydrobutyrine (Dhb), or involved in the formation of lanthionine (Ala-S-Ala) and methyllanthionine (aminobutyrate[Abu]-S-Ala), respectively, with cysteine residues, are shaded in grey.
Lastly, given that S. suis 65 may have potential probiotic and protective applications, it was of interest to perform MLST to determine to what sequence type (ST) it belongs. This analysis revealed that S. suis 65 is an ST28, that includes strains with a low or no virulence in a mouse model [20]. We also evaluated the susceptibility of S. suis 65 to antibiotics. S. suis 65 was found to be highly sensitive to amoxicillin with a MIC of 0.0049 μg/ml as well as to penicillin G and ceftiofur with a MIC of 0.0098 μg/ml.
Discussion
Antibiotics are widely used in animal agriculture to treat and/or prevent bacterial diseases in order to reduce economic losses. Moreover, with the objective to maintain health and increase productivity, animal production is sometimes associated with the regular use of antibiotics, thus promoting the emergence of antibiotic-resistant bacteria. In the United States, it has been estimated that ~80% of the nation’s annual antimicrobial consumption accounts for livestock production [22]. One strategy to overcome the fact that bacterial pathogens become increasingly resistant to several antibiotics is to look for new antimicrobial agents or alternative therapeutic strategies. In this regard, bacteriocins, which are ribosomally synthesized antimicrobial peptides of bacterial origin, have been proposed to represent a promising option to antibiotics in livestock production [9, 10]. Recently, our laboratory purified and characterized two bacteriocins of the lantibiotic family produced by S. suis and active against this swine pathogen [16, 17]. In this study, we report the purification and characterization of a novel class I type B lantibiotic produced by S. suis 65.
While the two previously reported S. suis lantibiotics (suicin 90–1330 and suicin 3908) were purified from a bacterial culture supernatant, the one produced by S. suis 65 was not detected in broth media and consequently has been purified from solid culture media. Lantibiotics produced by Streptococcus mutans [23] and Lactobacillus plantarum [24] were also reported to be only produced following growth on solid media. As observed for bacteriocins of the lantibiotic family, the purified suicin 65 was found to be stable to heat, pH and proteases. The bacteriocin produced by S. suis 65 was found to be active against all S. suis strains tested, including strains possessing resistance to erythromycin and/or tetracycline.
Suicin 65 is a class I type B lantibiotic made of 26 amino acid residues (mature peptide) with a linear portion in N-terminal and a globular portion in C-terminal. The previously reported suicin 3908 produced by S. suis 3908 was also a class I type B lantibiotic (33 amino acid residues); however, the two bacteriocins exibit a poor identity (20.6%) for the unmodified mature peptide. Mature unmodified suicin 65 showed a high homology with macedocin ACA-DC 198 (84.6%), streptococcin FF22 (84.6%), and lacticin 481 (74.1%) produced by S. macedonicus [25], S. pyogenes [26], and L. lactis subsp. lactis [27], respectively.
A genetic locus containing ten open reading frames has been identified in S. suis 65 and is most likely responsible for suicin 65 regulation, biosynthesis, and immunity, although a direct proof for its involvement would be to correlate an absence of activity with a mutation created in the structural gene. Surprisingly, the locus shows the presence of a duplicate of the structural gene. More specifically, immediately downstream of the sssA structural gene, there is another open reading frame named sssA’. The mature peptides of sssA and sssA’ possess a 100% homology. Analysis by RT-PCR provided evidence that both structural genes are cotranscribed and part of the same operon (data not shown). The presence of duplicate structural genes, also cotranscribed, has been reported in the genetic locus of macedocin ACA-DC 198 [25]. In addition, the sss genetic locus showed the presence of a third structural gene (sssA1) that appears unrelated to suicin 65 and that exhibit 65% homology with similar genes found in the genetic locus of macedocin ACA-DC 198 [25] and streptococcin FF22 [26]. Further studies are required to determine whether the peptide encoded by suiA1 corresponds to a functional lantibiotic and how its expression is regulated in S. suis 65.
Most of the S. suis serotype 2 strains from North America have been found to belong to ST25 or ST28, which are significantly less virulent than ST1 strains [20]. More specifically, ST28 isolates are essentially avirulent in a mouse model, while ST25 strains are moderately virulent and able to induce severe infections [20]. MLST performed on S. suis 65 showed that it is an ST28 isolate. Moreover, S. suis 65, which has been isolated from tonsils of a healthy pig, did not induce clinical signs when inoculated in pathogen-free piglets [28]. We also showed that this strain is highly susceptible to antibiotics (amoxicillin, ceftiofur, penicillin G) currently used in the swine industry. Taken together, these characteristics suggest that S. suis 65 may be safe, although additional animal studies will have to be carried out to confirm this. Future research should evaluate whether the suicin 65 or the producing strain S. suis 65 could be used as a therapeutic agent targeting S. suis.
Acknowledgments
We wish to thank Taryn Athey for technical assistance.
Data Availability
All relevant data are within the paper.
Funding Statement
This research was supported by Ministère de l’Agriculture, des Pêcheries et de l’Alimentation du Québec, Grant #811326, to DG MF MG. This study made use of the Streptococcus suis Multilocus Sequence Typing Database hosted at Imperial College and whose development is funded by the Wellcome Trust. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Gottschalk M. Streptococcosis In: Karriker L, Ramirez A, Schwartz KJ, Stevenson G, Zimmerman J, editors. Diseases of swine, NJ: Wiley Publishers, 2012; 841–855. [Google Scholar]
- 2. Fittipaldi N, Segura M, Grenier D, Gottschalk M. Virulence factors involved in the pathogenesis of the infection caused by the swine pathogen and zoonotic agent Streptococcus suis. Future Microbiol 2012; 7: 259–279. 10.2217/fmb.11.149 [DOI] [PubMed] [Google Scholar]
- 3. Baums CG, Valentin-Weigand P. Surface-associated and secreted factors of Streptococcus suis in epidemiology, pathogenesis and vaccine development. Anim Health Res Rev 2009; 10: 65–83. 10.1017/S146625230999003X [DOI] [PubMed] [Google Scholar]
- 4. Gottschalk M, Xu J, Calzas C, Segura M. Streptococcus suis: a new emerging or an old neglected zoonotic pathogen? Future Microbiol 2010; 5: 371–391. 10.2217/fmb.10.2 [DOI] [PubMed] [Google Scholar]
- 5. Khadthasrima N, Hannwong T, Thammawijaya P, Pingsusean D, Akkanij B, Jaikhar A, et al. Human Streptococcus suis outbreak in Phayao province, Thailand 2007. OSIR 2009; 1: 4–9. [Google Scholar]
- 6. Goyette-Desjardins G, Auger JP, Xu J, Segura M, Gottschalk M. Streptococcus suis, an important pig pathogen and emerging zoonotic agent—an update on the worldwide distribution based on serotyping and sequence typing. Emerg Microbes Infect 2014; 3: e45 10.1038/emi.2014.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Segura M. Streptococcus suis vaccines: candidate antigens and progress. Expert Rev Vaccines 2015; 12: 1587–1608. [DOI] [PubMed] [Google Scholar]
- 8. Cromwell GL. Why and how antibiotics are used in the swine production. Anim Biotechnol 2002; 13: 7–27. [DOI] [PubMed] [Google Scholar]
- 9. Varela NP, Gadbois P, Thibault C, Gottschalk M, Dick P, Wilson J. Antimicrobial resistance and prudent use for Streptococcus suis. Anim Health Res Rev 2013; 20: 1–10. [DOI] [PubMed] [Google Scholar]
- 10. Callens BF, Haesebrouck F, Maes D, Butaye P, Dewulf J, Boyen F. Clinical resistance and decreased susceptibility in Streptococcus suis isolates from clinically healthy fattening pigs. Microb Drug Resist 2013; 19: 146–151. 10.1089/mdr.2012.0131 [DOI] [PubMed] [Google Scholar]
- 11. Arthur TD, Cavera VL, Chikindas ML. On bacteriocin delivery systems and potential applications. Future Microbiol 2014; 9: 235–248. 10.2217/fmb.13.148 [DOI] [PubMed] [Google Scholar]
- 12. Lin J. Novel approaches for Campylobacter control in poultry. Foodborne Pathog Dis 2009; 6: 755–765. 10.1089/fpd.2008.0247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cotter PD, Ross RP, Hill C. Bacteriocins—a viable alternative to antibiotics? Nat Rev Microbiol 2013; 11: 95–105. 10.1038/nrmicro2937 [DOI] [PubMed] [Google Scholar]
- 14. Bierbaum G, Sahl HG. Lantibiotics: mode of action, biosynthesis and bioengineering. Curr Pharm Biotechnol 2009; 10: 2–18. [DOI] [PubMed] [Google Scholar]
- 15. De Arauz LJ, Jozala AF, Mazzola PG, Penna TCV. Nisin biotechnological production and application: a review. Trends Food Sci Technol 2009; 20: 146–154. [Google Scholar]
- 16. LeBel G, Vaillancourt K, Frenette M, Gottschalk M, Grenier D. Suicin 90–1330 from a non-virulent strain of Streptococcus suis: a nisin-related lantibiotic active on Gram-positive swine pathogens. Appl Environ Microbiol 2014; 80: 5484–5492. 10.1128/AEM.01055-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vaillancourt K, LeBel G, Frenette M, Gottschalk M, Grenier D. Suicin 3908, a new lantibiotic produced by a strain of Streptococcus suis isolated from a healthy carrier pig. PLoS One 2015; 10: e0117245 10.1371/journal.pone.0117245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Meyer HE, Heber M, Eisermann B, Korte H, Metzger JW, Jung G. Sequence analysis of lantibiotics: chemical derivatization procedures allow a fast access to complete Edman degradation. Anal Biochem 1994; 223: 185–190. [DOI] [PubMed] [Google Scholar]
- 19. Uniprot Consortium. The universal protein resource (UniProt). Nucleic Acids Res 2008; 36: 190–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fittipaldi N, Xu JG, Lacouture S, Tharavichitkul P, Osaki M, Sekizaki T, et al. Lineage and virulence of Streptococcus suis serotype 2 isolates from North America. Emerg Infect Dis 2011; 17: 2239–2244. 10.3201/eid1712.110609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mélançon D, Grenier D. Production and properties of bacteriocin-like inhibitory substances from the swine pathogen Streptococcus suis serotype 2. Appl Environ Microbiol 2003; 69: 4482–4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Food and Drug Administration. CVM Updates—CVM Reports on antimicrobials sold or distributed for food-producing animals (Food Drug Admin, Silver Spring. MD). 2010; Available at www.fda.gov/AnimalVeterinary/NewsEvents/CVMUpdates/ucm236143.htm.
- 23. Nicolas GG, LaPointe G, Lavoie MC. Production, purification, sequencing and activity spectra of mutacins D-123.1 and F-59.1. BMC Microbiol 2011; 11: 69 10.1186/1471-2180-11-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Maldonado-Barragan A, Ruiz-Barba JL, Jimenez-Diaz R. Knockout of three component regulatory systems reveals that the apparently constitutive plantaricin-production phenotype shown by Lactobacillus plantarum on solid medium is regulated via quorum sensing. Int J Food Microbiol 2009; 130: 35–42. 10.1016/j.ijfoodmicro.2008.12.033 [DOI] [PubMed] [Google Scholar]
- 25. Papadelli M, Karsioti A, Anastasiou R, Georgalaki M, Tsakalidou E. Characterization of the gene cluster involved in the biosynthesis of macedocin, the lantibiotic produced by Streptococcus macedonicus . FEMS Microbiol Lett 2007; 272: 75–82. [DOI] [PubMed] [Google Scholar]
- 26. McLaughlin RE, Ferretti JJ, Hynes WL. Nucleotide sequence of the streptococcin A-FF22 lantibiotic regulon: model for production of the lantibiotic SA-FF22 by strains of Streptococcus pyogenes . FEMS Microbiol Lett 1999; 175: 171–177. [DOI] [PubMed] [Google Scholar]
- 27. Dufour A, Rincé A, Hindré T, Haras D, Le Pennec JP. Lacticin 481: an antimicrobial peptide of the lantibiotic family produced by Lactococcus lactis . Recent Res Dev Bacteriol 2003; 1: 219–234. [Google Scholar]
- 28. Berthelot-Hérault F, Cariolet R, Labbé A, Gottschalk M, Cardinal J-Y, Kobisch M. Experimental infection of specific pathogen free piglets with French strains of Streptococcus suis capsular type 2. Can J Vet Res 2001; 65: 196–200. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.