Abstract
Regenerative dental therapies for bone tissues rely on efficient targeting of endogenous and transplanted mesenchymal stem cells (MSCs) to guide bone formation. Amelogenin is the primary component of Emdogain, which is used to regenerate periodontal defects; however, the mechanisms underlying the therapeutic effects on alveolar bone remain unclear. The tetracycline (Tet)-dependent transcriptional regulatory system is a good candidate to investigate distinct roles of genes of interest during stem cell differentiation. Here, we investigated amelogenin-dependent regulation of osteogenesis in MSCs by establishing a Tet-controlled transcriptional activation system. Clonal mouse bone marrow-derived MSCs were lentivirally transduced with the Tet repressor (TetR) expression vector followed by drug selection to obtain MSCs constitutively expressing TetR (MSCs-TetR). Expression vectors that contained the Tet operator and amelogenin-coding (Amelx) cDNA fragments were constructed using the Gateway system and lentivirally introduced into MSCs-TetR to generate a Tet regulation system in MSCs (MSCs-TetR/Amelx). MSCs-TetR/Amelx significantly overexpressed the Amelx gene and protein in the presence of the tetracycline derivative doxycycline. Concomitant expression of osterix, bone sialoprotein (BSP), osteopontin, and osteocalcin was modulated by addition or removal of doxycycline under osteogenic guidance. During osteogenic induction, MSCs-TetR/Amelx treated with doxycycline showed significantly increased gene expression of osterix, type I collagen, BSP, and osteocalcin in addition to increased alkaline phosphatase activity and mineralized nodule formation. Enhanced extracellular matrix calcification was observed when forced Amelx expression commenced at the early stage but not at the intermediate or late stages of osteogenesis. These results suggest that a Tet-controlled Amelx gene regulation system for mouse MSCs was successfully established, in which transcriptional activation of Amelx was associated with enhanced osteogenic differentiation, especially in the early stage of biomineralization.
Introduction
After tooth loss, alveolar bone resorption unavoidably occurs; therefore, bone augmentation is often essential in esthetic implant treatment [1]. Mesenchymal stem cells (MSCs), which are multipotent stem cells traditionally found in the bone marrow and subsequently isolated from many other adult tissues, are currently an excellent candidate for regenerative medicine [2]. Recent progress in regenerative approaches using MSCs may provide considerable benefits in dental implant and prosthodontic treatments, facilitating regeneration of atrophic alveolar bone [3]. In addition, activation of MSCs by growth factors is considered to be a promising strategy for efficient oral tissue regeneration [4].
Amelogenin is the primary component of Emdogain, which is used clinically to regenerate periodontal tissues in intrabony defects [5,6]. Amelogenin is secreted by ameloblasts, and it comprises more than 90% of extracellular enamel matrix proteins in developing teeth. During tooth development, amelogenin is known to play a crucial role in the biomineralization and structural organization of enamel [7,8]. Although amelogenin has traditionally been considered an enamel protein, its biological activity in the process of cell differentiation has recently been widely recognized.
Amelogenin is comprised of three domains: an N-terminal tyrosine-rich domain, a central hydrophobic proline-rich domain, and a C-terminal hydrophilic telopeptide [9]. A previous study reported signaling effects of specific amelogenin gene splicing products in cells in an in vivo implantation model. These effects included induction of mineralization accompanied by the presence of bone matrix proteins such as bone sialoprotein (BSP, also known as integrin-binding sialoprotein; Ibsp), suggesting a signaling role of amelogenin gene products in preodontoblast maturation [10]. Viswanathan et al. demonstrated that recombinant amelogenin regulated BSP expression in cementoblasts in vitro and also observed a dramatic reduction in the expression of BSP in cementoblasts and surrounding osteoblasts in amelogenin-null mice, indicating that amelogenin is a potential regulator of cementum-associated genes [11].
Low levels of amelogenin expression have been reported in non-dental cell types, including stem cells and cells present in bone, brain, and other soft tissues [12], suggesting additional functions of amelogenin such as signal transduction in these cells. Histological observations have shown that amelogenin is expressed at low levels in normal alveolar bone tissues, and that its expression increases at sites of high bone activity and remodeling [13]. In addition, several studies have suggested that amelogenin has a unique function of modulating the osteogenic differentiation of stem cells. In mouse embryonic stem (ES) cells, administration of exogenous leucine-rich amelogenin peptides was demonstrated to rescue a partially amelogenin-null phenotype, and it significantly increased BSP and osterix expression during osteogenic differentiation [14]. In human bone marrow-derived MSCs, recombinant amelogenin was shown to increase the mRNA level of alkaline phosphatase (ALP), type I collagen and BSP, and also to enhance extracellular matrix (ECM) mineralization [15]. Genome-wide expression profiling of amelogenin-overexpressing MSCs showed up-regulation of several osteogenesis-associated genes [16]. However, the mechanisms by which amelogenin expression contributes to the osteogenic differentiation of MSCs, particularly the effects of amelogenin expression on mineralization during MSC osteogenesis, remain unclear.
Forced expression of amelogenin by lentiviral transduction is one of the most powerful and cost-effective methods to investigate the direct effect of amelogenin expression in stem cells [16]. In this system, the viral genome integrates into host chromosomes, and the inserted gene is maintained and expressed in the cells over multiple passages. These properties of lentiviral transduction enable permanent and efficient expression of the transgene in stem cells even after proliferation and differentiation. However, stem cells often lose their intrinsic stemness properties and differentiation capability after viral transduction because of uncontrollable expression of the transgene, thus resulting in heterogeneous populations of cells in different states of differentiation. This drawback makes it difficult to perform reproducible experiments on stem cells using such over-expression systems.
The tetracycline (Tet)-dependent transcriptional regulatory system is one of the best-studied transduction systems with established efficacy of controllable gene expression, where transcription is reversibly turned on or off in the presence or absence of a Tet derivative (doxycycline: Dox) [17]. This system is based on the binding of Dox to the Tet-repressor (TetR) and de-repression of the promoter controlling expression of the gene of interest. The Tet-dependent system has been successfully used to control gene expression in lentiviral transduction systems, including those applied to stem cells [18,19]. Therefore, Tet-inducible gene transcription using a lentiviral transduction system could be a powerful method to investigate and control the functions of amelogenin in MSCs.
In this study, we used on the Tet-dependent lentiviral transcriptional regulatory system to control forced expression of an exogenous amelogenin (Amelx: amelogenin, X-linked) gene in MSCs during osteogenic differentiation. The objective of this study was to highlight the in vitro mechanisms underlying amelogenin-dependent regulation of osteogenesis in MSCs by establishing a Tet-regulated system for amelogenin expression in MSCs.
Materials and Methods
Cell culture
Clonal mouse bone marrow-derived MSCs (mBMSC-4), which we previously established [20], were maintained in the growth medium consisting of modified Eagle minimal essential medium-alpha (α-MEM) (Nacalai Tesque, Kyoto, Japan) supplemented with 15% fetal bovine serum (FBS), 100 units/mL penicillin, 100 μg/mL streptomycin, and 250 ng/mL amphotericin B. For osteogenic induction, cells were cultured in the growth medium supplemented with 0.1 μM dexamethasone, 10 mM β-glycerophosphate, and 50 μM ascorbate-2-phosphate.
Human embryonic kidney (HEK) cell line-derived 293FT cells (Life Technologies, Carlsbad, CA) were maintained in Dulbecco’s modified Eagle medium (DMEM: 4.5 g/L glucose without sodium pyruvate, Nacalai Tesque) with 10% FBS, 0.1 mM MEM non-essential amino acids, 6 mM L-glutamine, 1 mM MEM sodium pyruvate, 50 units/mL penicillin, and 50 μg/mL streptomycin.
Production of lentiviral vectors carrying Amelx gene
The pCMV-SPORT6.1 plasmid vector containing the full-length cDNA of mouse amelogenin, X-linked (Amelx: GenBANK:BC059090.1), was purchased from Open Biosystems (Thermo Scientific). The open reading frame of the Amelx cDNA (660 bp) was PCR-amplified (forward primer: CAC CAT GGG GAC CTG GAT TTT GTT; reverse primer: TCA TTT TTC TGT TGT GCT TTC C) and cloned into the pENTR™/D-TOPO vector using the pENTR Directional TOPO cloning kit (Life Technologies) to obtain the entry vector (pENTR™/D-TOPO/Amelx) for the Gateway cloning system (Life Technologies). Using this entry vector, the expression vector for Amelx [pLenti6.3/Tet operator (TO)/V5/Amelx] was constructed through the LR recombination reaction of the Gateway cloning system.
293FT cells were cultured in 6-cm dishes to produce the lentivirus from the pLenti3.3/TetR (Life technologies) and pLenti6.3/TO/V5/Amelx expression vectors. The plenti6.3/V5-GW/green fluorescent protein (GFP) expression vector (Life Technologies) was used as a control vector to examine transduction efficiency. The pLenti3.3/TetR vector contains a neomycin resistance gene and the pLenti6.3/TO/V5/Amelx and Plenti6.3/V5-GW/GFP vectors contain a blasticidin resistance gene for stable selection in mammalian cells. Three micrograms of Virapower Packaging Mix, 1 μg of the expression vector, and 12 μl of Lipofectamine 2000 (Life Technologies) were mixed in 1 ml of OPTI-MEM I (Life Technologies). After 25 minutes of incubation, the mixture was added to the 293FT cells. After 48 hours, the virus-containing supernatant was collected and filtrated with a 0.45-μm cellulose acetate filter.
Lentiviral-mediated transduction of TetR and Amelx
MSCs were cultured in 6-cm dishes in the growth medium. When the cells reached 80% confluence, the medium was replaced with the lentiviral stock solution containing pLenti3.3/TetR, and the cells were cultured overnight at 37°C under 5% CO2 (Fig 1a). Then, the lentivirus-containing medium was replaced with fresh growth medium. After 3 days, the cells were treated with geneticin (500 μg/ml) (Life Technologies). After 5 days, the surviving cell colonies were picked up to generate MSC clones (MSC-TetR) that strongly expressed the TetR gene. Expression of TetR in MSCs-TetR was examined by RT-PCR analysis.
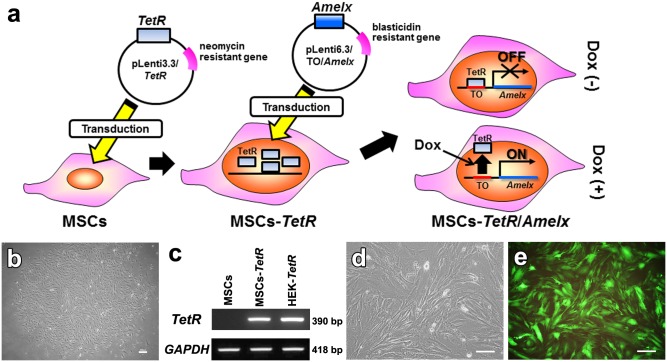
Fig 1. Establishment of a tetracycline (Tet)-controlled Amelx expression system in MSCs.
(a): Schematic diagram depicting the procedure to establish a Tet-controlled Amelx expression system in MSCs. TetR: Tet repressor, TO: Tet operator, Dox: doxycycline (tetracycline derivative). (b): MSC colony (MSC-TetR) in culture medium containing 500 μg/mL geneticin 10 days after transduction with the pLenti3.3/TetR expression vector (bar; 60 μm). (c): Expression of TetR repressor gene in MSCs (without transduction) and MSCs-TetR was determined by RT-PCR. HEK293 cells subjected to the same transduction procedure (HEK-TetR) were used as a positive control. (d, e): MSCs-TetR were lentivirally transduced with the expression vector pLenti6.3/TO/V5/Amelx or plenti6.3/V5-GW/GFP. MSCs-TetR/Amelx (d) and MSCs-TetR/GFP (e) were selected by 10 μg/mL blastcidin S (bar; 200 μm).
MSCs-TetR were seeded at a density of 3×105 cells in a 6-cm dish in the growth medium and incubated overnight at 37°C under 5% CO2. Then, the medium was replaced with the viral stock solution of pLenti6.3/TO/V5/Amelx or plenti6.3/V5-GW/GFP supplemented with 4 μg/ml polybrene (Nacalai Tesque). After 24 hours, the cells were washed once with phosphate-buffered saline (PBS) and cultured in fresh growth medium. After 5 days, the cells were treated with 10 μg/ml blasticidin S (Funakoshi, Tokyo, Japan) to select colonies of MSCs-TetR expressing Amelx (MSCs-TetR/Amelx) or GFP (MSCs-TetR/GFP). Tet-dependent expression of the Amelx gene in MSCs-TetR/Amelx in the presence or absence of Dox (2 μg/mL) was evaluated by RT-PCR and western blotting.
Assessment of osteogenic differentiation of MSCs-TetR/Amelx
MSCs-TetR/Amelx were cultured in the growth or osteogenic induction medium in the presence or absence of Dox (2 μg/mL) for 7–35 days. Tet-dependent expression of Amelx, osteogenic markers [Runx2, osterix, osteocalcin, osteopontin (also known as secreted phosphoprotein 1; Spp1), BSP, and type I collagen], and odontoblastic markers [dentin matrix protein 1 (DMP1) and dentin sialophosphoprotein (DSPP)] was evaluated by semi-quantitative RT-PCR and quantitative real-time RT-PCR. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control.
ALP activity was analyzed by ALP staining. The cells in 24-well plates were washed with PBS and fixed in 10% buffered formalin phosphate for the ALP assay. Cells were stained by incubation with 1.8 mM Fast Red TR (Sigma) and 0.9 mM naphthol AS-MX phosphate (Sigma) in 120 mM Tris buffer (pH 8.4) for 30 min at 37°C [21].
Mineralized nodule formation was evaluated by von Kossa staining. After washing with PBS and fixation with 10% formalin in phosphate buffer, the cells in 24-well plates were placed in 5% AgNO3 and exposed to ultraviolet (UV) light for 20 minutes. Subsequently, the cells were washed with distilled water and treated with 5% Na2S2O3 for 5 minutes [22].
Calcium deposition was evaluated by Alizarin Red S staining. After washing with PBS and fixation with 10% formalin in phosphate buffer, the cells were incubated in 40 mM Alizarin Red S (Sigma) in 24-well plates for 20 minutes with gentle shaking. After washing with distilled water, the samples were scanned to obtain digital images. Next, 400 μl of 10% acetic acid was added to the samples, which were then incubated at room temperature for 30 minutes for quantitative analysis [23]. Briefly, the stained samples were collected using a cell scraper and transferred into a 1.5-ml microcentrifuge tube. After heating at 85°C for 10 minutes and cooling on ice for 5 minutes, the samples were centrifuged at 20,000 g for 15 minutes. The colored supernatant (250 μl) was collected into a new tube and 100 μl of 10% ammonia was added to each tube. The optical density of the supernatant sample was measured at 405 nm.
RT-PCR analysis
RT-PCR analysis was performed as previously described [24]. Total RNA was extracted using an RNeasy Mini Kit (Qiagen, Hilden, Germany). After DNase I treatment (Ambion, Austin, TX), cDNA was synthesized from 1 μg of total RNA using Super Script III reverse transcriptase (Life Technologies). The cDNA target was amplified by PCR using Taq DNA polymerase (Promega, Madison, WI) following the manufacturer’s recommendations. The primer pairs used are described in Table 1. PCR products were subjected to 1.5% agarose gel electrophoresis with ethidium bromide staining and visualized under UV light illumination.
Table 1. Primers used for RT-PCR analyses.
| Gene | Primers (Fw, forward; Rv, reverse) | Ann Temp a | Product size (bp) |
|---|---|---|---|
| TetR | Fw: 5’-CGCCTTAGCCATTGGATGT-3’ | 61°C | 390 |
| Rv: 5’-TCTGCACCTTGGTGATCAAA-3’ | |||
| Amelx | Fw: 5’-CAGCAACCAATGATGCCAGTTCCT-3’ | 59.6°C | 293 |
| Rv: 5’-ACTTCTTCCCGCTTGGTCTTGTCT-3’ | |||
| BSP | Fw: 5’-AAAGTGAAGGAAAGCGACGA-3’ | 58°C | 214 |
| Rv: 5’-GTTCCTTCTGCACCTGCTTC-3’ | |||
| Runx2 | Fw: 5’-CCGCACGACAACCGCACCAT-3’ | 65°C | 289 |
| Rv: 5’-CGCTCCGGCCCACAAATCTC-3’ | |||
| osterix | Fw: 5’-CTTAACCCAGCTCCCTACCC-3’ | 59°C | 270 |
| Rv: 5’-TGTGAATGGGCTTCTTCCTC-3’ | |||
| osteocalcin | Fw: 5’-AAGCAGGAGGGCAATAAGGT-3’ | 60°C | 292 |
| Rv: 5’-AGCTGCTGTGACATCCATAC-3’ | |||
| osteopontin | Fw: 5’-TCACCATTCGGATGAGTCTG-3’ | 55°C | 437 |
| Rv: 5’-ACTTGTGGCTCTGATGTTCC-3’ | |||
| GAPDH | Fw: 5’-CACCATGGAGAAGGCCGGGG-3’ | 67°C | 418 |
| Rv: 5’-GACGGACACATTGGGGGTAG-3’ |
aAnn Temp: Annealing temperature
For quantitative real-time PCR analysis, SYBR Green and TaqMan assays were performed using Thunderbird SYBR qPCR Mix (Toyobo, Osaka, Japan) and TaqMan Gene Expression PCR Master Mix (Applied Biosystems, Foster City, USA), respectively, on an Applied Biosystems 7300 real-time PCR system (Applied Biosystems). The primer pairs used are shown in Table 2. Target gene expression was quantitatively analyzed using the ΔΔCt method [25].
Table 2. Primers used for quantitative real-time RT-PCR analyses.
| Gene | Primers (Fw, forward; Rv, reverse) | Ann Temp a | Product size (bp) |
|---|---|---|---|
| osterix | Fw: 5’-CTCGTCTGACTGCCTGCCTAG-3’ | 60°C | 84 |
| Rv: 5’-GCGTGGATGCCTGCCTTGTA-3’ | |||
| Collagen 1a1 | Fw: 5’-TGTCCCAACCCCCAAAGAC-3’ | 60°C | 92 |
| Rv: 5’-CCCTCGACTCCTACATCTTCTGA-3’ | |||
| osteocalcin | Fw: 5’-CCGGGAGCAGTGTGAGCTTA-3’ | 60°C | 68 |
| Rv: 5’-AGGCGGTCTTCAAGCCATACT-3’ | |||
| GAPDH | Fw: 5’-TGCACCACCAACTGCTTAG-3’ | 60°C | 177 |
| Rv: 5’-GGATGCAGGGATGATGTTC-3’ | |||
| Runx2 | Mm00501578_m1* | ||
| BSP | Mm00492555_m1* | ||
| osteopontin | Mm00436767_m1* | ||
| Amelx | Mm01166221_m1* | ||
| DMP1 | Mm01208363_m1* | ||
| DSPP | Mm00515666_m1* | ||
| GAPDH | NM_008084/ Mm99999915_g1* |
aAnn Temp: Annealing temperature
*PCR primers and a TaqMan probe (Applied Biosystem) for the TaqMan assay
Western blotting analysis
Cell pellets were collected when the cells reached 90% confluence, and the cells were lysed with RIPA buffer [50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Nonidet P-40 (NP-40), 1% sodium deoxycholate, and 0.1% SDS] supplemented with protease inhibitor cocktail (Nacalai Tesque). Proteins from the cell lysates were separated by 10% SDS-polyacrylamide gel electrophoresis and transferred to a polyvinylidine difluoride membrane (Millipore). The blots were blocked with TBST (10 mM Tris-HCl, pH 7.4, 100 mM NaCl, and 0.1% Tween-20) containing 5% skim milk, and then incubated with primary monoclonal antibodies against amelogenin (1:1000; clone F11, Santa Cruz Biotechnology, Santa Cruz, CA) or GAPDH (1:10000, clone 6C5, Millipore) at 4°C overnight. After washing with TBST, the membrane was incubated with anti-mouse IgG HRP-linked antibody (1:3000, Cell Signaling) for 1 hour at room temperature. Signals were detected using Immobilon Western Chemiluminescent HRP Substrate (Millipore) and the FPM 100 imaging system (Fujifilm, Tokyo, Japan).
Statistical analyses
One-way analysis of variance (ANOVA) with Tukey’s post hoc test was used for comparison of more than 2 groups. A significant difference was defined when P < 0.05.
Results
Establishment of a Tet-controlled Amelx expression system in MSCs
Ten days after the transduction of MSCs with the pLenti3.3/TetR expression vector, few colonies were observed in the culture containing 500 μg/mL geneticin (Fig 1b). The colonies were picked up and clonal cultures of MSCs (MSCs-TetR) were established. RT-PCR showed that MSCs-TetR strongly expressed TetR mRNA at levels comparable to those in control TetR-expressing HEK293 cells subjected to the same transduction procedure (Fig 1c).
The pLenti6.3/TO/V5/Amelx expression vector was lentivirally introduced into the MSCs-TetR to generate a Tet-controlled Amelx expression system. The pLenti6.3/V5-GW/GFP expression vector was also separately introduced into MSCs-TetR to determine the percentage of transduced cells in the population of surviving cells after drug selection. Seven days after the transduction of MSCs-TetR with each expression vector and culture in Blasticidin S, the surviving colonies were collected and referred to as MSCs-TetR/Amelx (Fig 1d) or MSCs-TetR/GFP. The percentage of transduced cells in the populations of MSCs-TetR/Amelx was estimated as >90%, as determined by GFP expression in MSCs-TetR/GFP (Fig 1e).
Induction of Amelx expression in MSCs-TetR/Amelx by Dox treatment
When MSCs-TetR/Amelx were cultured in the presence of Dox, enhanced gene expression of Amelx was confirmed after 24 hours (Fig 2a). Western blotting analysis showed that Dox stimulated MSCs-TetR/Amelx to express amelogenin protein after 48 hours (Fig 2b).
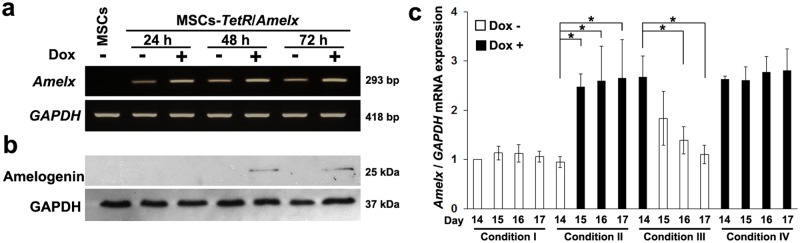
Fig 2. Induction of Amelx expression in MSCs-TetR/Amelx by Dox treatment.
MSCs-TetR/Amelx were cultured in the growth medium in the presence (+) or absence (-) of Dox for 24–72 hours. Inducible expression of the Amelx gene was detected by RT-PCR (a) and western blot (b) analyses. GAPDH was used as a loading control. (c) MSCs-TetR/Amelx were cultured in the osteogenic induction medium in the presence (+) (black bars) or absence (-) (white bars) of Dox for 17 days in four different conditions. (Condition I: day 0–17 Dox-; Condition II: day 0–14 Dox-, day 15–17 Dox+; Condition III: day 0–14 Dox+, day 15–17 Dox-; Condition IV: day 0–17 Dox+). Expression of Amelx was determined by quantitative real-time RT-PCR analysis. Significant differences (*P<0.01: ANOVA with Tukey’s multiple comparison test: n = 4) within each condition are shown.
MSCs-TetR/Amelx were cultured in osteogenic induction medium for 17 days in four different conditions, i.e., (I) in the absence of Dox for 17 days; (II) in the absence of Dox until day 14 and then in the presence of Dox from day 15 to 17; (III) in the presence of Dox until day 14 and then in the absence of Dox from day 15 to 17; and (IV) in the presence of Dox for 17 days. Quantitative real-time RT-PCR showed that expression of Amelx in cells from day 14 to 17 in Condition IV was markedly higher than that in Condition I (Fig 2c). In Condition II, expression of Amelx significantly increased on day 15 (one day after Dox addition). In contrast, in Condition III, expression of Amelx markedly decreased after day 15 (one day after Dox depletion), and significantly decreased after day 16.
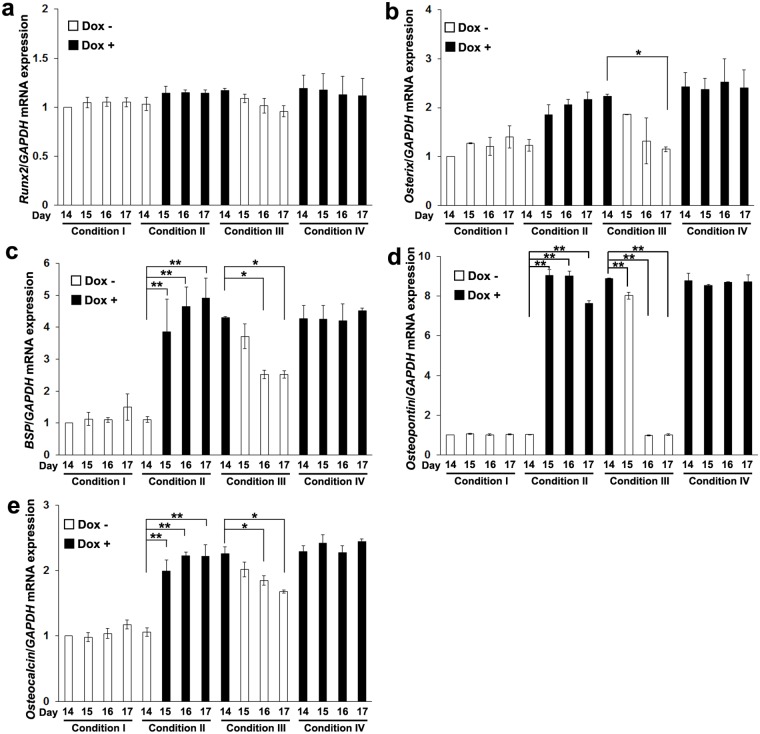
In these conditions, Runx2 expression was not significantly altered (Fig 3a). In contrast, expression of osterix was significantly decreased by the Dox depletion (Condition III) on day 17 (Fig 3b). Expression of BSP (Fig 3c), osteopontin (Fig 3d), and osteocalcin (Fig 3e) was significantly enhanced by addition of Dox (Condition II) and decreased by Dox depletion (Condition III).
Fig 3. Controllable expression of osterix, BSP and osteocalcin in MSCs-TetR/Amelx by Dox treatment.
MSCs-TetR/Amelx were cultured in the osteogenic induction medium in the presence (+) (black bars) or absence (-) (white bars) of Dox for 17 days in four different conditions. (Condition I: day 0–17 Dox-; Condition II: day 0–14 Dox-, day 15–17 Dox+; Condition III: day 0–14 Dox+, day 15–17 Dox-; Condition IV: day 0–17 Dox+). Expression of Runx2 (a), osterix (b), BSP (c), osteopontin (d) and osteocalcin (e) was determined by a quantitative real-time RT-PCR analysis. Significant differences (**P<0.01, *P<0.05: ANOVA with Tukey’s multiple comparison test: n = 3) within each condition are shown.
Effects of forced expression of Amelx in MSCs-tetR/Amelx on osteogenic differentiation
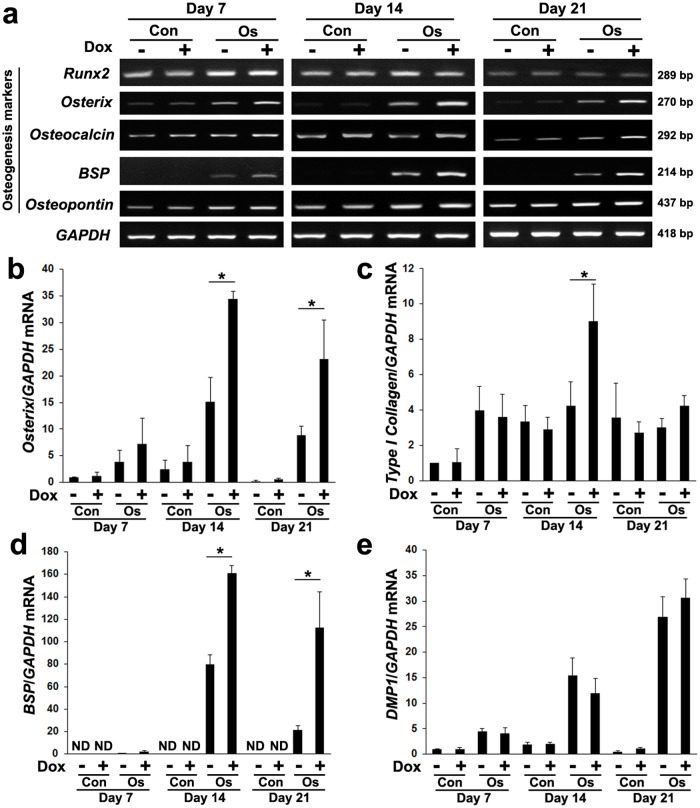
When MSCs-TetR/Amelx were cultured in osteogenic induction medium with Dox, enhanced expression of osterix, osteocalcin, and BSP genes was observed (Fig 4a). In contrast, expression of Runx2 was not significantly altered by the addition of Dox. Quantitative real-time RT-PCR demonstrated Dox-induced expression of osterix (Fig 4b), type I collagen (Fig 4c), and BSP (Fig 4d) genes after 14 days of osteogenic induction. Dox did not significantly alter the expression of DMP1 in MSCs-TetR/Amelx in the growth medium or in the osteogenic induction medium (Fig 4e). Expression of DSPP was not detected in MSCs-TetR/Amelx under any culture condition.
Fig 4. Effects of forced expression of Amelx in MSCs-tetR/Amelx on expression of osteogenic marker genes.
MSCs-TetR/Amelx were cultured in growth medium (Con) or osteogenic induction medium (Os) in the presence (+) or absence (-) of Dox for 21 days. (a) The expression of osteogenic marker genes (Runx2, osterix, osteocalcin, BSP, and osteopontin) was examined by RT-PCR analysis. GAPDH was used as an internal control. Quantitative real-time RT-PCR analysis was performed to examine expression of osterix (b), type I collagen (c), BSP (d), and DMP1 (e) genes. GAPDH was used as an internal control. Significant differences (*P<0.01: ANOVA with Tukey’s multiple comparison test: n = 3) are shown.
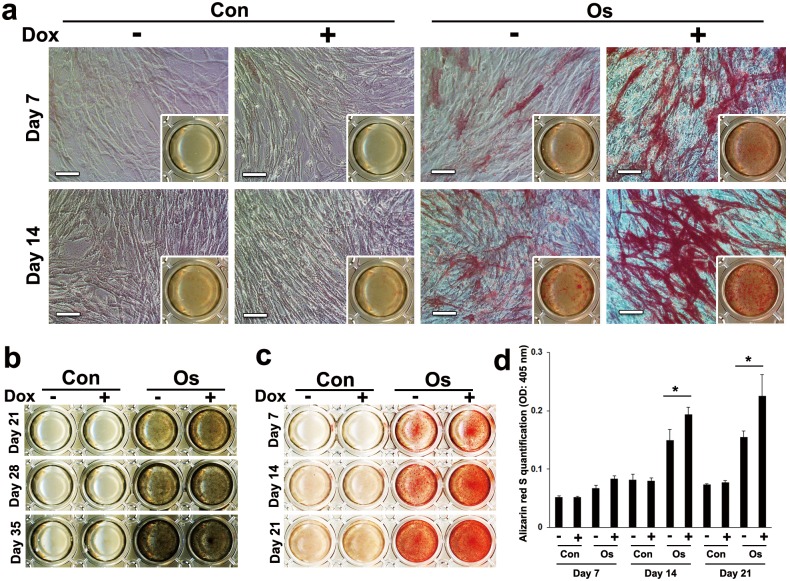
Dox induced ALP activity (Fig 5a) and mineralized nodule formation (Fig 5b) of MSCs-TetR/Amelx in osteogenic induction medium after 7 and 21 days, respectively. Alizarin Red S staining also supported the enhancement of MSC-TetR/Amelx matrix calcification by Dox treatment (Fig 5c). Quantitative analysis of Alizarin Red S staining showed that Dox treatment significantly induced calcium deposition of MSCs-TetR/Amelx after 14 days in the osteogenic induction medium (Fig 5d).
Fig 5. Effects of forced expression of Amelx on ALP activity and mineralized nodule formation.
MSCs-TetR/Amelx at 3 passages were cultured in the growth medium (Con) or osteogenic induction medium (Os) in the presence (+) or absence (-) of Dox in 24-well plates. (a) ALP activity on day 7 and 14 was examined by ALP staining (bars: 100 μm). (b) Mineralized nodule formation on day 21, 28 and 35 was detected by von Kossa staining. (c, d): Calcium deposition was determined by Alizarin Red S staining on day 7, 14 and 21 (c) and the staining intensity was quantitatively analyzed (d). Significant differences (*P<0.01: ANOVA with Tukey’s multiple comparison test: n = 9) are shown.
Effects of forced expression of Amelx on matrix calcification at different osteogenic differentiation stages
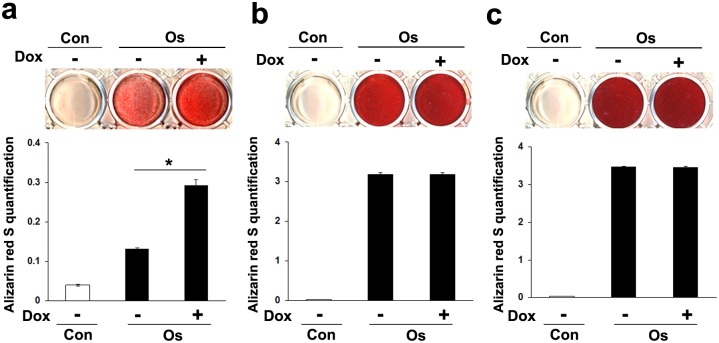
Alizarin Red S staining showed that addition of Dox at the early stage of osteogenic differentiation significantly induced matrix calcification of MSCs-TetR/Amelx on day 10 (Fig 6a). When Dox was applied to MSCs-TetR/Amelx on day 10–20 (intermediate stage) or day 20–30 (late stage) of osteogenic differentiation, matrix calcification of the MSCs-TetR/Amelx was not significantly altered (Fig 6b and 6c).
Fig 6. Effects of forced expression of Amelx at different osteogenic differentiation stages on matrix calcification of MSCs.
MSCs-TetR/Amelx at 8 passages were cultured in the growth medium (Con) or osteogenic induction medium (Os). Dox was applied to MSCs-TetR/Amelx on day 0–10 (a: early stage), day 10–20 (b: intermediate stage), or day 20–30 (c: late stage) of osteogenic differentiation, and Alizarin Red S staining was performed at day 10, day 20, or day 30, respectively. The staining intensity was also quantitatively analyzed. Significant differences (*P<0.01: ANOVA with Tukey’s multiple comparison test: n = 9) are shown.
Discussion
The amelogenin gene sequence is present in both the human X and Y chromosomes, whereas in the mouse it is found exclusively in the X chromosome [26]. Therefore, production of amelogenin protein in mice is encoded by the Amelx (amelogenin, X-linked) gene. In this study, we successfully established a Tet-controlled Amelx gene regulation system for mouse MSCs in which transcriptional activation of Amelx was associated with enhanced osteogenic differentiation.
Other than the Tet-dependent transcriptional regulatory system, recombinant adenoviruses that express site-specific recombinases such as Cre and FLP are widely used to provide a molecular switch to turn on/off transgene expression in cultured mammalian cells [27]. Although this system is attractive in that it shows little or no leakage of the target gene, control (on/off) of the target gene is accomplished by “transient” expression of the adenoviral recombinase; therefore, the duration of period in which gene expression can be controlled could be limited. In addition, recombinant adenovirus systems require transduction of adenovirus recombinase at each on/off regulation step, which is expected to be labor-intensive and time-consuming. In contrast, the Tet-dependent regulatory system has the advantage of maintaining the on/off regulation for a desired period of time, without limitation, simply by addition or removal of Dox.
It is known that primary mBMSC populations contain high proportions of non-MSCs and hematopoietic cells [28,29], which may lead to uneven and unpredictable behavior of individual cells during establishment of the Tet-dependent regulatory system. Therefore, in this study we used a clonal and immortalized population of MSCs (mBMSC-4 line) [20]. This cell line is multipotent, as demonstrated by its ability to differentiate specifically into osteoblast [30], chondrocyte [31], adipocyte, and myoblast [20] lineages and also to form ectopic bone in vivo [32].
According to the manufacturer’s information, the TetR expression vector and TO/target gene expression vector can be introduced into cells at the same time; however, in the present study we first established TetR-expressing MSCs, followed by transduction of the pLenti6.3/TO/V5/Amelx expression vector. The advantage of our method is that the established MSCs-TetR can be used for Tet regulation of not only Amelx but also any other target gene in future experiments. The expression vectors for such genes can be easily prepared because our system utilizes the Gateway cloning system as a template.
The established MSCs-TetR/Amelx enhanced expression of the Amelx gene and amelogenin protein in the presence of Dox. Although slight leakage of Amelx gene expression without Dox induction was detected by RT-PCR, there was no background expression of amelogenin protein when the cells were cultured in the absence of Dox. Many studies have indicated that leakage of transgene transcription in Tet-dependent regulatory systems is unavoidable and acceptable in most experimental cases [33,34]. Because MSCs-TetR and MSCs-TetR/Amelx were established from single colonies during the drug selection process, MSCs-TetR/Amelx appeared to be homogeneous. Indeed, the enhanced Amelx-induced calcification in MSCs-TetR/Amelx at early passages (Fig 5c and 5d) was reproducible in those cells after repeated passaging (Fig 6a), indicating that MSCs-TetR/Amelx maintained their homogeneity with osteogenic activity even after repeated passaging.
In this study, we focused on the effects of forced expression of Amelx on MSCs that were undergoing osteogenic differentiation. Hu et al. [16] evaluated the genome-wide expression profile of human MSCs without induced differentiation after lentiviral overexpression of amelogenin. In their approach, permanent expression of amelogenin should have been initiated immediately after lentiviral transduction, and undesired effects of the uncontrolled exogenous amelogenin expression may have occurred during drug selection. In this regard, the controllable expression system established in our study has the advantage of controlled initiation or cessation of amelogenin expression at desired time points, which we specifically utilized in this study of the time-dependent osteogenic differentiation of MSCs. Expression of Amelx in MSCs-TetR/Amelx was obviously enhanced or reduced by addition or removal of Dox during osteogenic differentiation. These results indicate that we successfully established a Tet-controlled Amelx gene regulation system for MSCs, in which the expression of Amelx could be regulated by Dox addition and removal even during the differentiation process.
Amelogenin has cell signaling properties [35–38]. We thus examined whether the controllable expression of Amelx would concomitantly affect the expression of these osteogenic genes. Interestingly, expression of osterix, BSP, osteopontin and osteocalcin was altered in parallel with the controlled expression of Amelx. In particular, BSP and osteopontin were extensively up-regulated by Amelx expression, implying that these phosphorylated sialoglycoproteins may be preferential target molecules for Amelx signaling during osteogenic differentiation of MSCs. Shimizu et al. demonstrated that amelogenin stimulates BSP expression in osteoblasts through the fibroblast growth factor 2 (FGF2) response element and transforming growth factor-β1 (TGF-β1) activation element in the promoter of the BSP gene [36]. Amelogenin promotes osteogenic differentiation of MSCs through the Wnt/beta-catenin signaling pathway by up-regulating Wnt10b [37]. Olivares-Navarrete et al. recently reported that both amelogenin and N-terminal amelogenin peptide (NTAP) induced osteogenic differentiation of human MSCs, and that the effects of the NTAP were mediated through PKC and ERK1/2 activation and β-catenin degradation [38]. In this study, the concomitant expression of osterix, BSP, osteopontin, and osteocalcin with the controlled expression of Amelx implies that the exogenous Amelx expression affects the transcriptional activation of these genes. In contrast, Runx2 expression was not significantly affected by the controlled expression of Amelx. Although Runx2 is a crucial transcriptional factor for osteoblast differentiation, osterix exerts its osteogenic function via Runx2-independent mechanisms [39–41]. The controlled expression of Amelx in our study may therefore have regulated osterix via Runx2-independent pathways, which in turn promoted expression of other osteogenic genes. However, our work appears to contradict recently published studies showing that recombinant amelogenin and its peptides promote expression of Runx2 mRNA in MSCs [37,38]. This discrepancy may have resulted from differences in the application of amelogenin, i.e., extracellular administration of amelogenin proteins/peptides vs. intracellular expression of Amelx mRNA. Further studies are necessary to elucidate these signaling pathways.
Osteogenic differentiation of MSCs is a well-orchestrated process that begins with activation of transcription factors including Runx2 and osterix [42]. In the late stage of the osteoblast developmental sequence, BSP, osteopontin, and osteocalcin serve as regulators of the mineralization process [43]. Many studies have shown that enamel matrix derivative (EMD), the active component of Emdogain that contains heterogeneous growth factors including amelogenin, stimulates mineralizing cell types to increase their ALP activity and production of osteopontin, BSP, and osteocalcin [44]. However, previous reports on the effects of recombinant amelogenin on osteogenic differentiation have presented relatively contradictory results. Matsuzawa et al. reported that mouse recombinant amelogenin increased type I collagen and osteocalcin mRNA levels in an osteoblast cell line (ROS17/2.8 cells) [45]. Zeichner-David et al. reported that mouse recombinant amelogenin induced expression of BSP and osteocalcin but down-regulated type I collagen in mouse periodontal ligament cells. In mouse cementoblasts, recombinant amelogenin and tyrosine-rich amelogenin peptide were shown to down-regulate BSP and osteocalcin and inhibit mineral nodule formation [11,46]. Therefore, the effects of amelogenin on osteogenic differentiation may depend on the cell type and on whether full-length or domain-derived peptides are used.
Regarding the effects of amelogenin on MSCs, increased osteogenesis was previously reported after treatment with recombinant amelogenin [15,38] or NTAP [38]. However, no data are available yet for the effects of forced expression of Amelx on MSCs under osteogenic guidance, especially for matrix mineralization of MSCs. In this study, MSCs-TetR/Amelx treated with Dox during osteogenic induction showed increased expression of the osteogenic marker genes osterix, BSP, type I collagen and osteocalcin. The up-regulation of these osteogenic genes resulted from expression of exogenous Amelx and not from a direct effect of Dox because Dox treatment did not significantly alter osteogenic gene expression in non-transduced MSCs (data not shown). Osterix is a typical transcription factor required for osteoblast differentiation and bone formation [47]. Type I collagen is a primary product of osteoblasts, and its gene expression increases at the early to intermediate stages and decreases gradually thereafter during bone matrix formation [48]. Expression of BSP is detected in more extensively differentiated osteoblasts, i.e., at a relatively late stage of differentiation [49]. BSP is a non-collagenous protein component of mineralized tissues, such as cementum and bone, and is believed to be a critical molecule for promoting biomineralization [49,50]. Thus, given the roles of these molecules in osteogenesis, their up-regulation by Amelx transduction may have contributed to the observed enhancement of MSC osteogenic differentiation. However, forced expression of Amelx alone appears to not be sufficient to induce osteogenic differentiation because Dox did not significantly stimulate osteogenic marker gene expression (Fig 4b–4d) under the non-induction condition. In addition, forced expression of Amelx appears to not induce dentin marker expression in MSCs under osteogenic induction because expression of DMP1 and DPSS in MSCs-TetR/Amelx was not stimulated by the presence of Dox.
Concomitantly with the up-regulation of osteogenic genes by transcriptional activation of Amelx, the ALP activity of MSCs-TetR/Amelx also clearly increased, and Dox treatment enhanced the matrix calcification of MSCs-TetR/Amelx in the osteogenic induction medium. These results provide further evidence that transcriptional activation of Amelx enhances the osteogenic differentiation of MSCs. However, the 2.5- (osterix and type I collagen) and 5.5- (BSP) fold increase in the expression of these genes (Fig 4b–4d) may not be sufficient to aggressively impart a mature osteoblastic phenotype to MSCs. Indeed, the matrix calcification of the MSCs was already significantly increased on day 10 after Amelx transduction (Fig 6a), although expression of the osteogenic marker genes did not markedly increase until day 7 (Fig 4a–4d). Full-length amelogenin has the capacity to stabilize the formation of amorphous calcium phosphate (ACP) [51], and during bone biomineralization, an abundant ACP phase serves as a precursor phase that later transforms into mature crystalline calcium phosphate [52,53]. Deshpande et al. recently demonstrated that amelogenin interacts with collagen fibrils and mineral particles to mineralize the collagen fibrils [54]. Therefore, the increased ECM mineralization observed after Amelx transduction may involve direct effects of the expressed amelogenin protein on ECM mineralization in addition to the effects mediated through up-regulation of osteogenic genes. The direct regulation of calcium phosphate mineral formation by amelogenin would thus be expected to mainly contribute to the precursor phase of biomineralization, which would explain why forced expression of Amelx initiated in the intermediate and late stages of the osteogenic differentiation did not significantly contribute to MSC calcification (Fig 6b and 6c). Information from the present study and the continued study of Tet-dependent Amelx regulatory systems in MSCs may help to clarify the mechanisms by which amelogenin regulates key molecules associated with bone mineralization.
Conclusions
This study established a Tet-controlled Amelx gene regulation system for MSCs that could be a beneficial tool to investigate novel functions of amelogenin in MSC osteogenesis. The present data show that transcriptional activation of Amelx enhances osteogenic differentiation of MSCs in vitro by up-regulating osterix, BSP, osteocalcin and type I collagen. In particular, Amelx expression was suggested to affect the osteogenic differentiation of MSCs through cell signaling roles associated with expression of osterix, BSP, osteopontin, and osteocalcin. Forced Amelx expression was also suggested to enhance ECM mineralization through a direct effect of amelogenin on calcium phosphate mineral formation at the early stage of biomineralization. These findings represent an important step toward the optimal application of amelogenin therapy for periodontal/bone regeneration.
Data Availability
All relevant data are within the paper.
Funding Statement
This investigation was supported in part by a Grant-in Aid for the 2013-2015 Bilateral Joint Research Projects from the Japan Society for the Promotion of Science (http://www.jsps.go.jp/english/index.html) (HE) and the National Research Council of Thailand (http://www.nrct.go.th/) (PP) (no numbers for the grants).
References
- 1. Masaki C, Nakamoto T, Mukaibo T, Kondo Y, Hosokawa R (2015) Strategies for alveolar ridge reconstruction and preservation for implant therapy. J Prosthodont Res 59: 220–228. 10.1016/j.jpor.2015.04.005 [DOI] [PubMed] [Google Scholar]
- 2. Egusa H, Sonoyama W, Nishimura M, Atsuta I, Akiyama K (2012) Stem cells in dentistry—Part I: Stem cell sources. J Prosthodont Res 56: 151–165. 10.1016/j.jpor.2012.06.001 [DOI] [PubMed] [Google Scholar]
- 3. Kaku M, Akiba Y, Akiyama K, Akita D, Nishimura M (2015) Cell-based bone regeneration for alveolar ridge augmentation—Cell source, endogenous cell recruitment and immunomodulatory function. J Prosthodont Res 59: 96–112. 10.1016/j.jpor.2015.02.001 [DOI] [PubMed] [Google Scholar]
- 4. Egusa H, Sonoyama W, Nishimura M, Atsuta I, Akiyama K (2012) Stem cells in dentistry—Part II: Clinical applications. J Prosthodont Res 56: 229–248. 10.1016/j.jpor.2012.10.001 [DOI] [PubMed] [Google Scholar]
- 5. Termine JD, Belcourt AB, Christner PJ, Conn KM, Nylen MU (1980) Properties of dissociatively extracted fetal tooth matrix proteins. I. Principal molecular species in developing bovine enamel. J Biol Chem 255: 9760–9768. [PubMed] [Google Scholar]
- 6. Brookes SJ, Robinson C, Kirkham J, Bonass WA (1995) Biochemistry and molecular biology of amelogenin proteins of developing dental enamel. Arch Oral Biol 40: 1–14. [DOI] [PubMed] [Google Scholar]
- 7. Fincham AG, Moradian-Oldak J, Simmer JP, Sarte P, Lau EC, Diekwisch T, et al. (1994) Self-assembly of a recombinant amelogenin protein generates supramolecular structures. J Struct Biol 112: 103–109. [DOI] [PubMed] [Google Scholar]
- 8. Robinson C, Kirkham J, Weatherell JA, Richards A, Josephsen K, Fejerskov O (1988) Mineral and protein concentrations in enamel of the developing permanent porcine dentition. Caries Res 22: 321–326. [DOI] [PubMed] [Google Scholar]
- 9. Margolis HC, Beniash E, Fowler CE (2006) Role of macromolecular assembly of enamel matrix proteins in enamel formation. J Dent Res 85: 775–793. [DOI] [PubMed] [Google Scholar]
- 10. Veis A, Tompkins K, Alvares K, Wei KR, Wang L, Wang XS, et al. (2000) Specific amelogenin gene splice products have signaling effects on cells in culture and in implants in vivo. J Biol Chem 275: 41263–41272. [DOI] [PubMed] [Google Scholar]
- 11. Viswanathan HL, Berry JE, Foster BL, Gibson CW, Li Y, Kulkarni AB, et al. (2003) Amelogenin: A potential regulator of cementum-associated genes. J Periodontol 74: 1423–1431. [DOI] [PubMed] [Google Scholar]
- 12. Gibson CW (2008) The amelogenin "enamel proteins" and cells in the periodontium. Crit Rev Eukaryot Gene Expr 18: 345–360. [DOI] [PubMed] [Google Scholar]
- 13. Haze A, Taylor AL, Haegewald S, Leiser Y, Shay B, Rosenfeld E, et al. (2009) Regeneration of bone and periodontal ligament induced by recombinant amelogenin after periodontitis. J Cell Mol Med 13: 1110–1124. 10.1111/j.1582-4934.2009.00700.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Warotayanont R, Zhu DH, Snead ML, Zhou Y (2008) Leucine-rich amelogenin peptide induces osteogenesis in mouse embryonic stem cells. Biochem Biophys Res Commun 367: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tanimoto K, Huang YC, Tanne Y, Kunimatsu R, Michida M, Yoshioka M, et al. (2012) Amelogenin Enhances the Osteogenic Differentiation of Mesenchymal Stem Cells Derived from Bone Marrow. Cells Tissues Organs 196: 411–419. 10.1159/000335912 [DOI] [PubMed] [Google Scholar]
- 16. Hu JC, Shu R, Song ZC, Chen L (2011) Human amelogenin up-regulates osteogenic gene expression in human bone marrow stroma cells. Biochem Biophys Res Commun 408: 437–441. 10.1016/j.bbrc.2011.04.042 [DOI] [PubMed] [Google Scholar]
- 17. Naidoo J, Young D (2012) Gene regulation systems for gene therapy applications in the central nervous system. Neurol Res Int 2012: 595410 10.1155/2012/595410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kafri T, van Praag H, Gage FH, Verma IM (2000) Lentiviral vectors: regulated gene expression. Mol Ther 1: 516–521. [DOI] [PubMed] [Google Scholar]
- 19. Zhou BY, Ye Z, Chen G, Gao ZP, Zhang YA, Cheng L (2007) Inducible and reversible transgene expression in human stem cells after efficient and stable gene transfer. Stem cells 25: 779–789. [DOI] [PubMed] [Google Scholar]
- 20. Egusa H, Kobayashi M, Matsumoto T, Sasaki J, Uraguchi S, Yatani H (2013) Application of cyclic strain for accelerated skeletal myogenic differentiation of mouse bone marrow-derived mesenchymal stromal cells with cell alignment. Tissue Eng Part A 19: 770–782. 10.1089/ten.TEA.2012.0164 [DOI] [PubMed] [Google Scholar]
- 21. Egusa H, Schweizer FE, Wang CC, Matsuka Y, Nishimura I (2005) Neuronal differentiation of bone marrow-derived stromal stem cells involves suppression of discordant phenotypes through gene silencing. J Biol Chem 280: 23691–23697. [DOI] [PubMed] [Google Scholar]
- 22. Egusa H, Iida K, Kobayashi M, Lin TY, Zhu M, Zuk PA, et al. (2007) Downregulation of extracellular matrix-related gene clusters during osteogenic differentiation of human bone marrow and adipose tissue-derived stromal cells. Tissue Eng 13: 2589–2600. [DOI] [PubMed] [Google Scholar]
- 23. Gregory CA, Gunn WG, Peister A, Prockop DJ (2004) An Alizarin red-based assay of mineralization by adherent cells in culture: comparison with cetylpyridinium chloride extraction. Anal Biochem 329: 77–84. [DOI] [PubMed] [Google Scholar]
- 24. Egusa H, Doi M, Saeki M, Fukuyasu S, Akashi Y, Yokota Y, et al. (2011) The small molecule harmine regulates NFATc1 and Id2 expression in osteoclast progenitor cells. Bone 49: 264–274. 10.1016/j.bone.2011.04.003 [DOI] [PubMed] [Google Scholar]
- 25. Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat protoc 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- 26. Lau EC, Mohandas TK, Shapiro LJ, Slavkin HC, Snead ML (1989) Human and mouse amelogenin gene loci are on the sex chromosomes. Genomics 4: 162–168. [DOI] [PubMed] [Google Scholar]
- 27. Nakano M, Odaka K, Ishimura M, Kondo S, Tachikawa N, Chiba J, et al. (2001) Efficient gene activation in cultured mammalian cells mediated by FLP recombinase-expressing recombinant adenovirus. Nucleic Acids Res 29: E40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Soleimani M, Nadri S (2009) A protocol for isolation and culture of mesenchymal stem cells from mouse bone marrow. Nat Protoc 4: 102–106. 10.1038/nprot.2008.221 [DOI] [PubMed] [Google Scholar]
- 29. Sun S, Guo Z, Xiao X, Liu B, Liu X, Tang PH, et al. (2003) Isolation of mouse marrow mesenchymal progenitors by a novel and reliable method. Stem cells 21: 527–535. [DOI] [PubMed] [Google Scholar]
- 30. Egusa H, Kayashima H, Miura J, Uraguchi S, Wang F, Okawa H, et al. (2014) Comparative analysis of mouse-induced pluripotent stem cells and mesenchymal stem cells during osteogenic differentiation in vitro. Stem Cells Dev 23: 2156–2169. 10.1089/scd.2013.0344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sasaki J, Matsumoto T, Egusa H, Matsusaki M, Nishiguchi A, Nakano T, et al. (2012) In vitro reproduction of endochondral ossification using a 3D mesenchymal stem cell construct. Integr Biol 4: 1207–1214. [DOI] [PubMed] [Google Scholar]
- 32. Sasaki J, Matsumoto T, Imazato S (2015) Oriented bone formation using biomimetic fibrin hydrogels with three-dimensional patterned bone matrices. J Biomed Mater Res A 103: 622–627. 10.1002/jbm.a.35212 [DOI] [PubMed] [Google Scholar]
- 33. Sun Y, Chen XG, Xiao D (2007) Tetracycline-inducible expression systems: New strategies and practices in the transgenic mouse modeling. Acta Biochimica et Biophysica Sinica 39: 235–246. [DOI] [PubMed] [Google Scholar]
- 34. Piper SL, Wang MQ, Yamamoto A, Malek F, Luu A, Kuo AC, et al. (2012) Inducible immortality in hTERT-human mesenchymal stem cells. J Orthop Res 30: 1879–1885. 10.1002/jor.22162 [DOI] [PubMed] [Google Scholar]
- 35. Boabaid F, Gibson CW, Kuehl MA, Berry JE, Snead ML, Nociti FH, et al. (2004) Leucine-rich amelogenin peptide: A candidate signaling molecule during cementogenesis. J Periodontol 75: 1126–1136. [DOI] [PubMed] [Google Scholar]
- 36. Shimizu E, Saito R, Nakayama Y, Nakajima Y, Kato N, Takai H, et al. (2005) Amelogenin stimulates bone sialoprotein (BSP) expression through fibroblast growth factor 2 response element and transforming growth factor-beta 1 activation element in the promoter of the BSP gene. J Periodontol 76: 1482–1489. [DOI] [PubMed] [Google Scholar]
- 37. Wen X, Cawthorn WP, MacDougald OA, Stupp SI, Snead ML, Zhou Y (2011) The influence of Leucine-rich amelogenin peptide on MSC fate by inducing Wnt10b expression. Biomaterials 32: 6478–6486. 10.1016/j.biomaterials.2011.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Olivares-Navarrete R, Vesper K, Hyzy SL, Almaguer-Flores A, Boyan BD, Schwartz Z (2014) Role of the N-terminal peptide of amelogenin on osteoblastic differentiation of human mesenchymal stem cells. Eur Cell Mater 28: 1–10. [DOI] [PubMed] [Google Scholar]
- 39. Lee MH, Kwon TG, Park HS, Wozney JM, Ryoo HM (2003) BMP-2-induced Osterix expression is mediated by Dlx5 but is independent of Runx2. Biochem Biophys Res Commun 309: 689–694. [DOI] [PubMed] [Google Scholar]
- 40. Matsubara T, Kida K, Yamaguchi A, Hata K, Ichida F, Meguro H, et al. (2008) BMP2 regulates Osterix through Msx2 and Runx2 during osteoblast differentiation. J Biol Chem 283: 29119–29125. 10.1074/jbc.M801774200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee DS, Choung HW, Kim HJ, Gronostajski RM, Yang YI, Ryoo HM, et al. (2014) NFI-C regulates osteoblast differentiation via control of osterix expression. Stem cells 32: 2467–2479. 10.1002/stem.1733 [DOI] [PubMed] [Google Scholar]
- 42. Komori T (2010) Regulation of osteoblast differentiation by Runx2. Adv Exp Med Biol 658: 43–49. 10.1007/978-1-4419-1050-9_5 [DOI] [PubMed] [Google Scholar]
- 43. Bonucci E (2012) Bone mineralization. Front Biosci 17: 100–128. [DOI] [PubMed] [Google Scholar]
- 44. Grandin HM, Gemperli AC, Dard M (2012) Enamel matrix derivative: a review of cellular effects in vitro and a model of molecular arrangement and functioning. Tissue Eng Part B Rev 18: 181–202. 10.1089/ten.TEB.2011.0365 [DOI] [PubMed] [Google Scholar]
- 45. Matsuzawa M, Sheu TJ, Lee YJ, Chen M, Li TF, Huang CT, et al. (2009) Putative signaling action of amelogenin utilizes the Wnt/beta-catenin pathway. J Periodontal Res 44: 289–296. [DOI] [PubMed] [Google Scholar]
- 46. Swanson EC, Fong HK, Foster BL, Paine ML, Gibson CW, Snead ML, et al. (2006) Amelogenins regulate expression of genes associated with cementoblasts in vitro. Eur J Oral Sci 114 Suppl 1: 239–243. [DOI] [PubMed] [Google Scholar]
- 47. Nakashima K, Zhou X, Kunkel G, Zhang ZP, Deng JM, Behringer RR, et al. (2002) The novel zinc finger-containing transcription factor Osterix is required for osteoblast differentiation and bone formation. Cell 108: 17–29. [DOI] [PubMed] [Google Scholar]
- 48. Kaku M, Yamauchi M (2014) Mechano-regulation of collagen biosynthesis in periodontal ligament. J Prosthodont Res 58: 193–207. 10.1016/j.jpor.2014.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ganss B, Kim RH, Sodek J (1999) Bone sialoprotein. Crit Rev Oral Biol Med 10: 79–98. [DOI] [PubMed] [Google Scholar]
- 50. Bianco P, Riminucci M, Silvestrini G, Bonucci E, Termine JD, Fisher LW, et al. (1993) Localization of Bone Sialoprotein (Bsp) to Golgi and Post-Golgi Secretory Structures in Osteoblasts and to Discrete Sites in Early Bone-Matrix. J Histochem Cytochem 41: 193–203. [DOI] [PubMed] [Google Scholar]
- 51. Margolis HC, Beniash E (2010) The role of amelogenin in dental enamel formation: A universal strategy for protein-mediated biomineralization In: Goldberg M, editor. Amelogenins: Multifaceted Proteins for Dental & Bone Formation & Repair. Sharjah, United Arab Emirates: Bentham e Books; pp. 133–142. [Google Scholar]
- 52. Crane NJ, Popescu V, Morris MD, Steenhuis P, Ignelzi MA Jr (2006) Raman spectroscopic evidence for octacalcium phosphate and other transient mineral species deposited during intramembranous mineralization. Bone 39: 434–442. [DOI] [PubMed] [Google Scholar]
- 53. Mahamid J, Sharir A, Addadi L, Weiner S (2008) Amorphous calcium phosphate is a major component of the forming fin bones of zebrafish: Indications for an amorphous precursor phase. Proc Natl Acad Sci U S A 105: 12748–12753. 10.1073/pnas.0803354105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Deshpande AS, Fang PA, Simmer JP, Margolis HC, Beniash E (2010) Amelogenin-collagen interactions regulate calcium phosphate mineralization in vitro. J Biol Chem 285: 19277–19287. 10.1074/jbc.M109.079939 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.