Abstract
Contamination of platelet units by bacteria has long been acknowledged as a significant transfusion risk due to their post-donation storage conditions. Products are routinely stored at 22 °C on an agitating shaker, a condition that can promote bacterial growth. Although the total number of bacteria believed to be introduced into a platelet product is extremely low, these bacteria can multiply to a very high titer prior to transfusion, potentially resulting in serious adverse events. The aim of this study was to evaluate a riboflavin based pathogen reduction process against a panel of bacteria that have been identified as common contaminants of platelet products. This panel included the following organisms: S. epidermidis, S. aureus, S. mitis, S. pyogenes, S. marcescens, Y. enterocolitica, B. neotomae, B. cereus, E. coli, P. aeruginosa and K. pneumoniae. Each platelet unit was inoculated with a high bacterial load and samples were removed both before and after treatment. A colony forming assay, using an end point dilution scheme, was used to determine the pre-treatment and post-treatment bacterial titers. Log reduction was calculated by subtracting the post-treatment titer from the pre-treatment titer. The following log reductions were observed: S. epidermidis 4.7 log (99.998%), S. aureus 4.8 log (99.998%), S. mitis 3.7 log (99.98%), S. pyogenes 2.6 log (99.7%), S. marcescens 4.0 log (99.99%), Y. enterocolitica 3.3 log (99.95%), B. neotomae 5.4 log (99.9996%), B. cereus 2.6 log (99.7%), E. coli ≥5.4 log (99.9996%), P. aeruginosa 4.7 log (99.998%) and K. pneumoniae 2.8 log (99.8%). The results from this study suggest the process could help to lower the risk of severe adverse transfusion events associated with bacterial contamination.
Keywords: Immunology, Issue 102, Mirasol, bacteria, platelet, riboflavin, ultra-violet light, pathogen reduction technology
Introduction
Transfusion of platelets, plasma, packed RBCs and whole blood products play a major role in modern medicine and can be used to treat various medical conditions, replace vital fluids and ultimately save lives. Platelets are a cellular product that are either isolated from whole blood and pooled into a transfusable dose or are collected through the process of platelet apheresis. The main role of platelets in the body is to stop bleeding at wound sites and to help maintain hemostasis. Patients suffering from a low platelet cell count (thrombocytopenia) are susceptible to spontaneous bleeding events and are transfused with platelets to bring their platelet cell count back into a normal range. Collected platelets are stored for a maximum of 5-7 days and are stored at 22 ± 2 °C while under constant agitation.
Despite the life saving properties of platelet transfusions there remains a slight risk to transfusion recipients due to contamination of these products by parasites, bacteria and viruses11. The implementation of viral nucleic acid testing (NAT) has significantly decreased the risk of viral transmission of major blood borne agents, such as hepatitis C virus (HCV), hepatitis B virus (HBV) and human immuno-deficiency virus (HIV)5. A recent publication from the Canadian Blood Services estimates the residual risk for these agents to be 1 per 8 million donations for HIV, 1 per 6.7 million donations for HCV and 1 per 1.7 million donations for HBV15.
Although bacteria typically garner less attention in the general public, the frequency of bacterial contamination in platelet products has been estimated to be as high as 1:1,0007 and because millions of platelet products are transfused each year many recipients are exposed to potentially life threatening complications like sepsis6. Research suggests that the bacterial load at the time of contamination is low, <100 colony forming units (CFU)/product2,16, however the nutrient rich environment and room temperature storage allows contaminating bacteria to proliferate to dangerously high titers prior to transfusion. Currently, the only approved methods available to prevent bacterially contaminated products from reaching a platelet recipient is through the use of culture based systems and rapid point of care testing. Briefly, for culture based systems platelet products are stored on an agitator at 22 °C for 12-24 hr to allow bacteria to proliferate in the product, upon which a 4-8 ml sample is removed from the platelet product and inoculated into a bottle containing nutrient media. The bottle is placed into an instrument which monitors the bottle for bacterial growth. If the instrument detects bacterial growth in the bottle it is flagged and the corresponding platelet unit is discarded. While this process is reasonably successful at detecting fast growing organisms, many of the slow growing species do not grow to a high enough titer to be detected, thus creating the potential for false-negative units to be released for transfusion7,12,14,16,22. Unlike culture based detection systems, rapid point of care testing is typically performed later in the platelet storage period when the bacterial load has increased significantly. The higher titers are required since point of care tests are less sensitive than the culture based systems, and only reliably detect bacteria once they reach a titer ≥1 x 103 CFU/ml17. However such tests can provide results within 1 hour of sampling. Variability in the performance of these tests have led to the release of a false negative product, causing a fatal septic reaction in the recipient9.
An alternative way to combat the issue of bacterial contamination of platelet products is through the routine use of a pathogen reduction process that can inactivate contaminating bacteria instead of try to detect them. Using riboflavin as a photosensitizer, in combination with UV light, has been shown to reduce the infectivity of a broad range of pathogenic blood-borne contaminants, including bacteria3,4,8,10,19-21.The use of riboflavin and UV light for pathogen reduction is non-toxic and non-mutagenic, and riboflavin and UV light-treated components have been shown to be safe for transfusion recipients as well as for those handling blood products18. Briefly, riboflavin molecules can associate with the nucleic acids (DNA and RNA) of bacteria, parasites, viruses and any nucleated cell (e.g. white blood cells). Exposure to UV light activates riboflavin, causing a chemical alteration to functional groups of the nucleic acids (primarily guanine bases), thus preventing replication and/or transcription of the nucleic acids and leaving the organism inactivated13. Anucleated cells like platelets and red blood cells are not affected by the riboflavin chemistry due to the lack of nucleic acid.
Previous work with riboflavin and UV light technology evaluated a select group of bacteria using an experimental design intended to mimic a platelet product contaminated with a clinically relevant bacterial load (<20 CFU/product)8. The goal of this study was to evaluate the riboflavin and UV light process against high titer bacterial contamination (>1.0 x 105 CFU/ml) in platelet products treated in plasma in order measure the total bacterial reduction capacity of the system. Based on data collected from hemovigilance studies1, a panel of commonly occurring gram negative and gram positive organisms was selected for evaluation in this study and included the following species: Staphylococcus epidermidis, Staphylococcus aureus, Streptococcus mitis, Streptococcus pyogenes, Serratia marcescens, Yersinia enterocolitica, Brucella neotomae, Bacillus cereus (vegetative form), Esherichia coli, Pseudomonas aeruginosa and Klebsiella pneumoniae. All organisms, except B. cereus, were obtained from ATCC and a priority was placed on obtaining bacterial strains that had been identified as being isolated from blood components. The B. cereus tested in this study is a clinical strain isolated internally from a contaminated platelet product.
Protocol
1. Bacteria Suspension Preparation:
From a streaked culture plate, aseptically inoculate a single colony into a 250 ml sterile bottle containing 100 ml of the appropriate growth media (Table 1) by way of sterile inoculating loop.
Place the bottle into an orbital shaking incubator at the appropriate conditions (Table 1) for 1-2 days, checking for growth periodically. Set the shaker speed to 140 RPM.
Aseptically transfer the bacterial suspension into two sterile 50 ml conical tubes and centrifuge the tubes at 3,000 x g for 20 min at 23 °C.
Aspirate the supernatant and resuspend each bacterial pellet with 15 ml of sterile peptone water. Vortex as necessary. Combine the suspended bacteria culture into one tube/container.
Titer the stock culture in triplicate per section 4 to determine the stock culture titer.
Refrigerate for up to 2 weeks at 4 ± 2 °C until needed. Re-titer the culture at the time of use, see step 4.8.
2. Platelet Product Preparation:
- Collect a standard apheresis human platelet product at an accredited blood bank. The platelet product must meet the following collection specifications: 90–360 ml volume and 0.8 –3.8 x 106 platelets/µl concentration.
- Ensure that the platelet product has rested for a minimum of 2 hr prior to processing for pathogen reduction. Store the products at 22 ± 2 °C in a platelet incubator without agitation for these 2 hr. After two hours ensure the platelet product is agitated at 22 ± 2 °C in a platelet incubator. Use the product for up to 22 hr post collection.
Using a 3 ml syringe remove a 2 ml sample to measure the pH22C of the platelet product. Use a blood gas analyzer to ensure the product has a pH >6.6. Use the same sample to measure the platelet concentration of the platelet product using a hematological analyzer. Ensure the product meets the 0.80–3.8 x 106 platelets/µl collection concentration requirement.
Visually evaluate the swirl of the platelet product. If the product has a “0” swirl the product will be discarded.
Hold the platelet bag up under a direct white-light source and squeeze the platelet bag briefly. Visually observe the swirling pattern of the platelet sample. Score the product using the following scale. NOTE: 2 – Positive Swirl: Swirling inhomogeneity visible throughout the entire bag with strong contrast. 1 – Intermediate Swirl: Some inhomogeneity visible, but only in a few places and with poor contrast. 0 – Negative Swirl: Turbid homogeneity remaining before and after squeezing the platelet bag
3. Riboflavin and UV Light Process:
Transfer the platelet product to a riboflavin and UV light illumination and storage bag using a sterile tubing docking device. After transfer remove the empty platelet unit collection bag using a tubing sealer and discard the empty bag as biohazardous waste.
Remove a 35 ml pouch of 500 μM riboflavin stock solution from its light protective outer wrap. Sterile dock the riboflavin kit onto the inlet line of the treatment and storage bag. Allow the contents to drain into the platelet unit.
Hang the riboflavin pouch / treatment and storage bag set from the riboflavin pouch and gently express residual air out of the treatment and storage bag and into the riboflavin bag. Ensure a small amount of platelet product is also expressed into the riboflavin pouch to rinse any residual riboflavin into the platelet product. Allow the riboflavin and platelet solution drain back into the treatment and storage bag and then clamp the inlet line. Remove the empty riboflavin bag using a tubing sealer and discard the empty bag as biohazardous waste.
Sterile dock a 6” tubing line with female syringe connector onto the inlet line of the illumination and storage bag. Attach a 10 ml syringe barrel (remove the plunger) onto the female syringe connector.
Aseptically add 5 ml of stock bacteria into the syringe barrel via a pipette. Open the clamp on the inlet line and allow the bacteria to drain into platelet product. Rinse the syringe barrel by allowing some platelet product to flow back into the 10 ml syringe barrel. Remove the 10 ml syringe barrel.
- Aseptically attach a 30 ml syringe and flush the inlet port a minimum of two times to ensure the stock bacteria is mixed into the platelet product. After the product is well mixed, use a clean 5-10 ml syringe and remove a 2 ml pre-treatment sample. Using a new syringe ensure any air introduced from the inoculation step or sampling step is removed.
- Titer the pre-treatment sample per section 4.
Remove the inlet line from the treatment and storage bag and weigh the product.
- Place the product into the illuminator and follow the onscreen commands to start a standard platelet treatment.
- Enter in Operator ID.
- Secure and mount the bag to the Illuminator platen.
- Enter the Sample ID into the Illuminator using either the barcode reader or the manual entry keypad.
- Scan product type.
- Close the quartz clamp.
- The illumination device will automatically deliver 6.24 J/ml of energy. The typical treatment time takes 5-10 min.
- Following treatment sterile dock a 6” tubing line with female syringe connector onto the sample bulb line of treatment and storage bag. Ensure the product is well mixed and then attach a 5 ml syringe and remove a 2 ml post-treatment sample.
- Titer the post-treatment sample per section 4.
4. Bacterial Titer
Estimate the titer of the pre-treatment and post-treatment samples and prepare the appropriate number of 5 ml dilution tubes needed to perform an end point serial dilution of each sample. Aseptically fill the required tubes with 1.8 ml of peptone water.
- Titer each sample using a 10-fold serial dilution scheme. Aseptically transfer 0.2 ml from the undiluted sample to the first dilution tube (10-1). Vortex this dilution tube and transfer 0.2 ml to the next dilution tube. Continue the serial dilution through the required number of dilutions.
- Aseptically transfer 100 µl of each dilution to be plated to an appropriate agar plate (Table 1).
Using a sterile cell spreader, evenly distribute the sample onto each agar plate.
Invert and incubate the plates at the appropriate conditions (Table 1) for 1-2 days checking for growth periodically.
Count plates with 30–300 colonies. Plates containing greater than 300 colonies are considered too numerous to count (TNTC). Plates containing less than 30 colonies are considered to be below the limit of quantification (LOQ).
- Calculate the titer of each test sample and record the results as CFU/ml.
- Use the following equation to calculate CFU/ml:

- If there are no colonies on the lowest dilution plate, use the following equation to calculate the Limit of Detection (LOD):
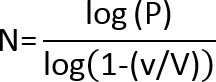 and
and 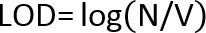 Where: N =lowest number of particles (CFU) in product that can be detected with confidence. P = probability that a microorganism will be undetected; for a 95% confidence of detecting a microorganism, P = .05. V = total product volume in ml v =volume plated (ml) x dilution level of plate
Where: N =lowest number of particles (CFU) in product that can be detected with confidence. P = probability that a microorganism will be undetected; for a 95% confidence of detecting a microorganism, P = .05. V = total product volume in ml v =volume plated (ml) x dilution level of plate
- For Negative Control, plate 0.5 ml of the peptone water that is used for the serial dilutions onto each of two agar plates. Use the same plate type as the samples to be tested. Invert and incubate the plate at the appropriate conditions for 1-2 days checking for growth periodically.
- If growth is observed on either agar plate, the bottle of peptone water used must be discarded; the results of the study are also invalidated.
For Positive Control, re-titer the stock culture of bacteria that was used. Invert and incubate the plates at the appropriate conditions for 1-2 days checking for growth periodically. The titer shall be ± 1.0 log CFU/ml of the recorded stock titer (See section 1.5) for the study to be considered valid.
Representative Results
The reduction of eleven different bacterial species was evaluated using the riboflavin and UV light based PRT process. These agents represent some of the more common contaminants of platelet products and include five Gram positive and six Gram negative organisms. At least six platelet products were evaluated per organism and each product was inoculated with enough bacteria to yield >1.0 x 105 CFU/ml final titer of bacteria in the platelet product. Prior to treatment a sample was removed to determine the pre-treatment titer, after which the platelet product was promptly treated with the riboflavin and UV process. Following treatment of the platelet products the following log reductions were observed: S. epidermidis 4.7 log (99.998%), S. aureus 4.8 log (99.998%), S. mitis 3.7 log (99.98%), S. pyogenes 2.6 log (99.7%), S. marcescens 4.0 log (99.99%), Y. enterocolitica 3.3 log (99.95%), B. neotomae 5.4 log (99.9996%), B. cereus 2.6 log (99.7%), E. coli ≥ 5.4 log (99.9996%), P. aeruginosa 4.7 log (99.998%) and K. pneumoniae 2.8 log (99.8%) (Figure 2). All units met the system specifications as stated above for the riboflavin and UV process. Additional reduction testing was performed with S. pyogenes using buffy coat derived platelets. No significant difference was observed in the reduction of S. pyogenes in buffy coat derived platelets versus apheresis platelets (data not shown).
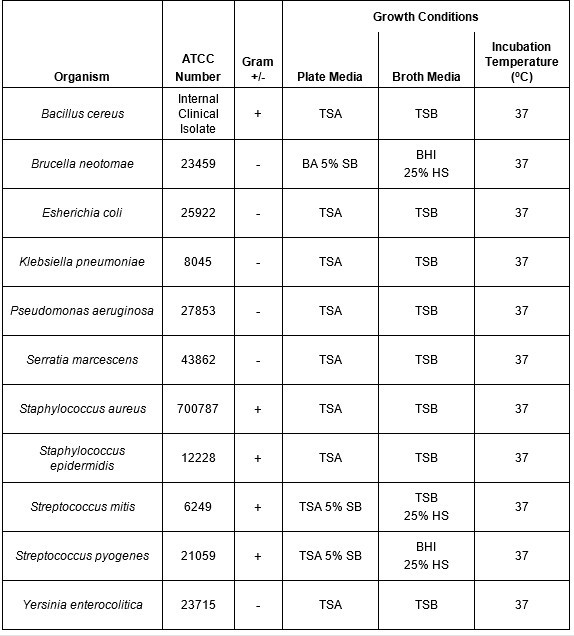 Table 1: Growth conditions for bacterial species evaluated using riboflavin and UV light. A panel of both Gram negative and Gram positive bacteria that have been shown to contaminate platelet products were selected for this study. The growth conditions (temperature and media type) used to propagate and titer the organisms are shown.
Table 1: Growth conditions for bacterial species evaluated using riboflavin and UV light. A panel of both Gram negative and Gram positive bacteria that have been shown to contaminate platelet products were selected for this study. The growth conditions (temperature and media type) used to propagate and titer the organisms are shown.
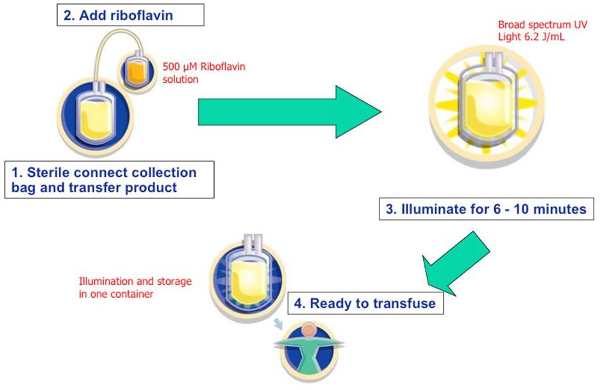 Figure 1: Riboflavin and UV light pathogen reduction process. Riboflavin was dissolved in 0.9% saline at a nominal concentration of 500 µM. The solution was sterile connected and drained into each platelet product. Bacteria was added aseptically to each platelet unit and then treated with 6.2 J/ml of UV energy (approximately 6–10 min). In this in vitro study the post-treatment platelet unit was sampled to determine the final bacterial titer.
Figure 1: Riboflavin and UV light pathogen reduction process. Riboflavin was dissolved in 0.9% saline at a nominal concentration of 500 µM. The solution was sterile connected and drained into each platelet product. Bacteria was added aseptically to each platelet unit and then treated with 6.2 J/ml of UV energy (approximately 6–10 min). In this in vitro study the post-treatment platelet unit was sampled to determine the final bacterial titer.
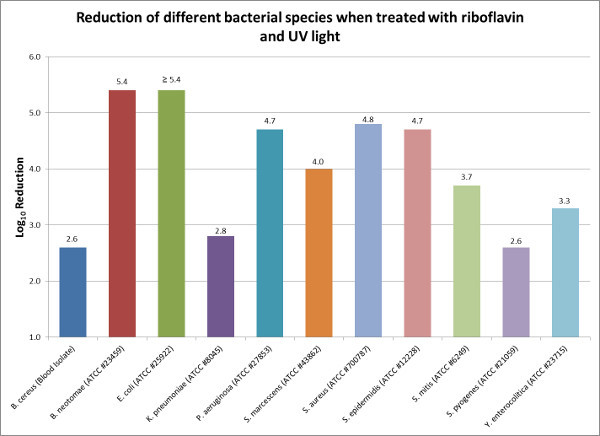 Figure 2: Reduction of different bacterial species when treated with riboflavin and UV light. Riboflavin and UV light was used to reduce the bacterial loads of common bacterial contaminants of platelet products. The log10 reduction is the difference between the average pre-treatment titer and the average post-treatment titer. Please click here to view a larger version of this figure.
Figure 2: Reduction of different bacterial species when treated with riboflavin and UV light. Riboflavin and UV light was used to reduce the bacterial loads of common bacterial contaminants of platelet products. The log10 reduction is the difference between the average pre-treatment titer and the average post-treatment titer. Please click here to view a larger version of this figure.
Discussion
Bacterial screening has become common practice in blood banking institutions and for many locations it serves as the primary method to prevent transfusion of bacterially contaminated platelets. Although bacterial screening typically interdicts the majority of fast growing Gram negative bacteria like S. marcescens, E. coli and E. cloacae, it struggles to detect slow growing Gram positive bacteria like S. aureus, Streptococcus spp. and S. epidermidis8. Unfortunately, these three Gram positive bacteria are implicated in upwards of 50% of all contaminated platelet units and transfusion of platelet products containing these three organisms has lead to patient mortality1.
Bacterial screening relies on the bacterial titer in a platelet product to be sufficiently high enough so that at least one organism is contained within the typical 4-8 ml screening sample. If the bacterial titer is not sufficiently high for a bacterium to be captured in the 4-8 ml sample volume, the unit is incorrectly flagged as negative and it is released for transfusion. An alternative approach to bacterial screening could be to employ a proactive process that can be performed on all platelet units and that could render the contaminating organisms as non-functional.
In this study, high titers of bacteria were purposely added to platelet units to test the total reduction capacity of the riboflavin and UV light pathogen reduction process. Although the bacterial loads tested in this study far exceed those normally found in blood products at the time of donation, this study helps to demonstrate that the riboflavin and UV light process is able to significantly reduce the titer of viable bacteria in a contaminated platelet unit by 2.6 to 5.4 log (99.7-99.9996% reduction of contaminating bacteria) (Figure 2). To obtain an accurate log reduction value, it is critical that the operator have good aseptic technique along with precise pipetting skills when performing the bacterial titer steps. Imprecise pipetting when performing serial dilutions can lead to an accumulation of errors, thus an inaccurate titer measurement.
The data collected in this study supports previous work with riboflavin and UV light, which challenged the system against low level, clinically relevant bacteremia (<20 CFU/unit). The previous study was designed to mimic an actual clinical contamination event and the results showed riboflavin and UV light was effective in preventing bacterial growth over the 7-day platelet storage period. Modeling based on the data from the previous study suggests that the current risk estimates of bacterial contamination of platelet products could be reduced by as much as 98% from current levels8. The riboflavin and UV light process compares favorably to bacterial screening, which only demonstrated a 66% effectiveness to detect bacterial contaminants in platelet units when challenged with a clinically relevant bacterial load8.
As with all technologies, PRT treatment of platelet products to prevent complications associated with bacterially contaminated platelet units has its limitations. PRT systems are not effective against bacterial spores and do not inactivate endotoxin, thus even if a PRT system can inactivate high titers of Gram negative bacteria the remaining endotoxin can still cause septic shock in the recipient. PRT is best performed early during storage before the bacteria can propagate to high titers.
In conclusion, as demonstrated by the combination of these two studies, the riboflavin and UV light process is effective at reducing the bacterial loads of both Gram negative and Gram positive organisms and may offer a viable alternative to bacterial screening. Routinely treating platelet products with riboflavin and UV light also has the added benefit of reducing the potential transmission of other blood borne agents like viruses and parasites.
Disclosures
All authors of this paper are currently employed by Terumo BCT, developer and manufacturer of the Mirasol PRT system. Publication fees for this video-article have been paid by Terumo BCT.
Acknowledgments
The authors have no acknowledgements.
References
- Brecher ME, Hay SN. Bacterial contamination of blood components. Clin. Microbiol. Rev. 2005;18(1):195–204. doi: 10.1128/CMR.18.1.195-204.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brecher ME, Holland PV, Pineda AA, Tegtmeier GE, Yomtovian R. Growth of bacteria in inoculated platelets: implications for bacteria detection and the extension of platelet storage. Transfusion. 2000;40(11):1308–1312. doi: 10.1046/j.1537-2995.2000.40111308.x. [DOI] [PubMed] [Google Scholar]
- Cardo LJ, et al. Pathogen inactivation of Leishmania donovani infantum in plasma and platelet concentrates using riboflavin and ultraviolet light. Vox Sang. 2006;90(2):85–91. doi: 10.1111/j.1423-0410.2005.00736.x. [DOI] [PubMed] [Google Scholar]
- Cardo LJ, Salata J, Mendez J, Reddy H, Goodrich R. Pathogen inactivation of Trypanosoma cruzi in plasma and platelet concentrates using riboflavin and ultraviolet light. Transfus. Apher. Sci. 2007;37(2):131–137. doi: 10.1016/j.transci.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Dodd RY. Current viral risks of blood and blood products. Ann. Med. 2000;32(7):469–474. doi: 10.3109/07853890009002022. [DOI] [PubMed] [Google Scholar]
- Dodd RY. Bacterial contamination and transfusion safety: experience in the United States. Transfus. Clin. Biol. 2003;10(1):6–9. doi: 10.1016/s1246-7820(02)00277-x. [DOI] [PubMed] [Google Scholar]
- Dumont LJ, et al. Screening of single-donor apheresis platelets for bacterial contamination: the PASSPORT study results. Transfusion. 2010;50(3):589–599. doi: 10.1111/j.1537-2995.2009.02460.x. [DOI] [PubMed] [Google Scholar]
- Goodrich RP, Gilmour D, Hovenga N, Keil SD. A laboratory comparison of pathogen reduction technology treatment and culture of platelet products for addressing bacterial contamination concerns. Transfusion. 2009;49(6):1205–1216. doi: 10.1111/j.1537-2995.2009.02126.x. [DOI] [PubMed] [Google Scholar]
- Greco-Stewart VS, et al. Serratia marcescens strains implicated in adverse transfusion reactions form biofilms in platelet concentrates and demonstrate reduced detection by automated culture. Vox Sang. 2012;102(3):212–220. doi: 10.1111/j.1423-0410.2011.01550.x. [DOI] [PubMed] [Google Scholar]
- Keil SD, et al. Inactivation of Plasmodium spp. in plasma and platelet concentrates using riboflavin and ultraviolet light. Transfusion. 2013;53(10):2278–2286. doi: 10.1111/trf.12079. [DOI] [PubMed] [Google Scholar]
- Kleinman S, Chan P, Robillard P. Risks associated with transfusion of cellular blood components in Canada. Transfus. Med Rev. 2003;17(2):120–162. doi: 10.1053/tmrv.2003.50009. [DOI] [PubMed] [Google Scholar]
- Larsen CP, Ezligini F, Hermansen NO, Kjeldsen-Kragh J. Six years' experience of using the BacT/ALERT system to screen all platelet concentrates, and additional testing of outdated platelet concentrates to estimate the frequency of false-negative results. Vox Sang. 2005;88(2):93–97. doi: 10.1111/j.1423-0410.2005.00596.x. [DOI] [PubMed] [Google Scholar]
- Mundt JM, Rouse L, Van den Bossche J, Goodrich RP. Chemical and Biological Mechanisms of Pathogen Reduction Technologies. Photochem. Photobiol. 2014. [DOI] [PMC free article] [PubMed]
- Murphy WG, et al. Screening platelet concentrates for bacterial contamination: low numbers of bacteria and slow growth in contaminated units mandate an alternative approach to product safety. Vox Sang. 2008;95(1):13–19. doi: 10.1111/j.1423-0410.2008.01051.x. [DOI] [PubMed] [Google Scholar]
- Brien SF, et al. Current incidence and residual risk of HIV, HBV and HCV at Canadian Blood Services. Vox Sang. 2012;103(1):83–86. doi: 10.1111/j.1423-0410.2012.01584.x. [DOI] [PubMed] [Google Scholar]
- Pearce S, Rowe GP, Field SP. Screening of platelets for bacterial contamination at the Welsh Blood Service. Transfus. Med. 2011;21(1):25–32. doi: 10.1111/j.1365-3148.2010.01037.x. [DOI] [PubMed] [Google Scholar]
- Ramirez-Arcos S, Kou Y, Perkins H. Evaluation of a universal point-of-issue assay for bacterial detection in buffy coat platelet components. Vox Sang. 2014;107(2):192–195. doi: 10.1111/vox.12148. [DOI] [PubMed] [Google Scholar]
- Reddy HL, et al. Toxicity testing of a novel riboflavin-based technology for pathogen reduction and white blood cell inactivation. Transfus. Med. Rev. 2008;22(2):133–153. doi: 10.1016/j.tmrv.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Ruane PH, et al. Photochemical inactivation of selected viruses and bacteria in platelet concentrates using riboflavin and light. Transfusion. 2004;44(6):877–885. doi: 10.1111/j.1537-2995.2004.03355.x. [DOI] [PubMed] [Google Scholar]
- Tonnetti L, Proctor MC, Reddy HL, Goodrich RP, Leiby DA. Evaluation of the Mirasol pathogen reduction technology system against Babesia microti in apheresis platelets and plasma. Transfusion. 2010;50(5):1019–1027. doi: 10.1111/j.1537-2995.2009.02538.x. [DOI] [PubMed] [Google Scholar]
- Vanlandingham DL, et al. Photochemical inactivation of chikungunya virus in plasma and platelets using the Mirasol pathogen reduction technology system. Transfusion. 2013;53(2):284–290. doi: 10.1111/j.1537-2995.2012.03717.x. [DOI] [PubMed] [Google Scholar]
- Walther-Wenke G, et al. Monitoring bacterial contamination of blood components in Germany: effect of contamination reduction measures. Vox Sang. 2011;100(4):359–366. doi: 10.1111/j.1423-0410.2010.01432.x. [DOI] [PubMed] [Google Scholar]


