Abstract
In the absence of specialized mobile immune cells, plants utilize their localized programmed cell death and Systemic Acquired Resistance to defend themselves against pathogen attack. The contribution of a specific Arabidopsis gene to the overall plant immune response can be specifically and quantitatively assessed by assaying the pathogen growth within the infected tissue. For over three decades, the hemibiotrophic bacterium Pseudomonas syringae pv. maculicola ES4326 (Psm ES4326) has been widely applied as the model pathogen to investigate the molecular mechanisms underlying the Arabidopsis immune response. To deliver pathogens into the leaf tissue, multiple inoculation methods have been established, e.g., syringe infiltration, dip inoculation, spray, vacuum infiltration, and flood inoculation. The following protocol describes an optimized syringe infiltration method to deliver virulent Psm ES4326 into leaves of adult soil-grown Arabidopsis plants and accurately screen for enhanced disease susceptibility (EDS) towards this pathogen. In addition, this protocol can be supplemented with multiple pre-treatments to further dissect specific immune defects within different layers of plant defense, including Salicylic Acid (SA)-Triggered Immunity (STI) and MAMP-Triggered Immunity (MTI).
Keywords: Infection, Issue 104, Arabidopsis thaliana, P. syringae pv. maculicola ES4326, infection, syringe infiltration, plant immunity, salicylic acid (SA), Enhanced Disease Susceptibility, Systemic Acquired Resistance, Microbe-Associated Molecular Patterns (MAMPs), MAMP-Triggered Immunity, SA-Triggered Immunity
Introduction
Due to their sessile nature, plants are constantly threatened by a plethora of pathogens exhibiting various lifestyles and nutritional strategies1. To a first approximation, biotrophic pathogens maintain their host alive to retrieve nutrients, while necrotrophic pathogens actively secret toxins and enzymes to kill host tissue and feed on the dead cells1. Another group of pathogens, termed hemibiotrophs, begins their infection course with the biotrophic stage and shifts to the necrotrophic stage upon reaching a certain threshold of pathogen accumulation2. In order to effectively defend themselves against these microorganisms, plants have evolved a complicated innate immune system equipped with multiple surveillance mechanisms to detect the pathogen attack and trigger localized programmed cell death3 as well as Systemic Acquired Resistance (SAR)4. Current research is focused on characterizing the essential signaling components and cross-talks within the plant immune system5.
As proposed in the “Zig-Zag” model5, the first layer of the plant innate immune response requires the presence of plasma membrane-localized Pattern Recognition Receptors (PRRs) to detect the invasion of a microbe. PRRs are able to recognize Microbe-Associated Molecular Patterns (MAMPs) and establish MAMP-Triggered Immunity (MTI)6. Besides inducing a transcriptional upregulation of genes encoding antimicrobial PR proteins7, MTI leads to a variety of events that arrest pathogen growth, including the production of Reactive Oxygen Species (ROS) and Reactive Nitrogen Species (RNS), deposition of callose to the cell wall as well as the activation of multiple kinase signaling pathways8.
Until now, several MAMPs have been identified to trigger MTI in Arabidopsis, including bacterial flg229 (a 22 amino acid fragment derived from flagellin), elf1810 (18 amino acids from the bacterial translation elongation factor Tu) and a structural cell wall component peptidoglycans11. To establish a successful infection, some specialized pathogens have evolved the ability to secret virulence effector proteins into the intracellular or intercellular spaces, and consequently repress MTI and trigger Effector-Triggered Susceptibility (ETS)12,13. For instance, virulence effectors can inactivate Mitogen-Activated Protein Kinase (MAPK) phosphorylation cascades of MTI to induce the disease development within the infected tissue14-16. During the dynamic co-evolution between hosts and pathogens, plants also developed the counterattack strategy to recognize the effector proteins and attenuate the pathogen virulence molecules17. This direct or indirect effector recognition is mediated by disease resistance (R) proteins18. Most of them are members of NB-LRR (Nucleotide Binding and Leucine-Rich Repeats) family19. The perception of an avirulent effector by an R protein elicits a stronger and broader immune response characterized as Effector-Triggered Immunity (ETI)20. Besides inducing the expression of defense genes21 and the production of defense metabolites22, ETI often leads to a rapid localized programmed cell death known as Hypersensitive Response (HR) to restrict the pathogen from spreading into the adjacent tissue3.
In addition to the localized programmed cell death23, plants are capable of initiating a long-term and system-wide immune response termed Systemic Acquired Resistance (SAR)4. Upon challenge with a biotrophic pathogen, plant cells trigger the biosynthesis and accumulation of an endogenous phytohormone salicylic acid (SA) and PR proteins in both local and systemic tissues24. Through this process, a heightened state of preparedness is achieved in the uninfected leaves that allows for mounting faster defense responses during a subsequent infection by a broad spectrum of pathogens24. SA and its synthetic analogs such as benzo-(1,2,3)-thiadiazole-7-carbothioic acid S-methyl ester (BTH) and 2,6-dichloroisonicotinic acid (INA) are capable of chemically inducing Salicylic Acid (SA)-Triggered Immunity (STI) upon external application24. Nonexpressor of Pathogenesis-Related genes 1 (NPR1) is proposed to be one of the SA receptors and functions as a major transcriptional regulator during SA-mediated defense response in both local and systemic tissues21,25,26. It has been conclusively demonstrated that NPR1 is required for SAR establishment and the loss of NPR1 leads to dramatic susceptibility towards Pseudomonas syringae25.
To extensively characterize the molecular contribution of plant components in the plant-pathogen interactions, multiple bioassays have been developed to measure specific defense events, including ROS burst27, callose deposition28, defense genes expression and accumulation of their protein products21. While these individual assays can provide insights into a specific form of the plant immune response, none of them, however, are able to represent the complete defense response on the whole plant level. Conversely, quantification of pathogen growth after infection provides an overall estimation of immune response at the organismal level. Therefore, the development and optimization of a precise and highly standardized pathogen inoculation assay is critical to fuel up the research and discoveries on the Arabidopsis immune responses.
Pseudomonas syringae, a Gram-negative bacterium, was identified as a phytopathogen capable of causing disease in a range of plant hosts including Arabidopsis29. As the model plant-pathogen system, the Arabidopsis - P. syringae interaction has been widely applied to understand the molecular mechanisms underlying plant defense responses29. Until now, over 50 P. syringae pathovars have been identified based on their ability to infect different plant species30. P. syringae pv. tomato DC3000(Pst DC3000)31and P. syringae pv. maculicola ES4326 (Psm ES4326)32 are the two most widely used and extensively characterized virulent strains. Aside from being recognized by the plant and triggering the MTI response, Pst DC3000 and Psm ES4326 are capable of secreting virulent effector proteins to suppress MTI and trigger ETS to favor the pathogen growth31,33.
To functionally dissect the interaction between Arabidopsis and P. syringae, multiple pathogen infection methods have been developed based on the pathogen delivery approach. For soil-grown plants, pathogen can be delivered by syringe infiltration, vacuum infiltration, dip inoculation and spray inoculation29,34. Recently, seedling flood-inoculation assay was developed to perform large-scale screens on tissue culture plates-grown young Arabidopsis plants35. Syringe infiltration, as one of the most commonly used approach, manually delivers the pathogen into the apoplast through the natural leaf openings termed stomata29. Through this approach, equal amounts of P. syringae can be infiltrated into the infected leaf and the strength of plant immune response is inversely correlated to the pathogen growth levels. Therefore, quantification of pathogen growth serves as an optimal approach to evaluate the immune function at the whole plant level. In addition, syringe infiltration can distinguish the local and systemic tissue, which can be applicable in characterizing the molecular mechanisms underlying SAR36.
In the following protocol, we describe an optimized syringe infiltration assay with Psm ES4326 to screen Arabidopsis mutants for enhanced disease susceptibility (EDS). This protocol will employ two Arabidopsis genotypes: a wild-type ecotype Columbia-0 (Col-0) plants (control) and npr1-1 loss-of-function mutants(hypersusceptible)that will be infected with virulent bacterial strain Psm ES432637. The npr1-1 mutant carries a point mutation within the ankyrin-repeat consensus sequence of the NPR1 molecule, which alters the highly conserved histidine to tyrosine and renders the protein non-functional25. In addition, a number of modifications of the syringe infiltration assay are described that allow quantification of defects in the specific layers of immune response, including MTI and STI.
Protocol
The following text describes a stepwise protocol to perform optimized Psm ES4326 syringe infiltration assay in Arabidopsis. Major procedures of this assay are represented in a simplified flowchart (Figure 1).
1. Plant Growth Conditions
- Sow seeds
- Prepare 2 pots (4 in diameter, 3.75 in tall) loosely filled with soil and water pots by soaking them from the bottom O/N before draining the excess water.
- Sow 50-100 Arabidopsis seeds, wild-type Col-0 or npr1-1 mutant, on each pot with a folded 70 mm weighing sheet or other paper.
- Cover the pot with a water-sprayed transparent dome to increase the relative humidity to 80-90% (Figure 2A).
Incubate the pot at 4 °C for 72 hr to allow for complete stratification and synchronous germination.
Transfer the pot to the standard growth conditions (12 hr light/12 hr dark, 21 °C, light intensity 100 µmol/m2/sec, relative humidity 40%) to allow the seed to germinate. Crack the dome to generate a 2-3 in opening immediately after transfer to the growth room, which will help avoid water condensation.
- When the first true leaf reaches the size of 2-3 mm in length, transplant the seedlings from the pot to a 72-wells flat filled with soil.
- Use a finger or a 1 ml pipette tip to create a 1-in deep depression in the middle of the soil surface on the flat.
- Gently separate individual seedlings with as little root damage as possible. Carefully pick up seedlings that have intact roots with forceps.
- Place a single separated seedling together with the adhering soil clump to minimize transplantation shock into the depression and gently pat down the surrounding soil to fill the depression. Discard the extra seedlings and soil into biohazard waste container and dispose in accordance with the local biohazard waste disposal guidelines.
- Water transferred seedlings from the top and completely cover the flat with a water-sprayed transparent dome to maintain 80-90% humidity. Keep the dome on for 3 days, then crack the dome in the morning of day 4 and completely remove it by the end of that day. Note: Sowing seeds can be alternatively performed by stratifying seeds in 1.5 ml centrifuge tubes containing 1 ml 0.1% agar for 48 hr at 4 °C followed by transferring 2-3 seeds with a glass pipette onto a 72-wells flat filled with soil. Flats need to be covered with a transparent dome that can be removed one week after the germination. Extra seedlings can be removed to leave only one seedling per well.
Water plants every two days by soaking flats in 1-in water for 20 min, then drain the excess water. Note: The time interval for watering plants depends on the growth room humidity and needs to be determined based on continuous observation of user’s plant growth facility. Check plants daily to make sure they are well watered. If only few spots are drying, water those plants from the top with a squirt bottle.
Perform pathogen infection assays (Step 3-5) on plants that are at or near developmental stage # 3.50 (when rosette size is 50% final size), which corresponds to 3-5 weeks old depending on growth conditions (photoperiod, temperature). Do not infect plants after inflorescence emergence (stage # 5) due to onset of age-related resistance38,39.
2. Preparation of Culture Media and Plates
Make 1 L of King’s B (KB) liquid medium. Gently stir 20 g proteose peptone and 2 g potassium phosphate dibasic trihydrate using a magnetic stir bar with 1,000 ml deionized H2O until there is no visible pellet. Aliquot 100 ml into 150 ml glass bottles and autoclave on a 20-min liquid cycle.
- Prepare culture plates with KB solid medium.
- Gently stir 20 g Proteose Peptone, 2 g Potassium Phosphate Dibasic Trihydrate and 15 g Agar with 1,000 ml deionized H2O. Autoclave.
- Cool down the medium on a magnetic stir plate until it is cool to touch to minimize future condensation and avoid degradation of antibiotics. Add 18 ml sterile 80% Glycerol and 5 ml sterile 1 M MgSO4 to the medium. Given that Psm ES4326 carries resistance against streptomycin, add streptomycin into the cooled media to a final concentration of 50 µg ml−1.
- Pour the plates. Prepare two different sizes of KB media plates: 100 mm x 15 mm and 150 mm x 15 mm Petri dishes. The smaller Petri dish containing medium serves as the primary bacteria culture plate, and the larger Petri dish serves as bacteria counting plate. Note: For the bacteria counting plates, rectangular-shaped plates can be used to reduce the consumables cost since twice as many treatments can be plotted per plate compared to traditional round plates.
- Pour the plates prior to the infection experiment. Store KB medium plates at 4 °C for up to 2-3 months since streptomycin sulfate gradually loses the antibiotic activity40.
3. Enhanced Disease Susceptibility (EDS)
- Day 1 - Streak bacteria on plate
- Two days before the EDS assay, streak Psm ES4326 from -80 °C glycerol stock on a KB-Strep bacterial culture plate and incubate at 28 °C for 24-48 hr.
- Day 2 - initiate liquid culture and water plants
- Initiate the liquid culture in a sterile test tube with 4 ml liquid KB medium containing 50 µg ml−1 streptomycin and shake at 250 rpm, 28 °C O/N. Note: For EDS infection assay, using 6 plants per genotype is recommended. For STI and MTI assays, 6 plants per genotype per treatment are recommended. For the bacteria inoculum, it is recommended to use a fresh liquid culture with optical density at λ600nm between 0.3 and 0.6.
- Mark the petioles of leaves number 5 and 6 with a blunt-end waterproof marker for easy identification of infected tissue at sampling (Figure 1). To enhance the opening of stomata and facilitate the entrance of pathogen solution into the leaf, water the plants well by soaking the flat from the bottom for 20 min, then drain the excess water. Note: Alternatively, cover the flat with a transparent dome and soak it in water for 2-4 hr to increase the stomatal opening.
- Day 3 - Pathogen dilution and syringe infiltration Note: Pathogen infection during morning hours is optimal for pathogen proliferation and development of the most pronounced disease symptoms on susceptible genotypes. Make every effort to keep the infection timing consistent to eliminate the effect of circadian rhythms and diurnal gene regulation41,42, which helps reduce the variation among experimental replications.
- Pellet the bacterial culture in a microcentrifuge 1.5 ml tube at 9,600 x g for 2 min at RT and discard the supernatant. Resuspend the bacterial pellet with 1 ml of sterile 10 mM MgCl2. Note: Magnesium cations can enhance the motility and adhesion of P. syringae43. Alternatively, use MgSO4 at the same concentration. Sterile water is an acceptable substitute for the Mg salt solutions.
- Dilute the bacteria suspension with 9 ml of 10 mM MgCl2 in a 50 ml centrifuge tube and measure the optical density (OD) of bacteria with a spectrophotometer at λ=600 nm. Dilute the bacteria with 10 mM MgCl2 to the final OD600nm=0.0002 for EDS infection assay.
- Infiltrate the leaf with a 1 ml blunt end needleless syringe (commonly known as the insulin syringe) that contains the diluted bacterial solution.
- Do not fill the syringe up to its full capacity. Fill it up with 0.5-0.6 ml to allow for a much better control during the infiltration process.
- Expose the lower surface of the leaf on the top of index finger, and then gently adjust the leaf position with the help of thumb. Position the syringe vertically against the leaf surface to ensure that pressure is evenly distributed. Try to avoid the midrib area during the infiltration to reduce leaf damage.
- Slowly push the plunger to infiltrate the bacterial solution; liquid entry into the leaf mesophyll will be visualized as indicated by the darker leaf color. Attempt to infiltrate the entire surface of the leaf. If this is not accomplished in a single attempt, choose another infiltration spot and repeat the actions described above until the entire leaf surface is covered.
- Once completed, gently blot the leaf with absorbent tissue to remove the extra pathogen solution. Visually inspect the infected leaf tissue for damage. Note: The circular syringe impression should not be visible after the infiltration is complete.
- Leave the infiltrated plants to dry for 1-2 hr before returning them into their original growth conditions. Next, spray a clear dome with water and cover the infected plants for 2 hr, then crack the dome to generate a 2-3 in opening and leave it on throughout the remainder of the infection experiment. Note: Since P. syringae is not an airborne pathogen, there is no risk of spreading the infectious agent to other plants grown in the same facility. However, as an added precaution, avoid physical contact between infected plants and other experimental plants located nearby. Covering the flat with a transparent dome will increase the pathogen virulence and accelerate disease progression. The need for this step and optimal duration of the covering period needs to be determined based on the humidity conditions of the user’s plant growth facility.
- Day 5 - Pre-dry the media plates
- Take the 150 mm x 15 mm KB plates out of the cold storage unit and dry any pre-existing water condensation on the plate. Dry plates by keeping them at RT for roughly 24 hr. To speed up this process, place them in a laminar flow hood with their lids cracked for 30-60 min. Note: This step is critical for the formation of circular droplet on the surface of the plate in the next step.
- Day 6 - Quantifying the pathogen growth Note: The following pathogen quantification procedure may be performed at earlier time points after the infection to confirm equal amounts of bacteria are delivered into the leaf, especially when plants have altered leaf morphology.
- Process the infected tissue after the emergence of chlorosis indicated by the yellowing of the infected tissue in the susceptible genotypes, but before the development of necrotic lesions (Figure 2B). Note: Suggested sampling time is three days after the pathogen inoculation. However, since pathogen growth depends on a number of environmental factors, the length of incubation may vary within a range of 2 ½-3 ½ days and needs to be determined by careful observations of the infection progression in user’s plant growth facility.
- Prepare 6 grinding tubes for each genotype (Figure 1). Place one stainless steel grinding ball and add 500 µl sterile 10 mM MgCl2 into the each tube.
- Detach the infected leaf from the plant and punch a leaf disc with a 1-hole paper punch (Figure 1). Note: To minimize the sampling error, try to punch each sampled leaf at the same position. Sampling of the leaf disc from the top of the leaf is recommended.
- Randomly place 2 leaf discs (from two different plants) into each grinding tube using forceps.
- Seal the tube and homogenize the tissue with a high-throughput homogenizer at maximum speed (1,600 strokes per minute) for 10 min. Repeat this process if needed until tissue is well homogenized and the solutions turn green due to chlorophyll release from the infected leaves.
- While waiting for the homogenization, fill the first 6 rows of a 96-well culture plate with 180 µl 10 mM MgCl2 using a multi-channel pipette and a pipetting reservoir.
- After grinding, transfer 20 µl of ground tissue suspension into the first row of the 96-well plate and mix by repeated pipetting the liquid up and down. If small fragments of tissue clog the tip, clip it by 2-3 mm to help acquire the correct volume of the solution.
- To provide enough space for the droplet on the top of the plate, space the tissue from different genotypes in alternative rows (Figure 1). To prepare a ten-fold serial dilution, transfer 20 µl liquid into the second row and repeat this procedure until the sixth dilution.
- Transfer 20 µl of the solution from the 96-well plate onto the 150 mm x 15 mm KB plate using divided pipette tips (Figure 1). Work from the most dilute suspension to the most concentrated. Note: Proceeding from most diluted to most concentrated makes it unnecessary to change pipette tips between the dilutions. If the plate is well pre-dried, the transferred droplet should stay intact on the top of the medium until absorbed (usually 15-30 min). Drying time varies with the RT and humidity.
- Dry the plate at RT with lid cracked. Once no more liquid can be observed on the plate surface, close the lid, invert and incubate the plate at RT or a 28 °C incubator.
- Incubate plates for 40-60 hr until the colonies become visible. Confirm that the growth on the plates reflects the predictable 10-fold drop in colony forming units (cfu) (Figure 2C). Count the bacteria before they overgrow and colonies fuse. Determine the number of bacteria in the lowest dilution that does not have overlapping colonies. Usually, the preferred dilution to be counted will contain between 10-50 colonies. Note: Variation may be present among technical replicates; therefore, the lowest dilution for each replicate should be determined separately.
- To calculate the levels of bacterial proliferation, document the number of the row (R) as well as the number of the bacteria within each technical replicate (T) in writing. Determine the number of colony forming unit – cfu/leaf disc through the formula: cfu/leaf disc = (T × 10R / 20) × 500 / 2
- Type the data in the following spreadsheet template to produce a graph (Figure 1) (for spreadsheet data analysis template file, see Table 2). Each data point is represented as the mean of six technical replicates on a logarithmic scale. Error bars represent 95% confidence interval of the mean (n = 6). Note: The calculation of 95% confidence intervals needs to be adjusted based on the number of technical replicates.
4. SA-Triggered Immunity (STI)
Day 1: Follow the same procedure as described in step 3.1.
- Day 2: In addition to watering the plants and initiating the liquid bacterial culture (Step 3.2), induce resistance by external application of SA derivative Sodium Salicylate21. Note: Pure SA has poor water solubility and requires prior pH adjusting, thus it’s not recommended for this assay.
- To induce SA-mediated defenses against P. syringae and prime the plants for future infection21,24, spray plants with a fine mist of 1 mM Sodium Salicylate or H2O (as mock) 16-24 h before the pathogen infection.
Day 3: Prepare pathogen dilution to OD600nm=0.001 and push the bacteria into the entire leaf surface by syringe infiltration (Step 3.3).
Day 6: For pathogen quantification and counting process, follow the procedure as described for the EDS assay (Steps 3.4 and 3.5). Plate and count the H2O and Sodium Salicylate – pretreated samples separately. For wild-type Arabidopsis, a 1-2 log reduction in pathogen growth is expected in the Sodium Salicylate – pretreated plants.
5. MAMP-Triggered Immunity (MTI)
Note: In this assay, we use Arabidopsis wild-type Col-0 and mutant fls2 and efr plants. FLS2 and EFR are plasma membrane-localized Pattern Recognition Receptors (PRRs) that can recognize flg22 and elf18, respectively9,10. Loss of each PRR results in the insensitivity to the specific type of MAMP, which is indicated by the unaltered pathogen growth in the mutant following external MAMP pre-treatment and syringe infiltration with Psm ES4326.
Day 1 and 2: Follow the same procedure as described in Step 3.1 and 3.2.
- Day 3: MAMP pre-treatment and Pathogen infection
- To establish the MAMP-Triggered Immunity, prepare 1 µM solution of flagellin epitope flg22 or EF-Tu epitope elf18 and syringe-infiltrate it into the entire leaf surface 4 h before the pathogen infection (Steps 3.3.3-3.3.6). This will allow for induction of MAMP-mediated defenses against P. syringae and prime the plants for future infection6,37. Note: Publicly available microarray data revealed the maximum transcriptional reprogramming of MAMP-responsive genes happens 4 h after flg22 or elf18 treatment37,44. Thus, 4 hr is the recommended duration of the MAMP pre-treatment that results in achieving a clear difference in the pathogen growth between MAMP-treated Col-0 and fls2 and efr mutants.
- Remove the extra MAMP solution with absorbent tissue and leave the plants uncovered to speed the liquid absorption process within the leaf. To account for the wounding effect from syringe pressure infiltration, infiltrate H2O into the leaves of control plants.
- Prepare pathogen dilution to OD600nm=0.001 and push the bacteria into the entire leaf surface of MAMP/H2O pre-treated leaf by syringe infiltration (Step 3.3).
Day 6: For pathogen quantification and counting process, follow the procedure as described in Step 3.4 and 3.5. Plate and count the H2O and MAMP–pretreated samples separately. In wild-type Arabidopsis, a 1-2 log reduction in pathogen growth is expected in the MAMP–pretreated plants, with somewhat stronger reduction mediated by flg22 compared to elf18.
Representative Results
The protocol we describe here represents an optimized P. syringae syringe infiltration assay to quantitatively evaluate the immune response in Arabidopsis plants. As illustrated in Figure 1, the syringe infiltration of Psm ES4326 is followed by pathogen extraction and quantification via serial dilutions and colonies enumeration.
As described in Step 3 within the protocol text, Enhanced Disease Susceptibility (EDS) against Psm ES4326 can be assessed by infection with an inoculum with OD=0.0002. As shown in Figure 3, the highly susceptible npr1-1 mutants have approximately 2.5 log (300 times) more pathogen growth compared to the wild-type Col-0 plants. Depending on the experimental conditions, the npr1-1 plants may support up to 3.0 log more bacterial growth than Col-0, with most common results within the range of 1.5-2.5 log. Therefore, this EDS assay provides a broad window of difference, in which the researchers may be able to place their Arabidopsis mutants and transgenics to identify candidate genes potentially involved in the plant immune response.
In addition to determining EDS, Psm ES4326 syringe infiltration assay can be modified to dissect different layers of immune response. To evaluate Salicylic Acid-Triggered Immunity, external application of the chemical Sodium Salicylate is used to trigger the immune response, which is quantified by the pathogen growth (Figure 4A). The loss of NPR1, which functions as the SA receptor and major transcriptional co-regulator of SA-dependent target genes, leads to insensitivity to exogenous application of salicylic acid derivative. This insensitivity to SA is demonstrated by the unaltered pathogen growth in the npr1-1 Sodium Salicylate pre-treated plants in contrast to a marked reduction in Col-0 plants (20 times less pathogen upon Sodium Salicylate application) (Figure 4B).
To characterize the MAMP-Triggered Immunity, pre-treatment with flg22 or elf18 was performed as demonstrated in Figure 5A. To characterize flagellin-triggered immunity, Col-0 and fls2 mutant plants are used. FLS2, a membrane-localized receptor-like kinase, recognizes flg22 and triggers MTI9. As demonstrated in Figure 5B, the flg22-treated Col-0 plants supported a ~ 1 log (10 times) reduction in the bacterial population, while the fls2 mutant plants failed to trigger the bacterial growth restriction effect. Similarly, the loss of EF-Tu receptor EFR in the efr mutant plants leads to insensitivity to elf18 pre-treatment as demonstrated by the unaltered pathogen growth following the elf18 pre-treatment (Figure 5B).
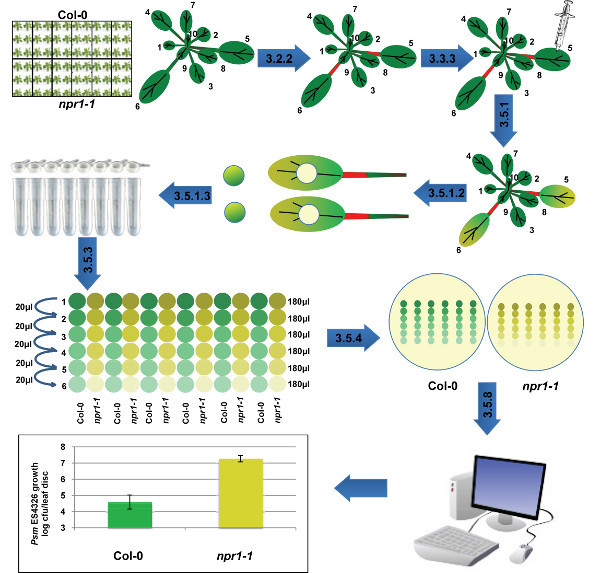 Figure 1. Schematic representation of Psm ES4326 syringe infiltration assay on Arabidopsis plants. Leaves number 5 and 6 of adult soil-grown Arabidopsis plants are marked and infected with Psm ES4326 through syringe infiltration. After the emergence of disease symptoms, detach leaves and harvest leaf discs for tissue homogenization. Well-homogenized tissue is serially diluted in 96-well plates before transferring onto a bacteria counting plate. After colonies emergence on the plate, count the number of bacteria and process the data to generate a graph. Please click here to view a larger version of this figure.
Figure 1. Schematic representation of Psm ES4326 syringe infiltration assay on Arabidopsis plants. Leaves number 5 and 6 of adult soil-grown Arabidopsis plants are marked and infected with Psm ES4326 through syringe infiltration. After the emergence of disease symptoms, detach leaves and harvest leaf discs for tissue homogenization. Well-homogenized tissue is serially diluted in 96-well plates before transferring onto a bacteria counting plate. After colonies emergence on the plate, count the number of bacteria and process the data to generate a graph. Please click here to view a larger version of this figure.
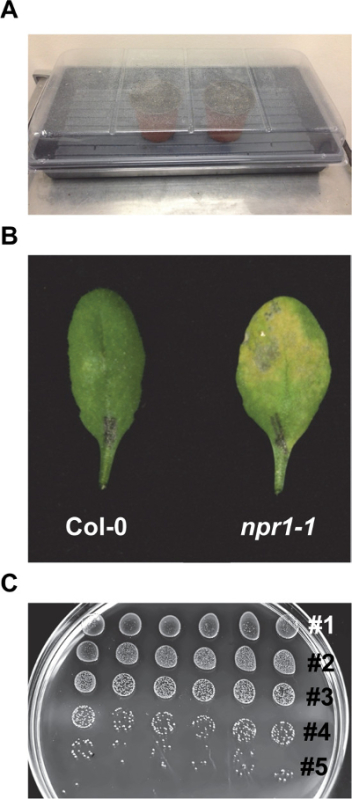 Figure 2. Representative procedures of the EDS assay. (A) Pots are covered with a water-sprayed transparent dome after sowing seeds. (B) Representative symptoms on Col-0 and npr1-1 leaves subjected to the EDS assay (OD600nm = 0.0002) 3 days post inoculation. Note severe chlorosis on npr1-1 and the nearly normal appearance of Col-0. (C) Serial dilutions of Psm ES4326 growing on KB (50 µg/ml streptomycin) media plate after ~ 45 hR of incubation. Five dilutions are visible. Please click here to view a larger version of this figure.
Figure 2. Representative procedures of the EDS assay. (A) Pots are covered with a water-sprayed transparent dome after sowing seeds. (B) Representative symptoms on Col-0 and npr1-1 leaves subjected to the EDS assay (OD600nm = 0.0002) 3 days post inoculation. Note severe chlorosis on npr1-1 and the nearly normal appearance of Col-0. (C) Serial dilutions of Psm ES4326 growing on KB (50 µg/ml streptomycin) media plate after ~ 45 hR of incubation. Five dilutions are visible. Please click here to view a larger version of this figure.
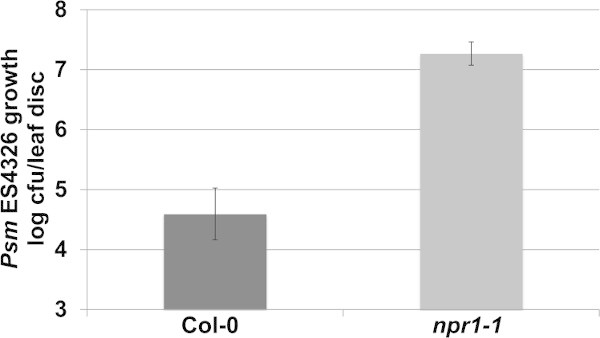 Figure 3. Representative results of the EDS assay. Psm ES4326 growth (colony forming units – cfu/leaf disc, expressed on a log scale) was quantified in 4-week-old Col-0 and npr1-1 plants 3 days post inoculation (OD600nm = 0.0002). Error bars represent 95% confidence intervals of the mean (n = 6). Please click here to view a larger version of this figure.
Figure 3. Representative results of the EDS assay. Psm ES4326 growth (colony forming units – cfu/leaf disc, expressed on a log scale) was quantified in 4-week-old Col-0 and npr1-1 plants 3 days post inoculation (OD600nm = 0.0002). Error bars represent 95% confidence intervals of the mean (n = 6). Please click here to view a larger version of this figure.
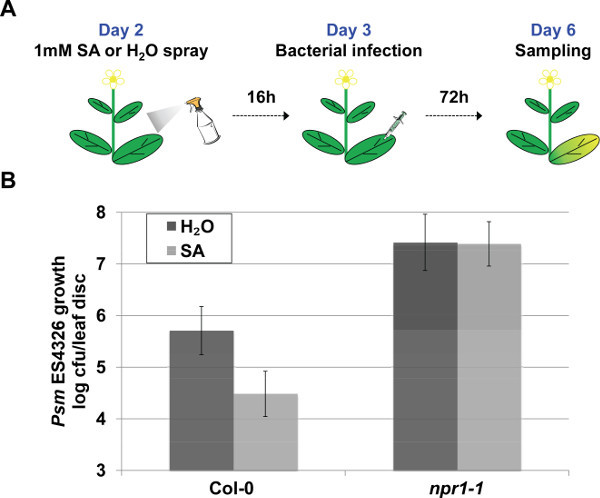 Figure 4. Representative procedure and results of the STI assay. (A) Schematic representation of the Salicylic Acid-Triggered Immunity (STI) assay. (B) Salicylic Acid-Triggered Immunity was quantified based on pathogen growth in plants pre-treated with 1mM Sodium Salicylate or H2O 16 hr prior to Psm ES4326 syringe infiltration (OD600nm = 0.001). Pathogen growth was quantified 3 days post inoculation. Error bars represent 95% confidence intervals of the mean (n = 6). Please click here to view a larger version of this figure.
Figure 4. Representative procedure and results of the STI assay. (A) Schematic representation of the Salicylic Acid-Triggered Immunity (STI) assay. (B) Salicylic Acid-Triggered Immunity was quantified based on pathogen growth in plants pre-treated with 1mM Sodium Salicylate or H2O 16 hr prior to Psm ES4326 syringe infiltration (OD600nm = 0.001). Pathogen growth was quantified 3 days post inoculation. Error bars represent 95% confidence intervals of the mean (n = 6). Please click here to view a larger version of this figure.
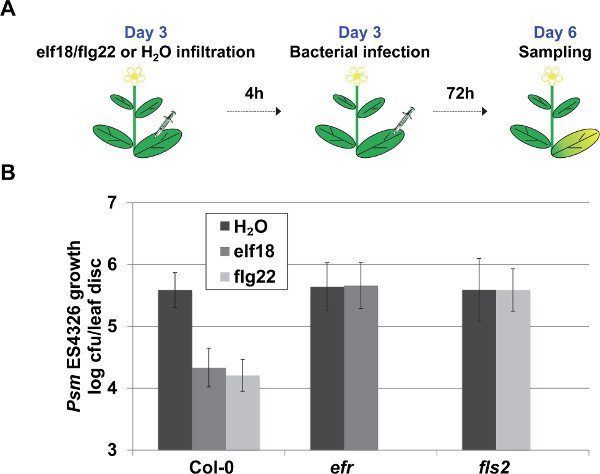 Figure 5. Representative procedure and results of the MTI assay. (A) Schematic representation of the MAMP-Triggered Immunity (MTI) assay. (B) MTI was quantified by pathogen growth in plants pre-treated with 1µM solution of flg22, elf18 or H2O (as control) 4 h prior to Psm ES4326 syringe infiltration (OD600 nm = 0.001). Pathogen growth was quantified 3 days post inoculation. Error bars represent 95% confidence intervals of the mean (n = 6). Please click here to view a larger version of this figure.
Figure 5. Representative procedure and results of the MTI assay. (A) Schematic representation of the MAMP-Triggered Immunity (MTI) assay. (B) MTI was quantified by pathogen growth in plants pre-treated with 1µM solution of flg22, elf18 or H2O (as control) 4 h prior to Psm ES4326 syringe infiltration (OD600 nm = 0.001). Pathogen growth was quantified 3 days post inoculation. Error bars represent 95% confidence intervals of the mean (n = 6). Please click here to view a larger version of this figure.
| Issue | Possible Causes | Recommended Actions |
| No bacteria growth in the liquid medium after O/N culture | Reduced bacteria activity | Initiate liquid culture from a newly streaked plate |
| Circular syringe impression presents on the leaf after syringe infiltration | Too much pressure during infiltration | Reduce the pressue during infiltration |
| Partial syringe impression presents on the leaf after syringe infiltration | Inappropriate positioning of the syringe | Adjust the syringe to be positioned vertical against the leaf surface |
| Leaf wilts wthtin few hours after infiltration | Too much pathogen solution is infiltrated into the leaf | Stop infiltrating immediately after the entire leaf surface turns darker green in color |
| No disease symptoms three days after infection | Humidity is too low for pathogen to proliferate | Increase the humidity where infected plants are maintained |
| Pathogen enters necrotrophic stage at or before 3 dpi | Incorrect concentration of pathogen solution | Use OD600nm = 0.0002 for EDS assay; confirm dilution using indepedent spectrophotometer |
| Droplets merge on the top of bacteria counting plate | Bacteria counting plate is not appropriately dried | Pre-dry the plate O/N before use |
| Pathogen dilution does not represent 10-fold reduction | Dilution is not accurately performed | Use a well calibrated multi-channel pipette for transferring liquid; confirm that all liquid is dispensed |
| Pathogen growth in the wild type exceeds 8 log cfu/leaf disc | Pathogen overgrew within the infected tissue - sampling performed too late | Sample infected tissue after the emergence of chlorosis |
| Big variation among technical replicates | Plants are not at the same developmental stage or infiltrated leaf number is inconsistent | Only use plants within the same developmental stage for infection. Confirm synchronous germination. Infect consistent leaf number among different plants |
Table 1. Troubleshooting of the syringe infiltration assay.
Please click here to view Table 2.
Table 2. Spreadsheet for statistical analysis of pathogen growth data.
Discussion
With decreasing available farmland and increasing population, researchers around the world are challenged with pressing needs for crop improvement. The yield can be greatly influenced by various biotic and abiotic stresses. Among them, pathogen infection is one of the leading causes of crop yield reduction, responsible for approximately 12% losses in the U.S. alone45. To resolve this issue, massive research has been conducted in the model Arabidopsis - P. syringae pathosystem to comprehensively characterize the components and regulatory mechanisms of the plant immune responses. Here, we present an optimized P. syringae ES4326 syringe infiltration assay to quantitatively assess the subtle differences in plant immune response on the whole plant level.
Though multiple pathogen infection assays have been developed to characterize the plant immune response29,35, the syringe infiltration provides a well-controlled system where equal amounts of pathogen are administered into the infected tissue. In other infection assays, including dip inoculation and spray inoculation, the pathogen is applied to the entire aerial tissue, including cotyledons as well as true leaves34. It has been reported that cotyledons from ecotype Landsberg erecta (Ler) are much more susceptible to the pathogen during dip inoculation assay, which may skew the data and lead to mistaken conclusions on the overall disease resistance34. In addition, the two aforementioned assays require pathogen to enter the leaf through stomata, which is controlled by various environmental factors, including humidity and light. Fluctuation of these factors contributes to a higher level of variation in the disease symptoms compared to the syringe infiltration assay35. For vacuum infiltration, the main drawbacks include the need for large volumes of pathogen solution, difficulty to effectively infiltrate all leaves while avoid damage, and considerable labor intensity29. Moreover, plants infected with spray, dip or vacuum inoculation are less likely to recover since the whole seedling is infected with pathogen. Given the numerous limitations of other methods, the syringe infiltration assay remains the method of choice to achieve uniform and highly predictable disease progression and accurate quantification of the pathogen to reliably characterize the immune response within the true leaf. In addition, this assay can be easily modified to characterize MAMP-Triggered Immunity and Systemic Acquired Resistance. Besides that, promising future applications of the technique could be applied for testing effects of multiple phytohormones, chemicals or inhibitors pre-treatments followed by pathogen syringe infiltration to dissect different layers of plant immune signaling network or identify roles of a specific pathway in the plant immune response. Infection with an increased dose of the pathogen (OD600nm=0.001) in the absence of any defense priming pre-treatments, known as the Enhanced Disease Resistance (EDR) test, is another useful modification of this method that can be used to identify mutants or transgenics with enhanced disease resistance.
It should be noted that syringe infiltration assay does not apply to the characterization of pre-invasive stage of defense response since the pathogen is delivered directly into the leaf tissue via stomata. Stomatal closure, which serves as the physical barrier for pathogen entrance, is often triggered to restrict pathogen entry upon the establishment of MAMP-Triggered Immunity 46. In addition, phytohormone abscisic acid contributes to stomatal closure during pre-invasive stage47. Therefore, the spraying or dipping methods may prove advantageous to characterize the stomatal layer of the immune response, despite caveats associated with those types of inoculations, discussed above29.
Since plant-pathogen interactions can be altered by various biotic and abiotic factors, it is critical to focus on the below listed critical steps in the protocol to achieve successful infection and obtain reliable outputs:
Plant status and bacteria activity
Arabidopsis plants can be easily stressed by various abiotic factors, including sub-optimal temperature, drought and osmotic stress, and trigger cellular responses that interfere with the immune output48. Therefore, it is crucial to grow plants under optimal growth conditions and protect them from the environmental stressors. In addition, since the developmental stage of the infected leaf may also alter the experimental outcome, it is necessary to consistently infect leaves number 5 and 6 to avoid artifacts. Progressing along developmental stages, elevated resistance against some virulent and avirulent pathogens correlates with the transition from vegetative stage to reproductive stage of Arabidopsis plants. This dynamic resistance over developmental stage is termed Age-Related Resistance (ARR)38. To avoid the ARR interference with the true immune response, it is critical to perform the infection at the correct developmental stage as indicated in the protocol and refrain from attempting infection experiments after the onset of the reproductive stage. Moreover, bacterial fitness and activity substantially affect the pathogen proliferation within the infected tissue. Thus, it is important to initiate the bacterial liquid culture from a freshly streaked culture plate that is no more than 2 days old.
Syringe infiltration skill
During the syringe infiltration, try to avoid any physical damage to the leaf since wounding effect is well known to interfere with the plant immune response, mainly via jasmonic acid-mediated signaling that can suppress SA-triggered defense pathways49. After the infection, a leaf that was infiltrated should fully recover to the normal physical appearance without any visual damage. For beginners, it is advised to practice this technique using non-experimental plants first before performing the Psm ES4326 syringe infiltration assay.
Pathogen extraction and dilution
To accurately measure the pathogen growth, it is vital to efficiently extract pathogen out of the infected plant tissue. Compared with other extraction methods, mechanical tissue homogenization through high-throughput homogenizers effectively extracts pathogen without the loss of tissue sample and cross-contamination. Subsequently, sample dilutions should be precisely carried out with a reliably calibrated multi-channel pipette to reduce the pipetting error. Deviation as little as ±1 µl in the dilution process would translate into a ~5% difference in the pathogen growth quantification. For more troubleshooting examples of the syringe infiltration assay, please see Table 1.
In conclusion, we describe the optimized the Psm ES4326 syringe infiltration assay on Arabidopsis to quantitatively and reliably assess the plant immune response. With the help of this pathosystem, understanding of plant-pathogen interaction will be accelerated in the laboratory and eventually be applied for crop protection in the field.
Disclosures
The authors have nothing to disclose.
Acknowledgments
We thank Dr. Shahid Mukhtar for critiquing the manuscript and Dr. Xinnian Dong for the sample data analysis file. This work is supported by a NSF-CAREER award (IOS-1350244) to KPM and the UAB Biology Department.
References
- Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack KE, Jones JD. Plant disease resistance genes. Annual review of plant biolog. 1997;48:575–607. doi: 10.1146/annurev.arplant.48.1.575. [DOI] [PubMed] [Google Scholar]
- Pontier D, Balague C, Roby D. The hypersensitive response. A programmed cell death associated with plant resistance. C R Acad Sci II. 1998;321:721–734. doi: 10.1016/s0764-4469(98)80013-9. [DOI] [PubMed] [Google Scholar]
- Ryals JA, et al. Systemic Acquired Resistance. Plant Cel. 1996;8:1809–1819. doi: 10.1105/tpc.8.10.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. The plant immune system. Natur. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Newman MA, Sundelin T, Nielsen JT, Erbs G. MAMP (microbe-associated molecular pattern) triggered immunity in plants. Front Plant Sc. 2013;4:139. doi: 10.3389/fpls.2013.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritig B, Heitz T, Legrand M. Antimicrobial proteins in induced plant defense. Curr Opin Immuno. 1998;10:16–22. doi: 10.1016/s0952-7915(98)80025-3. [DOI] [PubMed] [Google Scholar]
- Nicaise V, Roux M, Zipfel C. Recent advances in PAMP-triggered immunity against bacteria: pattern recognition receptors watch over and raise the alarm. Plant Physio. 2009;150:1638–1647. doi: 10.1104/pp.109.139709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, et al. Bacterial disease resistance in Arabidopsis through flagellin perception. Natur. 2004;428:764–767. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]
- Kunze G, et al. The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cel. 2004;16:3496–3507. doi: 10.1105/tpc.104.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gust AA, et al. Bacteria-derived peptidoglycans constitute pathogen-associated molecular patterns triggering innate immunity in Arabidopsis. J Biol Che. 2007;282:32338–32348. doi: 10.1074/jbc.M704886200. [DOI] [PubMed] [Google Scholar]
- Alfano JR, Collmer A. Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu Rev Phytopatho. 2004;42:385–414. doi: 10.1146/annurev.phyto.42.040103.110731. [DOI] [PubMed] [Google Scholar]
- Tyler BM. Entering and breaking: virulence effector proteins of oomycete plant pathogens. Cell Microbio. 2009;11:13–20. doi: 10.1111/j.1462-5822.2008.01240.x. [DOI] [PubMed] [Google Scholar]
- He P, et al. Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cel. 2006;125:563–575. doi: 10.1016/j.cell.2006.02.047. [DOI] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, et al. AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr Bio. 2009;19:423–429. doi: 10.1016/j.cub.2009.01.054. [DOI] [PubMed] [Google Scholar]
- Zhang Z, et al. Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microb. 2012;11:253–263. doi: 10.1016/j.chom.2012.01.015. [DOI] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: shaping the evolution of the plant immune response. Cel. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Jones DA, Takemoto D. Plant innate immunity - direct and indirect recognition of general and specific pathogen-associated molecules. Curr Opin Immuno. 2004;16:48–62. doi: 10.1016/j.coi.2003.11.016. [DOI] [PubMed] [Google Scholar]
- Elmore JM, Lin ZJ, Coaker G. Plant NB-LRR signaling: upstreams and downstreams. Curr Opin Plant Bio. 2011;14:365–371. doi: 10.1016/j.pbi.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann W, Bhattacharjee S. Effector-triggered immunity signaling: from gene-for-gene pathways to protein-protein interaction networks. Mol Plant Microbe Interac. 2012;25:862–868. doi: 10.1094/MPMI-01-12-0024-IA. [DOI] [PubMed] [Google Scholar]
- Wang D, Weaver ND, Kesarwani M, Dong X. Induction of protein secretory pathway is required for systemic acquired resistance. Scienc. 2005;308:1036–1040. doi: 10.1126/science.1108791. [DOI] [PubMed] [Google Scholar]
- Bednarek P. Chemical warfare or modulators of defence responses - the function of secondary metabolites in plant immunity. Curr Opin Plant Bio. 2012;15:407–414. doi: 10.1016/j.pbi.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Heath MC. Hypersensitive response-related death. Plant Mol Bio. 2000;44:321–334. doi: 10.1023/a:1026592509060. [DOI] [PubMed] [Google Scholar]
- Fu ZQ, Dong X. Systemic acquired resistance: turning local infection into global defense. Annu Rev Plant Bio. 2013;64:839–863. doi: 10.1146/annurev-arplant-042811-105606. [DOI] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cel. 1997;88:57–63. doi: 10.1016/s0092-8674(00)81858-9. [DOI] [PubMed] [Google Scholar]
- Wu Y, et al. The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Re. 2012;1:639–647. doi: 10.1016/j.celrep.2012.05.008. [DOI] [PubMed] [Google Scholar]
- Smith JM, Heese A. Rapid bioassay to measure early reactive oxygen species production in Arabidopsis leave tissue in response to living Pseudomonas syringae. Plant Method. 2014;10:6. doi: 10.1186/1746-4811-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura K, et al. A bacterial virulence protein suppresses host innate immunity to cause plant disease. Scienc. 2006;313:220–223. doi: 10.1126/science.1129523. [DOI] [PubMed] [Google Scholar]
- Katagiri F, Thilmony R, He SY. The Arabidopsis thaliana-Pseudomonas syringae interaction. The Arabidopsis book/American Society of Plant Biologist. 2002;1 doi: 10.1199/tab.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada H, Suzuki F, Matsuda I, Saitou N. Phylogenetic analysis of Pseudomonas syringae pathovars suggests the horizontal gene transfer of argK and the evolutionary stability of hrp gene cluster. J Mol Evo. 1999;49:627–644. doi: 10.1007/pl00006584. [DOI] [PubMed] [Google Scholar]
- Xin XF, He SY. Pseudomonas syringae pv. tomato DC3000: a model pathogen for probing disease susceptibility and hormone signaling in plants. Annu Rev Phytopatho. 2013;51:473–498. doi: 10.1146/annurev-phyto-082712-102321. [DOI] [PubMed] [Google Scholar]
- Dong X, Mindrinos M, Davis KR, Ausubel FM. Induction of Arabidopsis defense genes by virulent and avirulent Pseudomonas syringae strains and by a cloned avirulence gene. Plant Cel. 1991;3:61–72. doi: 10.1105/tpc.3.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey D, Holt 3rd BF, Wiig A, Dangl JL. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cel. 2002;108:743–754. doi: 10.1016/s0092-8674(02)00661-x. [DOI] [PubMed] [Google Scholar]
- Tornero P, Dangl JL. A high‐throughput method for quantifying growth of phytopathogenic bacteria in Arabidopsis thaliana. The Plant Journa. 2001;28:475–481. doi: 10.1046/j.1365-313x.2001.01136.x. [DOI] [PubMed] [Google Scholar]
- Ishiga Y, Ishiga T, Uppalapati SR, Mysore KS. Arabidopsis seedling flood-inoculation technique: a rapid and reliable assay for studying plant-bacterial interactions. Plant Method. 2011;7:32. doi: 10.1186/1746-4811-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XY, et al. Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microb. 2012;11:587–596. doi: 10.1016/j.chom.2012.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajerowska-Mukhtar KM, et al. The HSF-like transcription factor TBF1 is a major molecular switch for plant growth-to-defense transition. Curr Bio. 2012;22:103–112. doi: 10.1016/j.cub.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusterucci C, et al. Age-related resistance to Pseudomonas syringae pv. tomato is associated with the transition to flowering in Arabidopsis and is effective against Peronospora parasitica. Physiological and molecular plant patholog. 2005;66:222–231. [Google Scholar]
- Boyes DC, et al. Growth stage–based phenotypic analysis of Arabidopsis a model for high throughput functional genomics in plants. The Plant Cell Onlin. 2001;13:1499–1510. doi: 10.1105/TPC.010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regna PP, Wasselle LA, Solomons IA. The stability of streptomycin. Journal of Biological Chemistr. 1946;165:631–638. [PubMed] [Google Scholar]
- Wang W, et al. Timing of plant immune responses by a central circadian regulator. Natur. 2011;470:110–114. doi: 10.1038/nature09766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J. Modulation of plant immunity by light, circadian rhythm, and temperature. Current opinion in plant biolog. 2013;16:406–413. doi: 10.1016/j.pbi.2013.06.017. [DOI] [PubMed] [Google Scholar]
- Cuppels DA. Chemotaxis by Pseudomonas syringae pv. tomato. Appl Environ Microbio. 1988;54:629–632. doi: 10.1128/aem.54.3.629-632.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qutob D, et al. Phytotoxicity and innate immune responses induced by Nep1-like proteins. The Plant Cell Onlin. 2006;18:3721–3744. doi: 10.1105/tpc.106.044180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel D, Lach L, Zuniga R, Morrison D. Environmental and economic costs of nonindigenous species in the United States. BioScienc. 2000;50:53–65. [Google Scholar]
- Zeng W, Melotto M, He SY. Plant stomata: a checkpoint of host immunity and pathogen virulence. Current opinion in biotechnolog. 2010;21:599–603. doi: 10.1016/j.copbio.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton J, Flors V, Mauch-Mani B. The multifaceted role of ABA in disease resistance. Trends in plant scienc. 2009;14:310–317. doi: 10.1016/j.tplants.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Mahajan S, Tuteja N. Cold salinity and drought stresses: an overview. Archives of biochemistry and biophysic. 2005;444:139–158. doi: 10.1016/j.abb.2005.10.018. [DOI] [PubMed] [Google Scholar]
- León J, Rojo E, Sánchez‐Serrano JJ. Wound signalling in plants. Journal of Experimental Botan. 2001;52:1–9. doi: 10.1093/jexbot/52.354.1. [DOI] [PubMed] [Google Scholar]


