Abstract
High-grade serous ovarian cancer (HGSC), the cause of widespread peritoneal metastases, continues to have an extremely poor prognosis; fewer than 30% of women are alive 5 years after diagnosis. The omentum is a preferred site of HGSC metastasis formation. Despite the clinical importance of this microenvironment, the contribution of omental adipose tissue to ovarian cancer progression remains understudied. Omental adipose is unusual in that it contains structures known as milky spots, which are comprised of B, T, and NK cells, macrophages, and progenitor cells surrounding dense nests of vasculature. Milky spots play a key role in the physiologic functions of the omentum, which are required for peritoneal homeostasis. We have shown that milky spots also promote ovarian cancer metastatic colonization of peritoneal adipose, a key step in the development of peritoneal metastases. Here we describe the approaches we developed to evaluate and quantify milky spots in peritoneal adipose and study their functional contribution to ovarian cancer cell metastatic colonization of omental tissues both in vivo and ex vivo. These approaches are generalizable to additional mouse models and cell lines, thus enabling the study of ovarian cancer metastasis formation from initial localization of cells to milky spot structures to the development of widespread peritoneal metastases.
Keywords: Medicine, Issue 104, milky spots, omentum, ovarian cancer, peritoneal adipose, experimental metastasis, metastatic colonization
Introduction
Unlike most solid tumors, metastases from high-grade serous ovarian cancer (HGSC) are limited to the peritoneal cavity 1. Thus, effective peritoneal therapies could potentially control or eradicate HGSC. Currently, a standard therapeutic approach is surgical cytoreduction combined with chemotherapy 1-3. Unfortunately, the vast majority of patients experience and succumb to complications of disease recurrence. These dismal statistics show the need for improved understanding of metastatic colonization, the process by which cancer cells localize to, utilize, and proliferate within host tissues to form metastases 2.
The omentum is a preferred early site of HGSC metastasis 4-7. Unlike other peritoneal adipose, omental adipose tissue contains unusual immune structures known as milky spots, which contain B, T, and NK cells and macrophages, which play key roles in peritoneal homeostasis8,9. In addition to their physiologic functions, we found that milky spots play an active role in ovarian cancer metastatic colonization 4. In experimental metastasis assays, SKOV3ip.1, CaOV3, and HeyA8 (human) and ID8 (murine; C57BL/6) ovarian cancer cells rapidly home to milky spots, suggesting that the cells are moving toward a secreted chemotactic factor. Interestingly, cancer cells do not colonize peritoneal adipose lacking milky spots (i.e., gonadal and uterine fat) 4.
In order to identify mechanisms regulating milky spot colonization, we have optimized xenograft models that enable the interrogation of cellular and molecular events over the time course of metastatic colonization 4. A specific advantage of the approaches described herein is its emphasis on tissue architecture and function, which enables users to test hypotheses in fully integrated in vivo and ex vivo models of metastatic colonization 4,10. By comparing cancer cell localization and growth in peritoneal fat depots which either contain or lack milky spots, investigators can test the relative contribution(s) of adipocytes and cells within milky spots to the lodging and progressive growth of ovarian cancer cells in physiologically relevant tissues.
Protocol
All mice were housed, maintained and euthanized according to Institutional Animal Care and Use Committee (IACUC) guidelines and under the supervision of the University of Chicago Animal Resource Center.
1. Preparing Animals for Experimental Studies
Allow animals to acclimate to new housing and environment, to recover from potential physiologic effects of transport and handling. Note: The following technique is applicable to all commercially available strains of mice. The number of animals to be used for an experiment is dependent on the study design and should be done with consultation from a statistician.
2. Identification and Isolation of Peritoneal Fat Depots.
Euthanize animals via CO2 overdose and a secondary physical method to confirm death.
As harvesting peritoneal fat depots constitutes internal organ removal (secondary method), make a midline incision close to the inguinal papillae through the peritoneal wall to expose the internal organs (Figure 1A).
For the Omental Fat (OM), expose the omental/pancreatic complex by extending the spleen from the peritoneal cavity with forceps (Figure 1C). Release omentum from the pancreas and spleen by closely trimming all tissue connections. Ensure that any remaining pancreatic tissue is excised 4.
For the Gonadal Fat (GF), use forceps to lift the gonadal fat surrounding the ovaries and excise by cutting tissue connections (Figure 1A upper right) 4.
For the Uterine Fat (UF), use forceps to lift the uterine fat surrounding the uterine horns and excise by cutting tissue connections (Figure 1A lower right) 4.
For the Mesenteric Fat (MF), cut the junction between the small intestine and the pylorus. Use forceps to firmly grip the free end of the small intestine, and slowly “peel” it away from the mesenteric fat. Release the mesenteric fat from the mesenteric root using dissecting scissors (Figure 1A lower left) 4.
For the Splenoportal Fat (SF), lift the distal end of the spleen using forceps to expose the thin fatty band of tissue connecting the hilum of the spleen to the pancreas. Excise the splenoportal fat by first releasing it from the pancreas, and then carefully dissecting it from the spleen (Figure 1B) 4.
3. Milky Spot Identification Using Giemsa Staining
Excise omenta (refer to step 2.3) from C57BL/6 mice and fix in 5% neutral buffered formalin at 4 °C for 16 hr (O/N).
- For paraffin embedding the fixed tissue, choose a mold that is appropriate for the size of the tissue. Pour molten paraffin from the paraffin reservoir to fill the mold. Open the cassette and using warm forceps, transfer the tissue into the mold.
- Carefully orient the tissues during paraffin embedding to produce longitudinal sections. Place the labeled tissue cassette over the molding and press firmly in place. Allow the mold to cool for at least 30 min.
Cut serial sections (4 µm thick) from the formalin-fixed paraffin embedded (FFPE) tissue block. Place a precleaned slide under the sections as the ribbon of tissue comes off the paraffin block.
Serially section the whole omentum; collect and stain every third section for Giemsa stain (refer to 3.6). Allow the glass slide with the sections to air dry, preferably O/N at RT. Dried slides are ready for fixation. Stain the first section for hematoxylin and eosin for comparisons to Giemsa stained slides.
- Deparaffinize the slides by two sequential 5 min incubations in xylenes. Rehydrate slides by two sequential incubations in the following: 100% ethanol, 95% ethanol, and finally tap water.
- Take adequate precautions to avoid the slides from drying. Perform all steps at RT.
Completely immerse the slides in a Coplin jar containing 5% Giemsa solution (w/v prepared in tap water) and incubate for 4 min at RT. Rinse slides in tap water for 60 sec, air-dry and mount with coverslip using mounting media.
Image the stained slides using a Whole Slide Scanner and process with manufacturer software. Quantify the milky spot volume in the Giemsa-stained omental sections using ImageJ software4.
4. Propagation and Preparation of Ovarian Cancer Cells for In vivo and Ex vivo Studies
Pre-warm 15 ml of cell growth medium to 37 °C in a heat bath. Prepare an appropriately sized tissue culture flask (e.g., 75 cm2 surface area) by labeling with name of cell line, passage number, date and person who prepared the frozen stock, and the date and person who is thawing/plating the cells. Note: In studies of metastasis such detailed information is crucial, as the date of freeze down and passage number can affect the metastatic phenotype.
Using the sterile environment of the biological safety hood, pipette 10 ml of medium into the T-75 culture flask. Remove a vial of cells from the liquid nitrogen system and check the label to confirm the cell type. Thaw only one vial of cells at a time by warming in 37 °C heat bath.
- Using sterile technique, pipette 1x106 thawed cells into the culture flask. Place the flask in the incubator to allow cells to attach. Gently rock the flask back and forth to form a uniform cell suspension.
- Place flask in tissue culture incubator and maintain at 37 °C in 5% CO2 environment. Allow cells to adhere O/N.
Aspirate media, and replace with fresh pre-warmed growth media. Place the flask in the incubator and allow cells to grow. Monitor cell growth daily by visual inspection every 2-3 days using an inverted microscope. Use standard cell culture techniques to passage cells upon reaching 70-80% confluence. Note: For experimental assays harvest the cells at 70% confluence to ensure that cells are actively proliferating.
Aspirate the culture medium and rinse with 10 ml of sterile phosphate buffered saline (PBS). Aspirate PBS and add 0.25% trypsin/EDTA (2 to 3 ml for a 75 cm2 surface area) and incubate 5 min at 37 ºC. Visually confirm that the cells have detached and add an equal volume of growth medium, and gently pipette cells up and down to obtain a single cell suspension. Note: It is critical that the growth medium contain serum as it inactivates trypsin. Prepare at least 2- to 4-fold more cells than the number required for a given experiment to compensate for loss of cells during subsequent steps
Determine the percentage of viable cells in the suspension via trypan blue exclusion assay using a hemacytometer or an automated cell counter. Note: Here, only use cell preparations in which ≥ 96% of cells are viable.
Transfer the cell suspension from the flask to a 50 ml conical tube. Pellet cells by centrifuging 5 min at 1,100 x g, at 4 oC.
Aspirate the supernatant and resuspend the cell pellet in PBS to a concentration of 2x106 cells per ml.
5. Intraperitoneal Injection of Cells
Note: In our experiments, mice are handled in a laminar airflow or biosafety cabinet within our barrier facility in order to limit the risk of pathogen exposure, In order to demonstrate this technique clearly, the procedures in the accompanying video were conducted in a laboratory approved for animal work. This particular technique does not require the animals to be under anesthesia. Under approved protocols, perform this technique on live animals. Use appropriate strains of mice for this study. For example: use immunocompromised, athymic nude mice for the study of human ovarian cancer cell (SKOV3ip.1, and HeyA8) colonization; use immunocompetent C57BL/6 mice for study of mouse ovarian cancer cells (ID8) colonization.
Load 500 µl of the single cell suspension into the syringe and put in the sterile capped 25 G needle. This reduces cell shearing prior to injection.
Pick the mouse up by the scruff of the neck and hold the tail using the palm and forefinger and fix the left hind leg between the ring and little finger (when the mouse is restrained with the left hand). Note: To avoid traumatizing abdominal organs, restrain the mouse well so that it cannot move during the injection.
Imagine a line across abdomen just above the knees, and locate a point on the animal’s right side and close to the midline. The point of entry is cranial to and slightly medial of the last nipple. Note: Inserting the needle on the mouse’s right side avoids the cecum and reduces the risk of puncturing the intestines11.
- Insert the needle at the lower lateral region of the mice’s abdomen to a depth of approximately 0.5 cm. Pull back on the plunger to confirm that the needle has not penetrated a blood vessel or other peritoneal organs.
- If any fluid is aspirated discard the syringe (and sample)11. A greenish-brown or yellow aspirate indicates needle penetration into the intestines or bladder, respectively.
Inject the sample using slow, steady pressure. Withdraw the needle and return the mouse to its cage. Do not recap syringe before disposal in sharps container.
Sacrifice mice via CO2 asphyxiation and vital organ removal at specific time points post injection.
6. Milky spot colonization In vivo
- Quantitation of cancer cell localization to milky spots using flow cytometry
- Isolate peritoneal fat depots from mice (as described in step 2.3 - 2.7) injected with fluorescently tagged cancer cells and immediate place tissues in ice-cold PBS.
- For this particular technique, weight-match all adipose fat tissues to omentum. Transfer tissues to separate 5 ml tubes containing 1.5 ml of serum-free DMEM and 0.1% (w/v) bovine serum albumin. Pool tissues harvested from three independent mice to ensure a sufficient yield of cells for flow cytometry.
- Mince tissues using surgical scissors and add 1.5 ml of serum-free DMEM containing 0.4% (w/v) collagenase I. Incubate the tissue suspension at 37 °C for 30 min with rotational mixing.
- As an optional step, further disassociate tissue by mastication. In this case, transfer the tissue-collagenase suspension to a microstomacher bag, masticate for 10 min on low, rotating the orientation of the bags after 5 min.
- Filter samples through a nylon mesh filter (60 µm pore) to remove larger debris. Collect cells via centrifugation at 250 x g for 5 min at 4 °C and discard the supernatant fraction.
- Resuspend the cell pellet in 100 µl of PBS combined with 900 µl of ACK lysis buffer and incubate at RT for 1 min. Centrifuge cells at 250 x g for 5 min and discard supernatant.
- Resuspend the cell pellet in 250 µl of ice-cold PBS. Filter the samples through a 60 µm pore nylon mesh to ensure single cell suspension. Rinse the filter with 250 µl of ice-cold PBS.
- Quantitate the number of fluorescently tagged cells in the population using a flow cytometer equipped with a 561-nm yellow-green laser and a 585 nm/15 nm bandpass filter. Set the gate for tdTomato-positive cells based on analysis of parental cells and/or appropriate negative controls 4.
- Quantitation of microscopic and macroscopic metastases
- At the desired experimental endpoint(s), isolate, and immediately place tissues in the appropriate fixation media. Fix larger samples, such as intact gonadal, uterine, and mesenteric fat depots in 10% neutral buffered formalin for 48 hr at 4 °C. Fix smaller samples (omental and splenoportal fat, as well as tissue equivalents of uterine and mesenteric fat) in 5% neutral buffered formalin at 4 °C for 2-16 hr.
- Alternatively, snap freeze tissues by placing them in a freezing medium such as OCT and drop into an appropriate amount of liquid nitrogen.
- Transfer fixed tissues to 70% ethanol and store at 4 °C until embedding in paraffin. Embed and process tissue as described in 3.2 and 3.3.
- Use the H&E stained section to evaluate tissues for the presence or absence of microscopic metastases (i.e., clusters of 50 cells). Confirm the presence of microscopic metastases by a secondary method such as an IHC stain for cytokeratin 8/18 to detect ovarian cancer cells.
7. Milky Spot Colonization Ex Vivo
- Establishment of basic ex vivo omental organ culture conditions
- Place each omentum into a single culture plate insert and place within one well of a twelve-well tissue culture plate containing 500 µl of DME/F12 media containing 20% FCS. Maintain organ cultures at 37 °C in a 5% CO2 environment for 24 to 48 hr and/or additional time points. Use three independent omenta (triplicate samples) for each condition (e.g., media type/time point).
- To confirm the integrity of tissues at the experimental endpoints, fix and process samples as described in Step 3. Counting healthy versus necrotic adipocytes on H&E sections can assess tissue integrity. A minimum of ~120 cells from 5 biological samples is required to formulate a percentage of live tissue present10.
- To measure tissue function, place omenta (n = 3) in separate wells of a 24-well plate containing 500 µl of DME/F12 media with 20% FCS. Allow omenta to condition the media at 37 °C in a 5% CO2 environment for 24 hr, and then remove 250 µl for use in an IL-6 ELISA assay10.
- Ex vivo co-culture of the mouse omentum with SKOV3ip.1-GFP cells
- Grow and prepare fluorescently tagged cells as described in section 4 and resuspend at a concentration of 2 × 106 cells per ml.
- Apply ~6 µl of the tissue adhesive to the membrane of the culture insert and allow it to air dry. Wash the membrane twice with sterile water to remove any excessive adhesive. Air dry the membranes under a laminar hood.
- Carefully excise the omenta and attach it to the adhesive-coated membrane using sterile forceps. Allow the tissue to adhere to the membrane for 1 min prior to adding media. Add 500 µl of the cell suspension on top of each omentum in each culture insert. Fill the area around the transwell chamber with 2.5 ml of DME/F-12 10.
- Incubate omenta with cell suspension for 6 hr at 37 °C in a 5% CO2 environment. Carefully remove and wash the omenta with ~10 ml of PBS. Visualize fluorescent cancer cell foci using an appropriate fluorescent imaging system 10.
Representative Results
Identification and Histologic Examination of Peritoneal Fat Depots
Gross anatomic dissection allows identification of four of the five primary sources of peritoneal fat (Figure 1A). Moving clockwise from the top center are: the omentum (OM; outlined) located over the stomach and spleen, the gonadal fat (GF) surrounding the left ovary (ov), the uterine fat (UF) attached to the uterine horns (uh) and the mesentery (MY) attached to the small intestine (si). The ovary (ov), uterine horn (uh), and small intestine (si) are indicated as points of reference. The splenoportal fat (SP) surrounds the splenic artery and connects the hilum of the spleen to the celiac artery (Figure 1B). Finally, the omentum is attached to the stomach and pancreas (Figure 1C). The gross structure and relative size of these tissues is shown in Figure 1D. Histologic examination of the tissues shows that splenoportal and omental fat contain milky spot [MS] structures at the tissue periphery, surrounded by adipocytes [A]; uterine, gonadal, and mesenteric fat do not (Figure 1E).
Milky spot identification using Giemsa Staining
To enable the comparison of overall milky spot content between different mouse strains, milky spot volume and area present in individual omenta was quantitated using a novel method. As described in Methods, a “digital whole mount” of the omenta was constructed by staining every third section of tissue in a 5% Giemsa solution and capturing images of the stained tissues (Figure 2A). Milky spots appear as regions staining dark blue (Figure 2B). The identity of these regions as milky spots was confirmed in serial sections by both H&E staining and IHC for cells positive for CD45, a common lymphocyte marker (Figure 2C and D, respectively). As described in Clark and Krishnan et al4, images (Figure 2A) were compiled and processed to construct a “digital whole mount” which could then be used to calculate the milky spot volume and area in individual omenta.
Quantitative and qualitative assessment of cancer colonization of omentum
A flow cytometry-based method was developed in order to quantitate the number of cancer cells present in adipose depots after intraperitoneal injection. This protocol is especially useful to study early colonization steps of the metastatic process. For example, in Figure 3A, ID8-tdTomato cells were prepared and injected intraperitoneally into C57BL/6 mice. At 7 days post injection (dpi), the adipose organs were harvested and dissociated into a single-cell suspension as described in the Methods. The number of tdTomato cells was quantified using flow cytometry (Figure 3A, left). The omentum showed an approximately 12-fold increase in the number of tdTomato-positive events, relative to PBS- injected controls (Figure 3A, right), but there was no significant increase in cell preparations from the gonadal fat, uterine fat, or mesentery. A complementary qualitative IHC-based approach also showed that 7 days after i.p. injection of SKOV3ip.1 cells, comparable cancer cell lesions were observed in both omental and splenoportal fat (Figure 3B). IHC for human pan-cytokeratin (pan-CK) confirms the presence of cancer cells within the milky spots. The specificity of IHC staining was confirmed using an IgG control for the pan-cytokeratin antibody (Figure 3B). ID8 ovarian cancer colonization of peritoneal fat depots was evaluated using C57BL/6 mice at 7 dpi. Large cancer cell lesions in the milky spots of both the omentum and splenoportal fat are outlined. Small clusters of cancer cells were occasionally seen in other fat depots, as illustrated in the panel inset. Thus, one can access both the relative number and location of cells within a given tissue.
Ex vivo assay to study ovarian cancer colonization of milky spots
In order to address the limitations of in vivo conditions while maintaining tissue composition and architecture, we optimized an ex vivo approach to study ovarian cancer cell colonization of omental tissues. Whole omental tissues excised from mice are successfully cultured ex vivo to study ovarian cancer interaction with milky spots. SKOV3ip.1-GFP cells were seeded onto the ex vivo cultured omental tissues and maintained at 37 °C for a period of 6 hr. GFP-positive fluorescent cells were visible in discreet foci in both ex vivo and in vivo omenta harvested 6 hr post injection of SKOV3ip.1-GFP cells (Figure 4A). Under both conditions, histological verification using H&E staining confirmed cancer cell localization to milky spots (Figure 4B). Confirmatory stain using cytokeratin stain for cancer cells showed the presence of cancer cell foci within the milky spots (Figure 4C). These data demonstrate the feasibility of using ex vivo approaches for mechanism-based studies to complement in vivo findings.
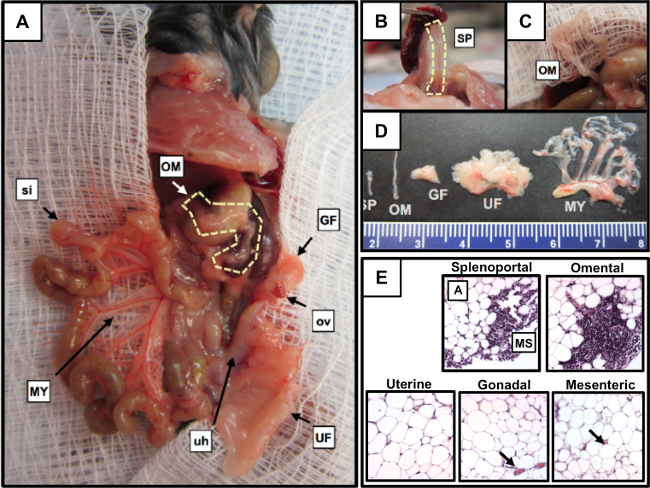 Figure 1. The relative locations and structures of the main peritoneal adipose depots. (A) Gross anatomic dissection showing the relative location of four of the five primary sources of peritoneal fat. The omentum (OM; outlined), gonadal fat (GF), uterine fat (UF), mesentery (MY), ovary (ov), uterine horns (uh), and small intestines (si) are shown. (B) Splenoportal fat (SP; outlined) exposed by lifting the spleen with forceps. (C) The mouse omentum shown dissected free from the pancreas to improve visualization. D: Relative sizes and structures of excised peritoneal fat; the mesentery is shown attached to the mesenteric root. E. Histologic evaluation of peritoneal fat for the presence of milky spots (A) adipocytes, (MS) milky spot. Please click here to view a larger version of this figure.
Figure 1. The relative locations and structures of the main peritoneal adipose depots. (A) Gross anatomic dissection showing the relative location of four of the five primary sources of peritoneal fat. The omentum (OM; outlined), gonadal fat (GF), uterine fat (UF), mesentery (MY), ovary (ov), uterine horns (uh), and small intestines (si) are shown. (B) Splenoportal fat (SP; outlined) exposed by lifting the spleen with forceps. (C) The mouse omentum shown dissected free from the pancreas to improve visualization. D: Relative sizes and structures of excised peritoneal fat; the mesentery is shown attached to the mesenteric root. E. Histologic evaluation of peritoneal fat for the presence of milky spots (A) adipocytes, (MS) milky spot. Please click here to view a larger version of this figure.
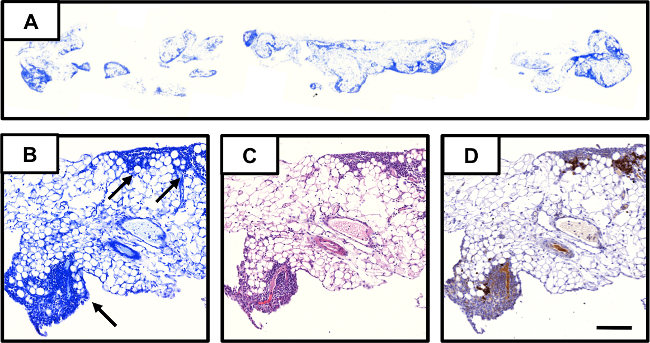 Figure 2. Giemsa-based approach for quantifying milky spots in omental tissue. (A) Image of section of whole omentum stained with Giemsa. Serial sections of omental tissue evaluated by: (B) Giemsa staining; milky spots are indicated with black arrows. (C) H&E staining; and, (D) IHC using anti-CD45 antibody to identify lymphocytes within the milky spot structure. The scale bar is the same for B-D and denotes 100 µm. Please click here to view a larger version of this figure.
Figure 2. Giemsa-based approach for quantifying milky spots in omental tissue. (A) Image of section of whole omentum stained with Giemsa. Serial sections of omental tissue evaluated by: (B) Giemsa staining; milky spots are indicated with black arrows. (C) H&E staining; and, (D) IHC using anti-CD45 antibody to identify lymphocytes within the milky spot structure. The scale bar is the same for B-D and denotes 100 µm. Please click here to view a larger version of this figure.
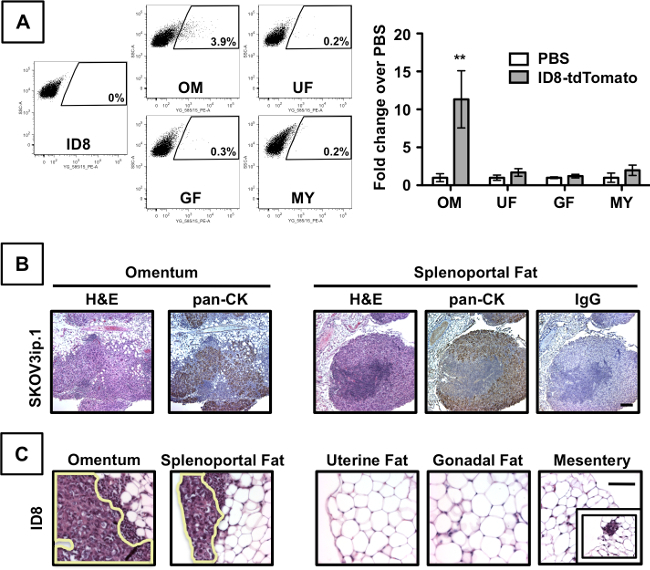 Figure 3. Ovarian cancer cells preferentially colonize peritoneal fat that contains milky spots. Flow cytometric analyses of omentum (OM), uterine fat (UF), gonadal fat (GF), and mesentery (MY) harvested from mice at 7 dpi of ID8-tdTomato cells (left). Data are presented as fold-change increase of tdTomato-postive cells over PBS-injected mice (right). Error bars indicate standard error of the mean. ** denotes p<0.01 compared to PBS controls. (B) Examination of tissues by both standard histology and IHC shows comparable colonization of milky spots in both omentum and splenoportal fat (after injection of 1x106 SKOV3ip.1 cells into Nude mice). Sections were evaluated by H&E staining. The presence of epithelial (cancer) cells within the lesions was confirmed by IHC detection of cytokeratin using a pan-cytokeratin (pan-CK) antibody. IHC using an IgG isotype antibody for pan-cytokeratin was used as a control for staining specificity. The scale bar is the same for all images and denotes 100 µm. (C) Evaluation of ID8 ovarian cancer colonization of peritoneal fat depots in C57BL/6 mice at 7 dpi. Please click here to view a larger version of this figure.
Figure 3. Ovarian cancer cells preferentially colonize peritoneal fat that contains milky spots. Flow cytometric analyses of omentum (OM), uterine fat (UF), gonadal fat (GF), and mesentery (MY) harvested from mice at 7 dpi of ID8-tdTomato cells (left). Data are presented as fold-change increase of tdTomato-postive cells over PBS-injected mice (right). Error bars indicate standard error of the mean. ** denotes p<0.01 compared to PBS controls. (B) Examination of tissues by both standard histology and IHC shows comparable colonization of milky spots in both omentum and splenoportal fat (after injection of 1x106 SKOV3ip.1 cells into Nude mice). Sections were evaluated by H&E staining. The presence of epithelial (cancer) cells within the lesions was confirmed by IHC detection of cytokeratin using a pan-cytokeratin (pan-CK) antibody. IHC using an IgG isotype antibody for pan-cytokeratin was used as a control for staining specificity. The scale bar is the same for all images and denotes 100 µm. (C) Evaluation of ID8 ovarian cancer colonization of peritoneal fat depots in C57BL/6 mice at 7 dpi. Please click here to view a larger version of this figure.
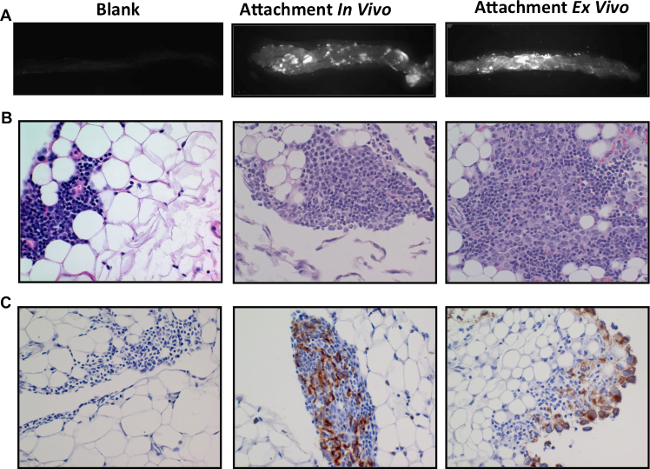 Figure 4. Ex vivo colonization of ovarian cancer cells to omentum.The columns represent a comparison of the three experimental conditions showing naïve omenta (left), omenta from mice 6 hr post i.p. injection of 1 x 106 SKOV3ip.1 cells (center) and omenta with SKOV3ip.1-GFP colonized under ex vivo conditions (right). Naïve omenta (Blank), omenta colonized by SKOV3ip.1-GFP from in vivo assay (attachment in vivo) or omenta colonized by SKOV3ip.1-GFP in ex vivo culture conditions (attachment ex vivo). Fluorescence imaging of whole omenta for GFP shows similar colonization pattern for both in vivo and ex vivo assays (A). H&E staining of serial sections of the omenta from panel A confirms the presence of cancer cells localized to the milky spots (B). Cytokeratin for ovarian cancer cells validates the H&E staining (C). Please click here to view a larger version of this figure.
Figure 4. Ex vivo colonization of ovarian cancer cells to omentum.The columns represent a comparison of the three experimental conditions showing naïve omenta (left), omenta from mice 6 hr post i.p. injection of 1 x 106 SKOV3ip.1 cells (center) and omenta with SKOV3ip.1-GFP colonized under ex vivo conditions (right). Naïve omenta (Blank), omenta colonized by SKOV3ip.1-GFP from in vivo assay (attachment in vivo) or omenta colonized by SKOV3ip.1-GFP in ex vivo culture conditions (attachment ex vivo). Fluorescence imaging of whole omenta for GFP shows similar colonization pattern for both in vivo and ex vivo assays (A). H&E staining of serial sections of the omenta from panel A confirms the presence of cancer cells localized to the milky spots (B). Cytokeratin for ovarian cancer cells validates the H&E staining (C). Please click here to view a larger version of this figure.
Discussion
Development of therapies to target disseminated cells requires a mechanistic understanding of metastatic colonization, the critical first step in the development of peritoneal disease. To address these issues we report approaches that can be used to discern how the omentum’s unique tissue composition and architecture promote ovarian cancer metastatic colonization. Distinguishing features of our approach are: 1) the focus on early events in the colonization process; 2) comparison of milky spot-containing and deficient adipose depots; 3) availability of a reproducible technique to quantitate milky spot volume in tissues; 4) availability of complementary in vivo and ex vivo approaches to evaluate ovarian cancer cell colonization of milky spot structures.
In conducting these studies we came to realize that investigators frequently are unaware of the location of the mouse omentum and the existence of the splenoportal fat band, which serves as a second source of milky spot-containing peritoneal fat. Although the techniques described herein enable studies of steps in metastatic colonization, they only offer “snapshots” of these processes, and not assessment of colonization in real time. If followed appropriately, the protocols as described should yield reproducible results to a range of investigators. Critical to the success of these techniques are: 1) Identifying and dissecting out specific peritoneal adipose tissues as contamination by other tissue types will skew quantitative data obtained from flow cytometry; and 2) Implementing appropriate strains of transgenic mice for the study in question. It should be noted that the time course studies should be determined for individual cell lines. Empirically as it depends on a variety of factors including proliferation rate, time to tumor take and metastatic potential.
In addition to dissecting the interactions of well-established ovarian cancer cell lines and their omental microenvironment that are important to metastasis formation, the technique described herein can also be used to evaluate the malignant potential of clinically derived cell lines that recapitulate early molecular alterations in serous tubal intraepithelial carcinomas in the fimbriae of the fallopian tubes 12-17. Although a pathway for the molecular progression of these lesions is emerging, in vivo studies are needed to determine the biological significance of specific alterations. Complementary in vivo and ex vivo assays can be used to assess specific steps in metastatic colonization of omental tissue by such model precursor cell lines Information gleaned from studies may be key to the generation of therapeutic hypotheses and the design of further preclinical studies.
Identifying mechanisms that control metastatic colonization may enable the prevention or control of metastasis formation. For example, women who are at increased risk for developing ovarian cancer 18-22 (i.e., BRCA1 or BRCA2 gene mutation carriers) often undergo prophylactic salpingo-oophorectomy to reduce the risk of developing peritoneal disease 23,24. Unfortunately, patients often still develop HGSC due to the outgrowth of ovarian cancer cells present within the omentum and other metastatic sites at the time of surgery. Identification of agents that disrupt omental tissue microenvironment could prevent cancer cell localization and/or growth within the omentum. Such approaches could potentially be used in combination with therapeutic agents that target ovarian cancer cells to prevent metastasis formation or suppress cancer regrowth (after surgical debulking).
Disclosures
The authors have nothing to disclose.
Acknowledgments
Supported by grants from the Department of Defense (W81XWH-09-1-0127), the NIH (2-R01-CA089569), the Elsa U. Pardee Foundation, a Marsha Rivkin Center for Ovarian Cancer Research Pilot Study Award, and generous philanthropic support from Section of Urology and Section of Research in the Department of Surgery, University of Chicago.
References
- Bast RC, Hennessy B, Mills GB. The biology of ovarian cancer: new opportunities for translation. Nature Reviews Cancer. 2009;9(6):415–428. doi: 10.1038/nrc2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowtell DDL. The genesis and evolution of high-grade serous ovarian cancer. Nature Reviews Cancer. 2010;10(11):803–808. doi: 10.1038/nrc2946. [DOI] [PubMed] [Google Scholar]
- Colgan TJ, Chang MC. Chapter 18 Familial Cancer and Prophylactic Surgery. Diagnostic Pathology of Ovarian Tumors. 2011. pp. 277–288.
- Clark R, et al. Milky Spots Promote Ovarian Cancer Metastatic Colonization of Peritoneal Adipose in Experimental Models. The American Journal of Pathology. 2013;183(2):576–591. doi: 10.1016/j.ajpath.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platell C, Cooper D, Papadimitriou JM, Hall JC. The omentum. World Journal Of Gastroenterology: WJG. 2000;6(2):169–176. doi: 10.3748/wjg.v6.i2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara A, et al. Milky spots as the implantation site for malignant cells in peritoneal dissemination in mice. Cancer Research. 1993;53(3):687–692. [PubMed] [Google Scholar]
- Gerber SA, et al. Preferential Attachment of Peritoneal Tumor Metastases to Omental Immune Aggregates and Possible Role of a Unique Vascular Microenvironment in Metastatic Survival and Growth. The American Journal of Pathology. 2010;169(5):1739–1752. doi: 10.2353/ajpath.2006.051222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebermann-Meffert D. The greater omentum. Anatomy, embryology, and surgical applications. The Surgical Clinics of North America. 2000;80(1):275–293. doi: 10.1016/s0039-6109(05)70406-0. [DOI] [PubMed] [Google Scholar]
- Collins D, Hogan AM, O’Shea D, Winter DC. The Omentum: Anatomical, Metabolic, and Surgical Aspects. Journal of Gastrointestinal Surgery. 2009;13(6):1138–1146. doi: 10.1007/s11605-009-0855-1. [DOI] [PubMed] [Google Scholar]
- Khan SM, et al. In vitro metastatic colonization of human ovarian cancer cells to the omentum. Clinical, & Experimental Metastasis. 2010;27(3):185–196. doi: 10.1007/s10585-010-9317-0. [DOI] [PubMed] [Google Scholar]
- Shimizu S. Routes of Administration. In: Hans H, editor. The Laboratory Mouse. Academic Press; 2004. pp. 527–542. [Google Scholar]
- Vang R, Shih I-M, Kurman RJ. Fallopian tube precursors of ovarian low- and high-grade serous neoplasms. Histopathology. 2012;62(1):44–58. doi: 10.1111/his.12046. [DOI] [PubMed] [Google Scholar]
- Folkins AK, Jarboe EA, Roh MH, Crum CP. Precursors to pelvic serous carcinoma and their clinical implications. Gynecologic Oncology. 2009;113(3):391–396. doi: 10.1016/j.ygyno.2009.01.013. [DOI] [PubMed] [Google Scholar]
- Li J, Fadare O, Xiang L, Kong B, Zheng W. Ovarian serous carcinoma: recent concepts on its origin and carcinogenesis. Journal of Hematology, & Oncology. 2012;5(1):8. doi: 10.1186/1756-8722-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurman RJ, Shih I-M. The origin and pathogenesis of epithelial ovarian cancer: a proposed unifying theory. The American Journal Of Surgical Pathology. 2010;34(3):433–443. doi: 10.1097/PAS.0b013e3181cf3d79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crum CP, et al. The distal fallopian tube: a new model for pelvic serous carcinogenesis. Current Opinion In Obstetrics, & Gynecology. 2007;19(1):3–9. doi: 10.1097/GCO.0b013e328011a21f. [DOI] [PubMed] [Google Scholar]
- Crum CP, et al. Lessons from BRCA: The Tubal Fimbria Emerges as an Origin for Pelvic Serous Cancer. Clinical Medicine, & Research. 2007;5(1):35–44. doi: 10.3121/cmr.2007.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes WD, Narod SA. Ovarian cancer risk and family history. The Lancet. 1997;349:878. doi: 10.1016/S0140-6736(05)61782-5. [DOI] [PubMed] [Google Scholar]
- Antoniou A, et al. Average Risks of Breast and Ovarian Cancer Associated with BRCA1 or BRCA2 Mutations Detected in Case Series Unselected for Family History: A Combined Analysis of 22 Studies. The American Journal of Human Genetics. 2003;72(5):1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M-C, Marks JH, Mandell JB. New York Breast Cancer Study Group Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302(5645):643–646. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- Kjær Krüger S, Gayther S. Ovarian cancer and genetic susceptibility in relation to the BRCA1 and BRCA2 genes. Occurrence, clinical importance and intervention. Acta Obstetricia et Gynecologica Scandinavica. 2006;85(1):93–105. doi: 10.1080/00016340500324621. [DOI] [PubMed] [Google Scholar]
- Easton DF, Ford D, Bishop DT. Breast and ovarian cancer incidence in BRCA1-mutation carriers. Breast Cancer Linkage Consortium. American Journal of Human Genetics. 1995;56(1):265–271. [PMC free article] [PubMed] [Google Scholar]
- Rebbeck TR, et al. Prophylactic Oophorectomy in Carriers of BRCA1or BRCA2Mutations. New England Journal of Medicine. 2002;346(21):1616–1622. doi: 10.1056/NEJMoa012158. [DOI] [PubMed] [Google Scholar]
- Olivier RI, et al. Clinical outcome of prophylactic oophorectomy in BRCA1/BRCA2 mutation carriers and events during follow-up. British Journal Of Cancer. 2004;90(8):1492–1497. doi: 10.1038/sj.bjc.6601692. [DOI] [PMC free article] [PubMed] [Google Scholar]


