Abstract
Patient derived xenograft (PDX) models are gaining popularity in cancer research and are used for preclinical drug evaluation, biomarker identification, biologic studies, and personalized medicine strategies. Circulating tumor cells (CTC) play a critical role in tumor metastasis and have been isolated from patients with several tumor types. Recently, CTCs have been used to generate PDX experimental models of breast and prostate cancer. This manuscript details the method for the generation of prostate cancer PDX models from CTCs developed by our group. Advantages of this method over conventional PDX models include independence from surgical sample collection and generating experimental models at various disease stages. Density gradient centrifugation followed by red blood cell lysis and flow cytometry depletion of CD45 positive mononuclear cells is used to enrich CTCs from peripheral blood samples collected from patients with metastatic disease. The CTCs are then injected into immunocompromised mice; subsequently generated xenografts can be used for functional studies or harvested for molecular characterization. The primary limitation of this method is the negative selection method used for CTC enrichment. Despite this limitation, the generation of PDX models from CTCs provides a novel experimental model to be applied to prostate cancer research.
Keywords: Medicine, Issue 104, Prostate cancer, patient derived xenograft, circulating tumor cells, intratumoral heterogeneity, preclinical models, human tissue samples
Introduction
Patient derived xenografts are increasingly popular experimental models used for cancer research. They can be used for characterization of biomarkers and biological pathways, pre-clinical evaluation of drug efficacy, and creation of avatars for personalized cancer therapies 1,2. Previously, other research groups have developed PDX models either by implanting or injecting single tumor cell suspensions or whole tumor explants into immunocompromised mice 1. These PDXmodels require surgical collection of fresh solid tumor, malignant ascites or pleural effusions from a patient undergoing a surgical procedure which is both costly and exposes the patient to increased risk of iatrogenic morbidity.
A significant recent development in cancer research has been the detection, isolation and characterization of circulating tumor cells. These tumor cells escape from the primary tumor mass and enter circulation where they play a critical role in metastasis and relapse, the most common cause of cancer related mortality 3. The evaluation and characterization of CTCs from several solid tumor types have provided clinical information for diagnosis, prognosis, and monitoring residual disease 3. A variety of currently used approaches relying on either the physical properties, expression of biomarkers, or functional characteristics of CTCs can be used to efficiently isolate CTCs 4. Existing macroscale CTC isolation methods include density gradient centrifugation, physical filtration with filter pores and separation against surface molecules. The most widely used CTC isolation methodologies are based on antibody-based capture of CTCs. Both positive and negative selection of cell surface markers can be employed to isolate CTCs from peripheral blood. Positive selection for CTCs in the peripheral circulation commonly uses epithelial markers (e.g., EpCAM) which are expressed on CTCs but not hematopoietic cells. The disadvantage of this method is that CTCs with metastatic potential have often undergone epithelial-to-mesenchymal transition (EMT), which downregulates epithelial surface markers 3. In order to isolate CTCs with metastatic potential, a negative selection methodology which employs the hematopoietic surface marker, CD45, to deplete the normal cell population of leukocytes can be used 5.
Prostate cancer is the most commonly diagnosed cancer and a major cause of cancer-related deaths in men 6. The mechanisms of tumor progression and aggressiveness are not completely understood and therefore the generation and characterization of experimental models that recapitulate the molecular heterogeneity of prostate cancer are of significant interest. PDX models of prostate cancer have beengenerated previously by engraftment of human prostate cancer cells into immunocompromised mice 7,8. However the generation of such models has been hampered by the low engraftment rate of prostate cancer into immunocompromised mice, which is primarily attributed to the indolent nature of the disease. Recently, CTCs have been used to generate breast cancer 9, lung cancer 10 and prostate cancer 11 PDX models. These proof-of-concept studies introduced the possibility of generating PDX models independent of the need for surgical sample collection. In this article we describe in detail a method for the generation of this novel experimental model.
Protocol
This protocol has been conducted at our institution with approval from the institutional research ethics board and is in compliance with all institutional, national, and international guidelines for human welfare.
1. Collection of Peripheral Blood from Patients with Advanced Prostate Cancer
Note: Select patients with metastatic prostate cancer. Obtain written patient consent and record clinical characteristics of patients, including age at isolation and previous chemotherapy and hormonal treatments. Patients with metastatic disease will potentially have highest concentration of CTCs in peripheral blood.
Collect blood samples after informed consent and under an appropriate Institutional Review Board approved protocol. Wear latex gloves and a laboratory coat throughout the procedure.
Connect 25 G butterfly needle and tubing to needle holder. Use an alcohol swab to clean area of antecubital vein between bicep and forearm, then apply tourniquet to patient’s upper arm.
Insert butterfly needle into antecubital vein and push blood collection tube into needle holder to start collection. Collect approximately 10 ml of blood in collection tubes that contain ethylenediaminetetraacetic acid (EDTA) to prevent clotting.
Remove butterfly needle from patient and apply pressure with sterile gauze pad over site of vein puncture for 1 min. Cover site with a sterile bandage.
2. Isolation of Mononuclear Cells
Note: The blood collected from step 1 will contain peripheral blood mononuclear cells (PBMCs) (e.g., lymphocytes and monocytes) in addition to the mononuclear CTCs.
Pipette whole blood into 50 ml polystyrene conical tube with a 5 ml serological pipette and add Hanks Balanced Salt Solution (HBSS) in a 1:1 ratio. Gently pipette mixture to homogenize.
Add 15 ml of a commercially prepared solution (Ficoll-Paque) to separate blood components by low speed density gradient centrifugation to an empty 50 ml polystyrene conical tube.
Gently pipette whole blood and HBSS mixture, from step 2.1, into the polystyrene tube from step 2.2, to form distinct top layer. Set pipette to lowest setting to minimize mixing of solutions, this will ensure efficient separation.
Centrifuge at 400 x g for 30 min at RT (25 °C). Set deceleration to lowest setting to prevent mixing of solutions after separation. Acceleration may be set to any speed.
After centrifugation, identify the thin white-grey stripe of PBMCs and CTCs sandwiched layer between plasma on top and separation solution at the bottom of the tube.
Collect the cells from the white-grey stripe in step 2.5 into a 50 ml polystyrene tube using a plastic transfer pipette.
Add HBSS to 50 ml polystyrene conical tube with PBMCs and CTCs and centrifuge again at 400 x g for 10 min RT to wash.
Decant liquid and resuspend pellet in 50 ml of HBSS and centrifuge again at 400 x g for 3 min at RT to wash.
Discard the supernatant, resuspend the remaining pellet with 5 ml of Red Blood Cell Lysis buffer and incubate the solution for 5 min at RT.
Remove the red blood cell lysis buffer by centrifugation at 400 x g for 3 min at RT.
Discard the supernatant and resuspend the pellet in 1 ml PBS 1x supplemented with 10% fetal bovine serum (FBS) to block prior to antibody staining.
3. Staining Mononuclear Cells with CD45-FITC Antibody for Fluorescence Activated Cell Sorting (FACS)
Quantify the number of viable (trypan blue-negative) cells (from step 2.11) using a hemocytometer or automated cell counter. Prepare a 1 x 106 cells/ml suspension (in PBS 1x with 10% FBS) and place it on ice for 1 hr.
Distribute the quantified cell suspension from the previous step into two tubes. Label tubes as control and CD45 staining.
In the control tube, add IgG1κ-FITC (2.5 µg/ml) at a dilution of 1:250. In the CD45 staining tube, add CD45 FITC (2.5 µg/ml) conjugated primary antibody at a dilution of 1:250 and incubate the cell suspensions on ice for 30 min. CD45 antigen labeling is used to exclude hematopoietic cells.
Centrifuge the cells at 400 x g for 3 min at 4 °C, and discard the supernatant.
Wash the cells twice, by suspending each pellet with sterile 1 x PBS supplemented with 10% FBS followed by centrifugation at 400 x g for 3 min at 4 °C.
Resuspend the cells in a 1 ml solution of PBS 1x containing 4',6-diamidino-2-phenylindole (DAPI) at a concentration of 10 µg/ml.
Filter the final solution through 35 µm strainer caps into 12 mm x 75 mm polystyrene tubes.
4. Isolation of Prostate CTCs by FACS
Note: Utilize a flow cytometer to collect live CD45 negative CTCs.
Set compensation controls using cells from the tube labeled, “Control.”
Create gates that exclude debris and clusters of cells using appropriate forward-scatter and side-scatter parameters.
Establish the FITC gates using the cell suspensions from the tube labeled, “CD45-FITC.”
Gate viable (DAPI-negative) cells using the pacific blue-A channel.
Collect CD45 negative population into a sterile 15 ml polystyrene conical tube containing 2 ml of Roswell Park Memorial Institute (RPMI) 1640 supplemented with 10% FBS.
Centrifuge the sorted prostate tumor cell suspension at 400 x g for 3 min, then resuspend the pellets in 200 - 500 µl of RPMI supplemented with 10% FBS.
5. Injection of CTCs into Mice and Monitoring of Xenograft Growth
Note: Conduct all animal procedures in compliance with protocols approved by the institutional animal care committee. This protocol has been conducted at our institution under a specific Animal Use Protocol approved by our Animal Care Committee in accordance and compliance with all relevant regulatory and institutional agencies, regulations and guidelines.
Mix the sorted prostate tumor cell suspension with extracellular matrix at a 1:1 ratio and place on ice.
Anesthetize mouse prior to subcutaneous implantation in a chamber supplying 5% (v/v) inhaled isoflurane in 1 L/min of oxygen. Ensure appropriate anesthesia by checking for loss of corneal and toe reflex in the mouse.
Using a 25 G needle and a 1 ml syringe, inject 250 µl of prostate tumor cell and extracellular matrix cell suspension subcutaneously into both upper flanks of a 8-10 week old male immunodeficient mouse. Inject 250 µl regardless of CTC concentration as the injection volume is the important value. Maximum SQ administration in mouse is no more than 1 ml.
Monitor mice by performing weekly palpation of mouse injection sites for growth of subcutaneous nodular densities and by measuring mice weight twice a week.
Euthanize mice by carbon dioxide asphyxiation followed by cervical dislocation and harvest subcutaneous human prostate cancer xenografts when tumor size is more than 0.5 cm and less than 1 cm in greatest diameter to ensure enough tumor tissue for molecular analyses and serial passage. Euthanize mice with signs of distress or weight loss of 15% despite no tumor is palpable.
Representative Results
This protocol will lead to the generation of PDX models from isolated CD45 negative prostate cancer CTCs. Based on the negative selection method used in our protocol it is necessary to exclude dead cells using DAPI stain. The percentage of CD45 negative cells detected by flow cytometry is variable and depends on the tumor burden of the patient (Figure 1A). Immunofluorescent staining of unsorted cells using CD45 and DAPI (to identify cell nuclei) reveals CD45 negative cells, as indicated by the white arrows (Figure 1B). In Figure 1C, subcutaneous nodular densities can be seen on both flanks of the mouse. Each nodular density represents xenograft growth and can be appreciated as distinct from healthy tissue as it protrudes from the dorsal musculature of the mouse and is firmer in texture. The time interval between implantation and xenograft development may vary greatly depending on the CTC burden of the patient sample, number of cells implanted and the biological aggressiveness of CTCs. Prostate PDX generated from CTCs recapitulate human prostate tumor as seen by Hematoxylin and Eosin staining, and by immunohistochemistry for prostate specific cellular markers, such as androgen receptor (AR) and prostate-specific membrane antigen (PSMA) (Figure 1D).
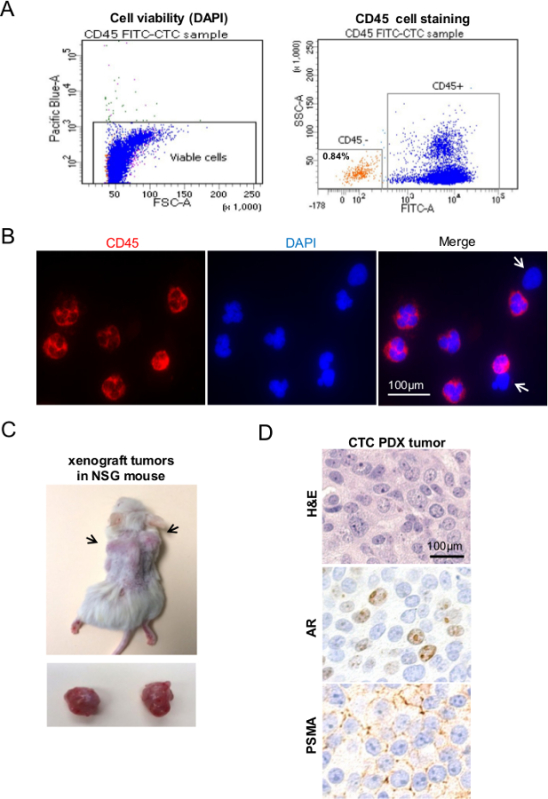 Figure 1. Generation of Prostate Cancer PDX Models from Human Prostate Cancer CTCs. (A) Flow cytometry plot illustrating specific CD45 staining cell populations from peripheral blood of a patient with prostate cancer. Cells with positive DAPI stain are dead and excluded by gating to ensure only viable cells are isolated. (B) CD45 immunofluorescence staining of unsorted population confirms presence of CD45- cells (white arrows). (C) Shows representative PDX tumor in vivo and after tumor harvest. (D) Analysis of CTC PDX models using conventional Hematoxylin and Eosin and the specific prostate markers; androgen receptor (AR) and prostate-specific membrane antigen (PSMA). Please click here to view a larger version of this figure.
Figure 1. Generation of Prostate Cancer PDX Models from Human Prostate Cancer CTCs. (A) Flow cytometry plot illustrating specific CD45 staining cell populations from peripheral blood of a patient with prostate cancer. Cells with positive DAPI stain are dead and excluded by gating to ensure only viable cells are isolated. (B) CD45 immunofluorescence staining of unsorted population confirms presence of CD45- cells (white arrows). (C) Shows representative PDX tumor in vivo and after tumor harvest. (D) Analysis of CTC PDX models using conventional Hematoxylin and Eosin and the specific prostate markers; androgen receptor (AR) and prostate-specific membrane antigen (PSMA). Please click here to view a larger version of this figure.
Discussion
This manuscript describes a method for the generation of prostate cancer PDX models from CTCs. The use of CTCs for the generation of PDXmodels has several potential important advantages when compared to existing methods. First, accessible collection of CTCs from peripheral blood enables the generation of experimental models from the same patient at different disease stages. Second, blood collection represents a safer and inexpensive method to isolate tumor cells when compared to existing methodologies that require surgical procedures for collection of tumor cells.
Multiple methods of CTC enrichment have been described however many are not accessible to every laboratory due to funding and resource limitations. We sought to develop an assay in our laboratory that could capitalize in the use of FACS, a convenient, affordable and accessible methodology for many laboratories. A potential limitation of this methodology is the inadequate sensitivity for detection of rare cells which could limit the use of this protocol to patients with high tumor burden. Several additional technologies which employ physical or biological properties of epithelial cells can be used to over mitigate this limitation and increase the detection and isolation of this rare cell population 4. The method for CTC enrichment described in this protocol employs negative selection; CD45+ leukocytes are discarded and CD45- CTCs are retained. It is important to remove potentially contaminating CD45- elements (e.g., non-viable cells) from the cell suspensions when sorting by flow cytometry. In particular, the use of red blood cell lysis buffer and the omission of nonviable cells by DAPI staining are critical considerations. While these measures may not guarantee that non-CTC elements do not contaminate the CD45- population, our previous studies suggest that the desired cell population is sufficiently enriched with these methods 11. It is possible that the purity of the CTC population could be improved through positive selection, using antibodies to cell surface markers unique to prostate cancer cells 3. In addition, CTC heterogeneity and number of epithelial cells present in the sample can be tested using EpCAM or other appropriate stains. However additional processing of the peripheral blood sample may ultimately increase purity while decreasing overall yield.
In this protocol we performed subcutaneous injection of isolated CTCs which allows for easy implantation and monitoring of the developing tumor. However, subcutaneous models have inadequate nutrition supply that may compromise tumor engraftment and loss of tumor cell subpopulations. Alternative injection sites such as the mouse prostate (orthotropic), which promotes a prostate microenvironment, and subrenal capsule injection, which enhances successful engraftment and preserves tumor heterogeneity, can be performed based on investigator preference 1,2. Optimally, NOD/SCID/IL2λ-receptor null (NSG) mouse models should be used to increase the rate of xenograft engraftment 12, however other strains of immunocompromised mice (NOD/SCID or, Nude, SCID/Beige) have also been successfully used in the generation of solid tumor derived PDX models 1. Generated PDX models can be serially passaged and stability of original tumor characteristics monitored using established genetic methods 9-11,13.
The recovery of high numbers of viable prostate cancer CTCs is dependent on disease stage. The successful generation of PDX models is best assured when CTCs are isolated from blood of patients with high metastatic burden. We have successfully generated PDX models from blood samples with a range of 100 to 10,000 CTCs. In our experience, it is necessary for well-trained laboratory personnel to perform the protocol frequently in order to assure the successful isolation of prostate CTCs from blood samples. We recommend that laboratory personnel be initially trained in this protocol by using blood samples from normal healthy individuals spiked with cells from tumor cell lines. The absence or low number of CTCs in primary prostate cancer may limit the use of this methodology to metastatic prostate cancer.
The method for the generation of CTC derived PDX models described in this protocol can be subsequently employed in many research applications including molecular characterization of prostate cancer, drug screening, monitoring therapy response, biomarker development, and other advancements in personalized cancer therapies.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgments
We thank Dr. Jordi Ochando from the Flow Cytometry Shared Resources at the Mount Sinai Medical Center for their assistance in flow cytometry analysis. We thank Dr. Rumana Huq from the Microscopy Shared Resource Facility at the Mount Sinai Medical Center for their imaging assistance. The authors thank the TJ Martell Foundation for its support in this project.
References
- Hidalgo M, et al. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer discovery. 2014;4:998–1013. doi: 10.1158/2159-8290.CD-14-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siolas D, Hannon GJ. Patient-derived tumor xenografts: transforming clinical samples into mouse models. Cancer research. 2013;73:5315–5319. doi: 10.1158/0008-5472.CAN-13-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosse SA, Gorges TM, Pantel K. Biology, detection, and clinical implications of circulating tumor cells. EMBO molecular medicine. 2014;1:1–11. doi: 10.15252/emmm.201303698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Stott S, Toner M, Maheswaran S, Haber DA. Circulating tumor cells: approaches to isolation and characterization. The Journal of cell biology. 2011;192:373–382. doi: 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, et al. Negative enrichment by immunomagnetic nanobeads for unbiased characterization of circulating tumor cells from peripheral blood of cancer patients. Journal of translational medicine. 2011;9:1–9. doi: 10.1186/1479-5876-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A. Cancer statistics , 2013. CA: a cancer journal for clinicians. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- Domingo-Domenech J, et al. Suppression of acquired docetaxel resistance in prostate cancer through depletion of notch- and hedgehog-dependent tumor-initiating cells. Cancer cell. 2012;22:373–388. doi: 10.1016/j.ccr.2012.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein KA, et al. Progression of metastatic human prostate cancer to androgen independence in immunodeficient SCID mice. Nature Medicine. 1997;3:402–408. doi: 10.1038/nm0497-402. [DOI] [PubMed] [Google Scholar]
- Yu M, et al. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014;345:216–220. doi: 10.1126/science.1253533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkinson CL, et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nature medicine. 2014;20:897–903. doi: 10.1038/nm.3600. [DOI] [PubMed] [Google Scholar]
- Vidal S, et al. A Targetable GATA2-IGF2 Axis Confers Aggressiveness in Lethal Prostate Cancer. Cancer cell. 2015;27:223–239. doi: 10.1016/j.ccell.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana E, et al. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRose YS, et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nature. 2011;17:1514–1520. doi: 10.1038/nm.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]


