Abstract
Recently infection biologists have employed stem cell derived cultures to answer the need for new and better models to study host-pathogen interactions. Three cellular sources have been used: Embryonic stem cells (ESC), induced pluripotent stem cells (iPSC) or adult stem cells. Here, culture of mouse and human gastric organoids derived from adult stem cells is described and used for infection with the gastric pathogen Helicobacter pylori. Human gastric glands are isolated from resection material, seeded in a basement matrix and embedded in medium containing growth factors epidermal growth factor (EGF), R-spondin, Noggin, Wnt, fibroblast growth factor (FGF) 10, gastrin and transforming growth factor (TGF) beta inhibitor. In these conditions, gastric glands grow into 3-dimensional organoids containing 4 lineages of the stomach. The organoids expand indefinitely and can be frozen and thawed similarly as cell lines. For infection studies, bacteria are microinjected into the lumen of the organoids. Infected organoids are processed for imaging. The described methods can be adapted to other organoids and infections with other bacteria, viruses or parasites. This allows the study of infection-induced changes in primary cells.
Keywords: Infection, Issue 105, Primary cell culture, Infection, Host pathogen interaction, Stomach, Microinjection, Helicobacter.
Introduction
The study of pathogens relies on adequate model systems to mimic the in vivo infection. For some infective agents, adequate model systems are lacking while some of the used systems are far from optimal. One example is the gastric bacterium Helicobacter pylori (H. pylori), which is causally related to development of gastric cancer. Yet in absence of a more suitable cell culture system, many studies that aim to analyze the molecular mechanisms underlying cancer development use cancer cell lines, which represent the endpoint of the cancerous cascade. Primary, non-transformed cells would be a better model for these studies. However, primary cells are only available from a small number of donors and cannot be cultured over longer periods of time. In recent years, stem cell research has made significant progress to provide new sources for primary cell cultures for the study of infection biology.
Cultures from three stem cell sources have been used: Embryonic stem cells (ESC), induced pluripotent stem cells (iPSC) or adult stem cells. They have been used to model infections with viruses, such as Cytomegalovirus1,2 or Hepatitis C Virus3–7, parasites, such as Plasmodium falciparum8 or Toxoplasma gondii9, and bacteria, such as Bacterioides thetaiotaomicron10 or Salmonella enterica11. Most recently, several approaches have been published to model infection with H. pylori with organoids derived from ESC or iPS cells12, mouse adult stem cells21,22 or human adult stem cells13–15.
The development of organoid cultures from adult stem cells originated from a study, in which single stem cells isolated from murine intestinal epithelium were seeded into a 3-dimensional matrix and embedded in medium that mimicked the environment of the intestinal stem cells containing EGF as mitogen, R-spondin to enhance Wnt signalling and Noggin to inhibit bone morphogenic protein (BMP) signalling16. Notably these cultures do not require co-culture with mesenchymal cells. In these conditions, the stem cells proliferate and form small structures with domains harboring cells of the intestinal crypts, and domains that contain the cells of the intestinal villus. The organoids thus self-organize to mimic the in vivo situation. Today, adult stem cells from many murine and humane tissues can be grown in vitro and self-organize into organoids that resemble their in vivo counterpart, such as small intestine and colon17, stomach13,18, liver19,20, pancreas21 and prostate22.
Here we provide a video protocol to culture mouse or human gastric organoids from adult stem cells and microinject them with H. pylori. This protocol is based on previous reports13,18. This method can be adapted for culturing and infecting other organoid cultures such as intestinal organoids.
Protocol
1. Establishment of Gastric Organoid Culture
Note: This protocol can be used for the isolation of gastric glands from mouse or human tissue. It is advised to use tissue of approximately 1 cm². Human tissue can be obtained from gastric resections or biopsies.
- Preparation of Material Note: The used basement matrix is Matrigel. Keep the basement matrix on ice at all times. Store the basement matrix at -20 °C and thaw on ice before use. Basal medium refers to Advanced DMEM/F12 supplemented with HEPES; an appropriate glutamine source such as Glutamax and appropriate antibiotics such as 1x Primocin. The incubator used is a standard cell culture incubator (37 °C, humidified atmosphere, 5% CO2)
- Sterilize preparation utensils by autoclaving or using 100% isopropyl alcohol: sharp forceps, scissors, a scalpel and a glass microscopy slide.
- The day before isolation, place a 24 well cell culture plate in the incubator.
- Prepare the basement matrix according to manufacturers’ recommendation. Prepare aliquots of 1 ml to avoid repeated freeze thaw cycles.
- Prepare 500 ml chelating buffer and cool on ice. Prepare 1x buffer from sterile distilled water with 5.6 mM Na2HPO4, 8.0 mM KH2PO4, 96.2 mM NaCl, 1.6 mM KCl, 43.4 mM sucrose, 54.9 mM D-sorbitol, 0.5 mM DL-dithiothreitol (CAUTION), pH 7. If planning several isolations, prepare a 5x concentrated solution without the DL-dithiothreitol. Sterile filter the buffer. Before isolation, dilute the 5x buffer with sterile water to 1x and add DL-dithiothreitol just before isolation.
- Prepare and dilute all growth factors according to manufacturers’ recommendations. Use small-sized aliquots and avoid freeze-thaw cycles. Functional growth factors are crucial for the results.
- Cool basal medium on ice and cool the centrifuge to 4 °C.
- Warm another aliquot of basal medium to 37 °C.
- Isolation of Glands
- Collect approximately 1 cm² of tissue in ice cold basal medium. Keep on ice until preparation. While it is preferable to process tissue as soon as possible, store tissue in medium on ice if necessary.
- Place the tissue in a 10 cm Petri dish and cover it with cold 1x chelation buffer. Wash by gently moving back and forth.
- Place the tissue in a dry 10 cm Petri dish. Using forceps, carefully remove the mucus as well as the muscle layer under a stereomicroscope. Wash the tissue in cold 1x chelating buffer by gently moving back and forth.
- Place 1 cm2 tissue on a new, dry 10 cm Petri dish. Using scissors cut it into 20-50 small pieces of approximately 2-5 mm² size.
- Using forceps, place all pieces into a 50 ml tube.
- Take a sterile 10 ml plastic pipette. Wet the pipette in 1x chelating buffer. Keep using this same pipette throughout the procedure.
- Add 10 ml cold 1x chelating buffer to the 50 ml tube. Wash the pieces by vigorously pipetting up and down 10 times. Let the pieces settle. Remove the supernatant.
- Repeat washing (step 1.2.7) until the supernatant is clear. This takes approximately 50-100 ml of 1x chelating buffer for 1 cm² of human tissue.
- Incubate the tissue pieces in 20 ml 1x chelating buffer supplemented with 10 mM EDTA at RT for 10 min with gentle inverting every 2 min. Note: Time and temperature of this incubation can be optimized for either speed and/or preservation of glandular architecture. They can range from several hours incubation at 4 °C on a rolling platform to 5 min at 37 °C in a shaker with 200 rpm. Good starting points for mouse stomach are 30 min at 4 °C on a rolling platform and for human stomach, 10 min at RT.
- Pipette once gently up and down using the 10 ml pipette. Allow the pieces to settle. Discard the supernatant.
- Using the pipette, transfer the pieces to a clean 10 cm Petri dish. Remove the liquid.
- Place a glass microscopy slide on top of the tissue pieces. Observe the intact tissue under an inverted microscope for cell culture with 10X magnification.
- Apply pressure to the glass slide and Petri dish. Hold both in hand (with sterile gloves) and pressing the glass with the thumb. The rim of the tissue will appear cloudy indicating that glands are released out of the tissue.
- Observe the released glands under an inverted microscope for cell culture with 10X magnification.
- Collect glands and tissue in 30 ml cold basal medium. Let large tissue fragments settle.
- Collect the supernatant containing glands in two 15 ml tubes.
- Centrifuge 5 min at 200 x g and 4 °C. Discard supernatant. The pellet contains isolated glands. Keep them on ice.
- Seeding of Glands
- For the seeding, keep basement matrix ice-cold at all times, so it will stay liquid.
- Alternative 1: Seed 6 wells of a dilution row.
- Suspend the pellet in 60 µl of basement matrix.
- Prepare 5 sterile 1.5 ml tubes on ice with each containing 50 µl of basement matrix. Transfer 10 µl of glands/basement matrix suspension into the first 1.5 ml tube. Suspend the glands and transfer 10 µl of this solution to the next 1.5 ml tube. Repeat for the next tubes.
- Alternative 2: Seed a calculated amount of glands per well. Start with 100 glands per 50 µl basement matrix in one well of a 24 well plate.
- Carefully suspend the pelleted glands in 10 ml basal medium. Place 50 µl on a clean 10 ml Petri dish. Count number of glands under an inverted microscope for cell culture with 10X magnification.
- Calculate the concentration of glands in the solution. Transfer the volume that contains 400 glands to a new 15 ml tube.
- Centrifuge 5 min at 200 x g and 4 °C. Discard supernatant and keep the tube on ice. Add 200 µl basement matrix and carefully resuspend. Keep on ice.
- Take the 37 °C warm 24-well plate (see step 1.1.2) from the incubator. Place one drop of 50 µl glands in basement matrix in the center of the well. The drop will form a dome.
- Carefully transfer the plate back to the cell culture incubator. Let the basement matrix solidify 10 min.
- Prepare warm medium with all growth factors.
- For human glands: Supplement the basal medium with the following growth factors for human gastric organoids: 50 ng/ml EGF, 10% noggin-conditioned medium, 10% R-spondin1–conditioned medium, 50% Wnt-conditioned medium, 200 ng/ml fibroblast growth factor (FGF)10, 1 nM gastrin, and 2 µM TGFbeta inhibitor and 10 µM rho-associated coiled coil forming protein serine/threonine kinase inhibitor (RHOKi).
- For murine glands: Supplement the basal medium with the following growth factors for mouse gastric organoids: 50 ng/ml EGF, 10% noggin-conditioned medium, 10% R-spondin1–conditioned medium, 50% Wnt-conditioned medium, 100 ng/ml FGF10, 10 nM gastrin, 0.5 mM TGFbeta inhibitor and 10 µM RHOKi.
- Take the plate from the incubator. Carefully add 500 µl medium with all growth factors (1.3.6) to each well without disturbing the basement matrix. Place it back to the incubator.
- Refresh the medium 3 times per week (every 2-3 days). For refreshing medium, remove the old medium with leaving the drop of basement matrix intact. Carefully add fresh, warm medium with all growth factors. Do not add RHOKi. Only add RHOKi directly after seeding of glands or after splitting of the organoids (step 2).
2. Passage of Gastric Organoid Culture Before Microinjection.
Note: Every type of organoid culture has its own doubling time. Mouse intestinal as well as gastric cultures are usually split 1:5 every 5-7 days. Human intestinal cultures are split 1:5 every 10-12 days. Human gastric cultures are split 1:5 every 14 days. If initiated from single cells, human gastric organoids may also take 20 days to form properly. It is a good sign if budding structures surround the central lumen. In this protocol, organoids are split in 4 well plates for microinjection. Maintenance of organoids follows the same protocol, but can use any other cell culture plate, such as standard 24 well cell culture plates.
- Preparation of Material
- Autoclave Pasteur pipettes.
- The day before passage, place a 4 well cell culture plate in the incubator at 37 °C. The 4 well plates have low edges and allow micromanipulation.
- Prepare the basement matrix according to manufacturers’ recommendation. On the day of passaging, thaw basement matrix on ice.
- Prepare and dilute all growth factors according to manufacturers’ recommendations.
- Cool basal medium on ice and cool the centrifuge to 4 °C.
- Passage of Organoid Cultures
- Take the plate with the organoids from the incubator. Remove medium from one well.
- Add 1 ml of cold basal medium to the basement matrix in the well. Using a 1 ml micropipette, vigorously pipette up and down until the basement matrix is broken up. Transfer to a 15 ml tube. Place on ice.
- Take a glass Pasteur pipette and a fire source such as a Bunsen burner.
- Hold the tip of the pipette into the fire until the opening of the tip has narrowed from 1.5 mm to about 0.5 mm (Figure 1). Let the pipette cool to RT.
- Wet the pipette in basal medium. An optimally narrowed pipette will take up the medium visibly slower than an un-narrowed pipette, but it will still allow pipetting well.
- Take up organoids in the pipette and vigorously pipette 10x up and down. The breaking of the organoids can be observed by eye.
- Add 9 ml of cold basal medium and centrifuge for 5 min at 200 x g and 4 °C.
- Carefully discard all supernatant. Suspend the pellet in 250 µl basement matrix. Keep on ice.
- Take the 37 °C warm 4 well culture plate and place a drop of 50 µl organoids/basement matrix in each well.
- Transfer the plate carefully back to the incubator. Let the basement matrix solidify at 37 °C for 10 min.
- Prepare appropriate medium for either mouse or human organoids as listed in step 1.3.6.
- Take the plate from the incubator and overlay each basement matrix drop with 500 µl medium.
- Transfer the plate to the incubator.
- Refresh the medium 3 times per week every 2-3 days (see 1.3.8.).
- For freezing of organoids, mechanically disrupt organoids as described for splitting in this protocol (Steps 2.2.1-2.2.5.). Suspend the organoid fragments in 500 ml freezing medium and freeze to -80 °C using cryotubes and a freezing container. For long-term storage, transfer the tubes to liquid nitrogen.
- For thawing of organoids, prepare the culture plate, basement matrix, growth factors, cold and warm medium as outlined in steps 1.1.2, 1.1.3., 1.1.5., 1.1.6., 1.1.7. Warm the frozen vial using a water bath, and transfer cells immediately after thawing to a 15 ml tube with 10 ml cold basal medium. Centrifuge and plate the cells as outlined in 2.2.5 to 2.2.12. Note: Microinjection of organoids is laborious, but allows the unique possibility to study interaction in 3D.
- If 3D studies are not needed, seed organoids in two dimensions. For this, disrupt the organoids as laid out in steps 2.2.1 to 2.2.6. Add 2,250 µl cold medium with all growth factors to 250 µl of basement matrix with organoid fragments.
- Seed in 5 wells of a 24 well plate with 500 µl per well. Change medium 3 times a week, with carefully replacing the upper 400 µl only, to keep the basement matrix at the bottom of the plate undisturbed. Note: Organoids will attach to the cell culture plate and expand in 2D. Electron microscopy has shown that the apical side of the cells faces the medium in the well (data not shown).
- Before infection, exchange all medium including basement matrix, to allow bacteria to attach to the cells. A similar protocol has also been described for infection with H. pylori14. 2D cell layers may contain not all cell types that are present in 3D organoids.
3. Microinjection of Organoid Culture.
Note: This protocol can be used to microinject bacteria into organoids. It may be helpful to start the injection with organoids that are more permissive to microinjection. For example, mouse gastric organoids can grow into very large cystic organoids that are easy to target.
- Preparation of Material
- Pull a glass capillary into two injection needles using a micropipette puller, according to manufacturers’ recommendation. The micropipette puller will heat the glass capillary in the middle and will pull the capillary into two parts, thereby generating two glass needles. It is practical to pull several needles and store them in a clean petri dish.
- To allow bacterial infection, remove the antibiotics from all media for at least 3 medium changes (=one week). This will dilute antibiotics from basement matrix. If using a bacterial strain that is resistant to a specific antibiotic, add this specific antibiotic to minimize chances for contamination.
- Prepare the work bench for microinjections. For work at different biosafety levels it may be necessary to place a stereomicroscope inside a sterile bench. In this protocol, inject H. pylori under biosafety level 2 conditions under a stereomicroscope inside a sterile bench.
- Culture the bacteria according to standard protocols13,23.
- To estimate the multiplicity of infection (MOI), estimate two parameters: The number of bacteria and the number of cells per organoid as outlined below. The MOI can correlate with results, for example with hosts IL-8 mRNA expression13.
- To calculate the bacterial density in the infection solution, first establish the standard curve of optical density and colony forming units (CFU) according to standard protocols24. An optical density of 0.1 at 550 nm should yield approximately 107 CFU/ml.
- To estimate the number of cells per organoid, count number of organoids in 1 well. Disrupt organoids as outlined in steps 2.2.1-2.2.4. After centrifugation 5 min at 200 x g and 4 °C, add 1 ml of enzymatic dissociation solution and resuspend.
- Incubate at 37 °C with repeated shaking for 5-10 min. Determine whether the cells have dissociated into single cells under the microscope (inverted microscope such as Leica DMIL at 10X magnification). If necessary prolong the incubation.
- Count cells per organoid using a hemocytometer and calculate the number of cells per organoid. Human gastric organoids with a diameter of about 200 µm have approximately 4,000 cells. Injection of 0.2 µl of a bacterial solution with 1 x 109 bacteria per ml results in an approximate MOI of 50.
- Injection of Organoids. Note: For microinjection, the glass needle is stabilized in a holder, which can be maneuvered in 3 dimensions by a micromanipulator. Organoids remain within the basement matrix within the well. Due to size and positioning in the well, not all organoids are equally amenable to injection. Usually, the 30 largest organoids in one well can be targeted in 5 min.
- Harvest bacteria and wash them in basal medium according to standard protocols24.
- Calculate the number of bacteria per ml according to the optical density (see step 3.1.5.1). Dilute the bacteria to 1 x 109 bacteria per ml in basal medium.
- Take a glass needle. Using forceps break the tip so that the opening will be approximately 10 µm wide.
- Insert the needle into the injection holder and fix it to the micromanipulator.
- Take up approximately 10 µl bacterial solution into the needle.
- Place a 4 well plate with low edges containing organoid cultures under the stereomicroscope.
- Navigating with the micromanipulator, target a single organoids with the needle. Position the needle close to the organoid and then insert the needle into the organoid with one swift movement. Inject approximately 0.2 µl bacterial solution into each organoid using the microinjector.
- Harvest Organoids for Any Analysis.
- As example, fix organoids after 4 hr. Remove supernatant and add 1 ml 2% formaldehyde and incubate 20 min at RT or O/N at 4 °C. Organoids can be processed for immunohistochemistry according to standard procedures25.
Representative Results
This protocol allows isolation of gastric glands (Figure 2). Glands are seeded into basement matrix, which solidifies as drop within a well, providing a 3 dimensional framework rich in laminin and collagen to allow the glands grow into organoids (Figure 3). Organoids typically start as small cysts and within 12-16 days, they expand into spheres with a diameter of 50-300 µm (Figure 4). Some organoids will stay cystic, some will develop small buddings. The latter is usually a sign of a healthy growing culture. In this protocol one well of a 24 well plate is used for 100 glands, 50 µl of basement matrix and 500 µl of medium. However, this can be up- or downscaled.
The success of the microinjection can readily be observed under the stereomicroscope as the cloudy, bacterial solution fills the organoid (Figure 5). After adequate incubation time, organoids can be processed for any analysis method desired. For example, organoids can be embedded in paraffin, sections can be cut and stained using standard immunohistochemistry techniques. Microscopic analysis of immunostained organoids demonstrate successful injection of the bacteria (Figure 6).
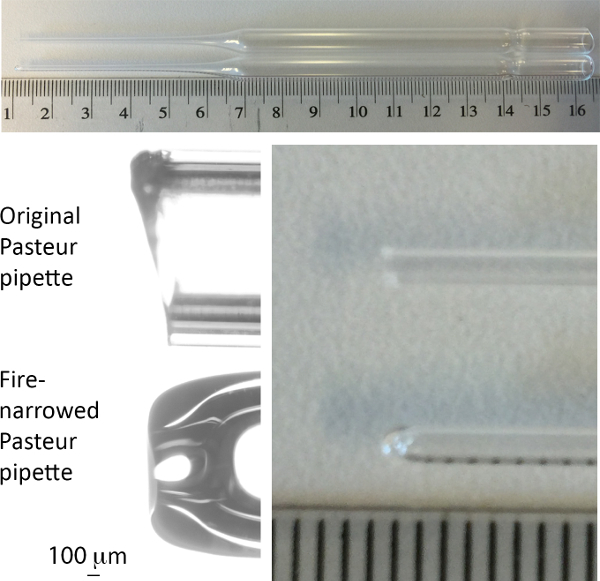 Figure 1. Image of Pasteur pipette used for passage of organoids. In each panel, the upper pipette is before, the lower pipette after narrowing by fire. Scale in upper and right panels is cm and mm. Please click here to view a larger version of this figure.
Figure 1. Image of Pasteur pipette used for passage of organoids. In each panel, the upper pipette is before, the lower pipette after narrowing by fire. Scale in upper and right panels is cm and mm. Please click here to view a larger version of this figure.
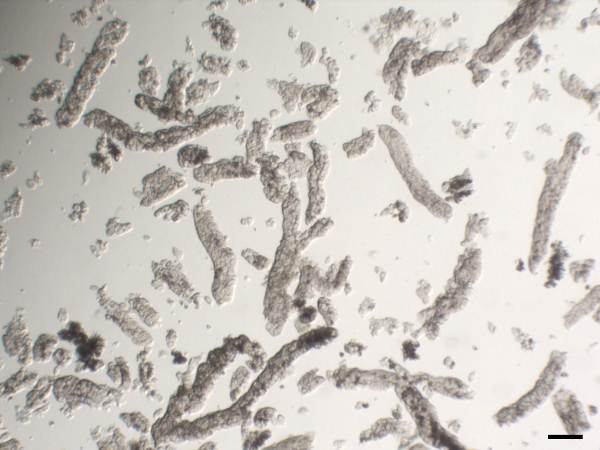 Figure 2. Representative image of isolated human gastric glands. Scale bar 100 µm. Please click here to view a larger version of this figure.
Figure 2. Representative image of isolated human gastric glands. Scale bar 100 µm. Please click here to view a larger version of this figure.
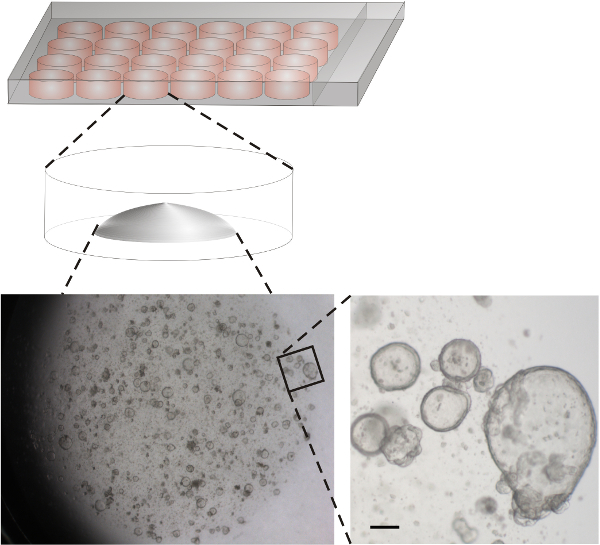 Figure 3. Scheme of wells and representative image of human gastric organoids. Isolated human gastric glands were dispensed in basement matrix and placed as drops into wells of a 24 well plate. Lower left: Overview of a representative well 11 days after seeding. Lower right: Enlargement of the indicated area. Scale bar 100 µm. Gastric mouse organoids expand faster18. Please click here to view a larger version of this figure.
Figure 3. Scheme of wells and representative image of human gastric organoids. Isolated human gastric glands were dispensed in basement matrix and placed as drops into wells of a 24 well plate. Lower left: Overview of a representative well 11 days after seeding. Lower right: Enlargement of the indicated area. Scale bar 100 µm. Gastric mouse organoids expand faster18. Please click here to view a larger version of this figure.
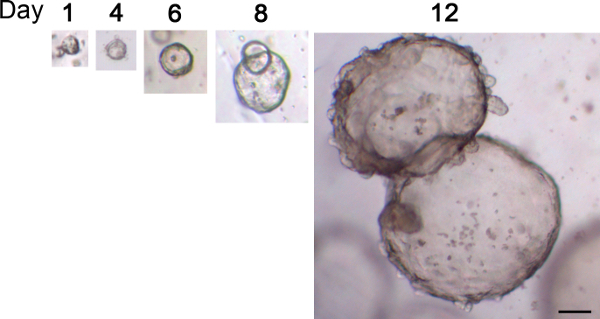 Figure 4. Typical growth of human gastric organoids. Glands were seeded into basement matrix and images of the same organoid were taken over a period of 12 days. Scale bar 100 µm Please click here to view a larger version of this figure.
Figure 4. Typical growth of human gastric organoids. Glands were seeded into basement matrix and images of the same organoid were taken over a period of 12 days. Scale bar 100 µm Please click here to view a larger version of this figure.
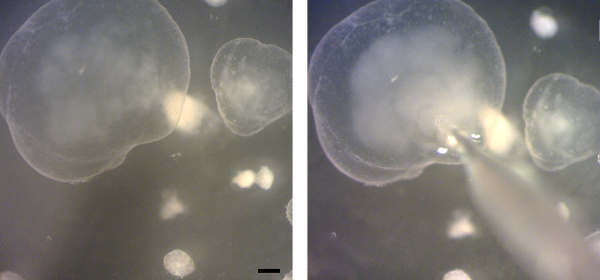 Figure 5. Microinjection of gastric organoids. Stereoscope images of a gastric organoid before (left) and during (right) microinjection of bacteria into the lumen. Bacteria are visible as cloud inside the organoid. Scale bar 200 µm. Please click here to view a larger version of this figure.
Figure 5. Microinjection of gastric organoids. Stereoscope images of a gastric organoid before (left) and during (right) microinjection of bacteria into the lumen. Bacteria are visible as cloud inside the organoid. Scale bar 200 µm. Please click here to view a larger version of this figure.
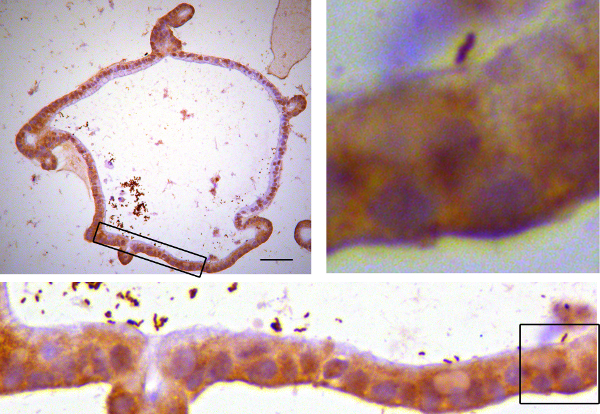 Figure 6. Immunostained organoids. Human gastric organoids were microinjected with H. pylori. After 4 hr, organoids were fixed in paraformaldehyde and embedded in paraffin. Sections were stained using antibodies targeting the bacterial protein Cytotoxicity associated gene A (CagA) according to standard histology methods25. Upper left: Image of a representative organoid. Lower panel: Higher magnification of the boxed area. Upper right: Higher magnification of the boxed area with a single bacterium close to the epithelial cells. Please click here to view a larger version of this figure.
Figure 6. Immunostained organoids. Human gastric organoids were microinjected with H. pylori. After 4 hr, organoids were fixed in paraformaldehyde and embedded in paraffin. Sections were stained using antibodies targeting the bacterial protein Cytotoxicity associated gene A (CagA) according to standard histology methods25. Upper left: Image of a representative organoid. Lower panel: Higher magnification of the boxed area. Upper right: Higher magnification of the boxed area with a single bacterium close to the epithelial cells. Please click here to view a larger version of this figure.
Discussion
This protocol describes the use of ever-expanding, untransformed primary organoids from adult stem cells for infection biology. Critical steps are i) the isolation of viable glands, ii) expansion of organoids and iii) the microinjection. Below are some suggestions for modifications, troubleshooting and technical considerations.
Compared to other isolation methods, which use vigorous shaking or pipetting to release glands and can be equally successful, the technique presented here has the advantage, that the release of the glands from the tissue can directly be observed under the microscope (step 1.2.14.). If glands are not released, incubation in EDTA can be varied (see step 1.2.9.). If preferred, try to isolate glands using vigorous shaking and control the presence of glands in the supernatant under the microscope as described for mouse intestinal crypts26. It is not critical, if isolated glands disintegrate during the washing steps. Organoids will still initiate from fragments or even single stem cells, as demonstrated previously13,18, 27.
When glands have been isolated and seeded, but organoids are not expanding as expected, troubleshooting should start with testing activity of Wnt and R-spondin. These factors are crucial for expansion of the organoids and it is advised to test the functionality of these two factors by an in vitro assay, for example the TOP flash reporter construct using Luciferase28. It is important that all growth factors are diluted and handled following manufacturers’ recommendation and kept in small aliquots to avoid freeze-thaw cycles.
The microinjection into organoids is comparable to injection of fish eggs or murine oocytes. Needles are pulled from glass capillaries and can be broken using tweezers directly before use to result in a tip end of about 10 µm. Smaller tips penetrate the organoids more easily but they also bear the risk that bacterial accumulations may clog the needle. For micromanipulation, different systems have specific advantages and limitations. It is practical to use a 3- dimensional manipulator instead of a 2 dimensional manipulator to allow maximal flexibility to reach into the wells. Oil-based microinjectors (such as Celltram, or IM-5B) provide a finer and slower manual control compared to air-based microinjectors (such as FemtoJet11, or Nanojet microinjector10). In turn, the latter have the important advantage that a precise injection volume can be programmed. If microinjection is initially difficult, it may be helpful practice with large mouse gastric organoids and a dye. With practice, also smaller organoids such as mouse intestinal organoids are amenable to microinjection.
Microinjection of organoids is technically limited to a small experimental size because every organoid has to be targeted manually. Positioning or size may not render all organoids equally amenable to injection, thus the technique is limited to applications that can tolerate some heterogeneity in a well. Reflux from the injection hole is minimal10,11, but should also be considered.
Comparing the different sources for new gastric stem cell derived cell cultures, it is evident that each of the systems has its own advantages. iPSC have been used to generate antral gastric organoids and allow following of the developmental steps towards the antral gastric lineages12. The cultures described here can be derived from antrum or corpus. While long-term corpus cultures as described here do not contain parietal cells13, very early passages as well as co-cultures with mesenchymal cells have been reported to contain parietal cells15,29.
The protocol described here has been developed for optimal long-term culture and allows unlimited maintenance of organoids (1 year tested). The organoids have typical advantages of cell lines: They can be expanded to allow larger experiments (split 1:5 every 2 weeks for human, every week for mouse organoids). They can be frozen and thawed. Human gastric organoids contain stem cells next to differentiated cells that express markers of mucous pit cells, mucous neck cells, chief cells and enteroendocrine cells. Differentiation to the lineages can be directed using Wnt and Nicotinamide. It is also possible to generate organoids from biopsies of cancers and thus organoids from healthy and cancerous tissue can be compared13,21,33. Generally, organoids are also amenable to genetic modification and intestinal or gastric organoids have been genetically modified using bacterial artificial chromosomes (BAC) transgenes30, retrovirus26,31, lentivirus14 and clustered regularly interspaced short palindromic repeats (CRISPR)/Cas932. Organoids have been used for various imaging techniques13,14,29, RNA analysis13, Western Blot33, Immunoprecipitation34 and drug screening33.
In summary, organoids grown from adult stem cells provide a valuable new tool for infection biology. The here provided protocol for gastric organoids may serve as basis to establish other new infection models.
Disclosures
Hans Clevers is an inventor on several patents for organoid culture. Otherwise the authors have nothing to disclose.
Acknowledgments
This work was supported by EU Marie Curie Fellowship (EU/300686-InfO) to S.B. and a Research Prize from the United European Gastroenterology Foundation to H.C. We would like to thank Harry Begthel, Jeroen Korving and the Hubrecht Imaging Center for technical assistance, Meritxell Huch for help with initial organoid culture and Yana Zavros for discussion.
References
- Aiuto L, et al. Human Induced Pluripotent Stem Cell-Derived Models to Investigate Human Cytomegalovirus Infection in Neural Cells. PLoS ONE. 2012;7(11):e49700. doi: 10.1371/journal.pone.0049700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkert RR, Kalejta RF. Human Embryonic Stem Cell Lines Model Experimental Human Cytomegalovirus Latency. mBio. 2013;4(3):e00298-13-e00298-13. doi: 10.1128/mBio.00298-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelandt P, et al. Human pluripotent stem cell-derived hepatocytes support complete replication of hepatitis C virus. J Hepatol. 2012;57(2):246–251. doi: 10.1016/j.jhep.2012.03.030. [DOI] [PubMed] [Google Scholar]
- Schwartz RE, Trehan K, et al. Modeling hepatitis C virus infection using human induced pluripotent stem cells. Proc Natl Acad Sci USA. 2012;109(7):2544–2548. doi: 10.1073/pnas.1121400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomai A, et al. Modeling host interactions with hepatitis B virus using primary and induced pluripotent stem cell-derived hepatocellular systems. Proc Natl Acad Sci USA. 2014;111(33):12193–12198. doi: 10.1073/pnas.1412631111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, et al. Productive Hepatitis C Virus Infection of Stem Cell-Derived Hepatocytes Reveals a Critical Transition to Viral Permissiveness during Differentiation. PLoS Pathogens. 2012;8(4):e1002617. doi: 10.1371/journal.ppat.1002617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, et al. Use of human hepatocyte-like cells derived from induced pluripotent stem cells as a model for hepatocytes in hepatitis C virus infection. Biochem Biophys Res Commun. 2011;416(1-2):119–124. doi: 10.1016/j.bbrc.2011.11.007. [DOI] [PubMed] [Google Scholar]
- Ng S, et al. Human iPSC-Derived Hepatocyte-like Cells Support Plasmodium Liver-Stage Infection In Vitro. Stem Cell Report. 2015;4(2) doi: 10.1016/j.stemcr.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz C, Aebischer T, Seeber F. Stem cell-derived cell cultures and organoids for protozoan parasite propagation and studying host-parasite interaction. Int J Med Microbiol. 2012;302(4-5):203–209. doi: 10.1016/j.ijmm.2012.07.010. [DOI] [PubMed] [Google Scholar]
- Engevik MA, et al. Loss of NHE3 alters gut microbiota composition and influences Bacteroides thetaiotaomicron growth. AJP: GI. 2013;305(10):G697–G711. doi: 10.1152/ajpgi.00184.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SS, Tocchi A, Holly MK, Parks WC, Smith JG. A small intestinal organoid model of non-invasive enteric pathogen-epithelial cell interactions. Mucosal Immunol. 2015;8(2):352–361. doi: 10.1038/mi.2014.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken KW, et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature. 2014;516(7531):400–404. doi: 10.1038/nature13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartfeld S, et al. In Vitro Expansion of Human Gastric Epithelial Stem Cells and Their Responses to Bacterial Infection. Gastroenterology. 2014;148(1) doi: 10.1053/j.gastro.2014.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaermann P, Toelle B, et al. A novel human gastric primary cell culture system for modelling Helicobacter pylori infection in vitro. Gut. 2014. [DOI] [PMC free article] [PubMed]
- Bertaux-Skeirik N, et al. CD44 Plays a Functional Role in Helicobacter pylori-induced Epithelial Cell Proliferation. PLOS Pathogens. 2015;11(2):e1004663. doi: 10.1371/journal.ppat.1004663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, et al. Single Lgr5 stem cells build crypt villus structures in vitro without a mesenchymal niche. Nature. 2009;459(7244):262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Sato T, et al. Long-term Expansion of Epithelial Organoids From Human Colon, Adenoma, Adenocarcinoma, and Barrett's Epithelium. Gastroenterology. 2011;141(5):1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- Barker N, et al. Lgr5+ve Stem Cells Drive Self-Renewal in the Stomach and Build Long-Lived Gastric Units In Vitro. Cell Stem Cell. 2010;6(1):25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- Huch M, et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494(7436):247–250. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M, et al. Long-Term Culture of Genome-Stable Bipotent Stem Cells from Adult Human Liver. Cell. 2015;160(1-2):299–312. doi: 10.1016/j.cell.2014.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boj SF, et al. Organoid Models of Human and Mouse Ductal Pancreatic Cancer. Cell. 2015;160(1-2):324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthaus WR, et al. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell. 2014;159(1):163–175. doi: 10.1016/j.cell.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartfeld S, et al. High-throughput and single-cell imaging of NF-kappaB oscillations using monoclonal cell lines. BMC cell. 2010;11:21. doi: 10.1186/1471-2121-11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard TG, Nedrud JG. Laboratory Maintenance of Helicobacter Species. Curr Protoc Microbiol. 2006. [DOI] [PMC free article] [PubMed]
- Van Es JH, de Geest N, van de Born M, Clevers H, Hassan BA. Intestinal stem cells lacking the Math1 tumour suppressor are refractory to Notch inhibitors. Nat Commun. 2010;1(2):1–5. doi: 10.1038/ncomms1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson-Rolf A, Fink J, Mustata RC, Koo B-K. A Video Protocol of Retroviral Infection in Primary Intestinal Organoid Culture. J Vis Exp. 2014. [DOI] [PMC free article] [PubMed]
- Stange DE, Koo B-K, et al. Differentiated Troy+ Chief Cells Act as Reserve Stem Cells to Generate All Lineages of the Stomach Epithelium. Cell. 2013;155(2):357–368. doi: 10.1016/j.cell.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Wetering M, Sancho E, et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111(2):241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- Schumacher MA, Aihara E, et al. The use of murine-derived fundic organoids in studies of gastric physiology. Journal Physiol. 2015;593(8):1809–1827. doi: 10.1113/jphysiol.2014.283028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwank G, Andersson-Rolf A, Koo B-K, Sasaki N, Clevers H. Generation of BAC Transgenic Epithelial Organoids. PLoS ONE. 2013;8(10):e76871. doi: 10.1371/journal.pone.0076871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo B-K, et al. Controlled gene expression in primary Lgr5 organoid cultures. Nat Meth. 2012;9(1):81–83. doi: 10.1038/nmeth.1802. [DOI] [PubMed] [Google Scholar]
- Schwank G, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13(6):653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Li VSW, Ng SS, et al. Wnt Signaling through Inhibition of β-Catenin Degradation in an Intact Axin1 Complex. Cell. 2012;149(6):1245–1256. doi: 10.1016/j.cell.2012.05.002. [DOI] [PubMed] [Google Scholar]
- Van de Wetering M, et al. Prospective derivation of a 'Living Organoid Biobank' of colorectal cancer patients. Cell. 2015;161(4):933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]


