Abstract
Here we provide novel convergent evidence across three independent cohorts of healthy adults (n=531) demonstrating that a common polymorphism in the gene encoding the α2 subunit of neuronal voltage-gated type II sodium channels (SCN2A) predicts human general cognitive ability or “g.” Using meta-analysis, we demonstrate that the minor T allele of a common polymorphism (rs10174400) in SCN2A is associated with significantly higher “g” independent of gender and age. We further demonstrate using resting-state fMRI data from our discovery cohort (n=236) that this genetic advantage may be mediated by increased capacity for information processing between the dorsolateral prefrontal cortex and dorsal anterior cingulate cortex, which support higher cognitive functions. Collectively, these findings fill a gap in our understanding of the genetics of general cognitive ability and highlight a specific neural mechanism through which a common polymorphism shapes inter-individual variation in “g.”
Keywords: SCN2A, general cognitive ability, “g” prefrontal cortex, functional magnetic resonance imaging (fMRI)
Introduction
The vast capacity of the human mind to engage in complex thought can be captured by a single factor representing general cognitive ability or “g” (Johnson, Bouchard, Krueger, McGue, & Gottesman, 2004; Spearman, 1904). There is, however, considerable inter-individual variability in “g,” which manifests in multiple domains of functioning including academic and occupational achievement as well as physical and mental health (Deary, 2012). There is clear evidence that a substantial proportion (30–80%) of inter-individual variability in “g” reflects heritable genetic differences (Deary, Penke, & Johnson, 2010; Plomin, Haworth, Meaburn, Price, & Davis, 2013). However, no specific genes have been reliably identified as contributing to this heritable variability. Here we provide convergent evidence that a common polymorphism in the human SCN2A gene reliably predicts inter-individual variability in “g,” and that genotype effects on prefrontal cortex physiology mediate this relationship.
The SCN2A gene, located on 2q24.3, encodes the α2 subunit of voltage-gated type II sodium channels, which contribute to the generation and propagation of action potentials throughout the adult central nervous system (Eijkelkamp et al., 2012; LITT, 1989). The α2 subunit encoded by SCN2A (Nav1.2) specifically mediates the conformational change of sodium channels based on voltage differences across cell membranes (Ahmed et al., 1992). Thus, variation in SCN2A may impact nervous system function by altering Nav1.2-modulated sodium channel regulation of neuronal activity and signaling. In fact, early studies in mice (Kearney et al., 2001) and humans (Sugawara et al., 2001) have linked mutations in SCN2A with abnormal electrical activity in the form of epileptic seizures (Heron et al., 2010; Kamiya et al., 2004). Furthermore, antiepileptic drugs that inhibit Nav1.2 activity (e.g., topiramate) are associated with diminished intellectual function.
Subsequent research has identified de novo mutations in SCN2A as primary genetic candidates in neurodevelopmental disorders including intellectual disabilities as well as autism spectrum disorders and schizophrenia, where impaired cognitive performance is often observed (Hoischen, Krumm, & Eichler, 2014). Recently, a genome-wide association study of cognitive function in patients with schizophrenia identified a common single nucleotide polymorphism in SCN2A (rs10174400; C/T) that explained 10.4% of the variance in “g” in a sample of patients with schizophrenia and 3.4% of the variance in their unaffected siblings (Dickinson et al., 2014). Specifically, the minor T allele was associated with poorer performance in both cases and siblings. Association was also found in two further small samples of patients with schizophrenia, though with smaller overall effect. Moreover, in postmortem brain samples from patients with schizophrenia, the minor T allele was associated with relatively decreased SCN2A mRNA in dorsolateral prefrontal cortex (dlPFC). It was also associated with relatively increased or “inefficient” dlPFC activity during a working memory task (Dickinson et al., 2014). Interestingly, a trend in the opposite direction was observed in data from healthy controls wherein the minor T allele was associated with higher “g” as well as relatively increased SCN2A mRNA in postmortem dlPFC and greater DLPFC efficiency (Dickinson et al., 2014).
In the present study, we further investigated the relationship between SCN2A rs10174400 and “g” in three independent cohorts of healthy individuals (total n = 531), including control participants from the cohort originally reported by Dickinson et al. Given the critical importance of the dlPFC and interconnected cortical structures in supporting “g” (Barbey, Colom, & Grafman, 2013; Duncan et al., 2000), we further tested if effects of SCN2A rs10174400 on “g” were mediated by differences in dlPFC functional coupling at rest using fMRI data available in 236 participants from our discovery sample. Based on prior research, we hypothesized that there would be relatively increased “g” associated with the SCN2A minor T allele, and that this genotype effect would be mediated by increased dlPFC intrinsic coupling with other cortical regions.
Methods
Discovery Cohort
Participants
Participants were recruited as part of the Duke Neurogenetics Study, an ongoing study investigating biological mechanisms of individual differences in brain function and behavior. Informed consent was obtained for all participants as approved by the Duke University School of Medicine Institutional Review Board. All participants were healthy, young adult volunteers free of the following exclusion criteria including: (1) medical diagnoses of cancer, stroke, head injury with loss of consciousness, untreated migraine headaches, diabetes requiring insulin treatment, chronic kidney or liver disease, or lifetime history of psychotic disorder; (2) use of psychotropic, glucocorticoid, or hypolipidemic medication; and (3) conditions affecting cerebral blood flow and metabolism (e.g. hypertension). The DNS seeks to establish broad variability in multiple behavioral phenotypes related to psychopathology, so other than psychotic disorders, participants were not excluded based on diagnosis of any other past or current DSM-IV Axis I or select Axis II (borderline and anstisocial personality) disorder. No participants were taking psychotropic medication at the time or at least 10 days prior to study participation. All reported analyses were restricted to 236 self-reported non-Hispanic Caucasian participants who completed the DNS by January 1, 2014.
Genotyping
Genotyping was conducted by 23andMe, Inc. Genomic DNA from all participants was isolated from buccal cells derived from Oragene DNA self-collection kits (DNA Genotek, Inc., Kanata, Canada) customized for 23andMe. DNA extraction and genotyping were performed at the National Genetics Institute, a CLIA-certified clinical laboratory and subsidiary of Laboratory Corporation of America. The Illumina Omni Express Plus chip and a custom array containing an additional ~300,000 SNPs were used to provide genome-wide data (Eriksson et al., 2010; Joëls, Fernandez, & Roozendaal, 2011; Tung et al., 2011). The Illumina Omni Express Plus chip included SCN2A rs10174400, and genotypes were extracted from the master database using the ‘extract’ command in Plink (Purcell et al., 2007). Standard quality control steps were applied using the following parameters: SNP call rate >90%; sample call rate >90%; minor allele frequency >5%; SNP Hardy–Weinberg equilibrium P > 0.05. Population stratification was examined using identity-by-state (IBS) analysis in PLINK of the whole-genome SNPs, extracting the first four multidimensional scaling (MDS) components.
Derivation of “g”
All participants were administered a neuropsychological battery from which the following cognitive measures were utilized in the discovery cohort to derive a “g” score based on Dickinson et al. (2014): 1) California Verbal Learning Test (CVLT-II) Trials 1–5 total, 2) Symbol Digit Modalities Test (SDMT), 3) Trail Making Test, 4) Digit Span, 5) Verbal Fluency, and 6) Wechsler Abbreviated Scale of Intelligence (WASI) subscales of Vocabulary, and Matrix Reasoning. Summary performance scores from each measure were converted to z-scores. These standardized scores were entered into a principal components analysis (PCA) and the first principal component was used as the index of “g” (Ree & Earles, 1991; Spearman, 1904).
BOLD fMRI Data Acquisition
Each participant was scanned using a research-dedicated GE MR750 3T scanner at the Duke-UNC Brain Imaging and Analysis Center. This scanner is equipped with high-power, high-duty-cycle 50 mT/m gradients at 200 T/m/s slew rate and an 8-channel head coil for parallel imaging at high bandwidth up to 1 MHz. A semi-automated high-order shimming program was used to maximize global field homogeneity. To allow for spatial registration of each participant’s data to a standard coordinate system, high-resolution 3-dimensional structural images were acquired in 34 axial slices coplanar with the functional scans [repetition time (TR)/echo time (TE)/flip angle = 7.7 s/3.0 ms/12°; voxel size = 0.9 × 0.9 × 4 mm; field of view (FOV) = 240 mm; interslice skip = 0]. For the 4:16 resting-state scan, a series of 34 interleaved axial functional slices aligned with the anterior commissure–posterior commissure plane were acquired for whole-brain coverage using an inverse-spiral pulse sequence to reduce susceptibility artifact (TR/TE/flip angle = 2000 ms/30 ms/60°; FOV = 240 mm; voxel size = 3.75 × 3.75 × 4 mm; interslice skip = 0). Four initial radiofrequency excitations were performed (and discarded) to achieve steady-state equilibrium. Participants were shown a blank gray screen and instructed to lie still with their eyes open, think about nothing in particular, and remain awake.
BOLD fMRI Data Preprocessing
Preprocessing of all resting-state fMRI data was conducted at the Laboratory of NeuroGenetics at Duke University using SPM8 (www.fil.ion.ucl.ac.uk/spm). Images for each subject were slice-time-corrected, realigned to the first volume in the time series to correct for head motion, spatially normalized into a standard stereotactic space (Montreal Neurological Institute template) using a 12-parameter affine model (final resolution of functional images = 2 mm isotropic voxels), and smoothed to minimize noise and residual differences in gyral anatomy with a Gaussian filter, set at 6 mm full-width at half-maximum. Voxel-wise signal intensities were ratio normalized to the whole-brain global mean.
Resting-State fMRI Data Analysis
A seed region in left dorsolateral prefrontal cortex (l-dlPFC) was used in the analyses [−42 16 36] and right dorsolateral prefrontal cortex (r-dlPFC) [42 16 36]. Mean time series were extracted from each ROI and subsequently subjected to a temporal band-pass filter, retaining frequencies between 0.008 and 0.09 Hz. These time series were entered into regression models including individual head motion realignment parameters as well as mean global gray matter, ventricular, and white matter signals. The statistical images produced (i.e., Z-score images) were then used in second-level random effects models to determine intrinsic coupling with the dlPFC using multiple regression, covarying for SCN2A genotype. Whole brain correlations were examined in SPM8. Correction for multiple comparisons was conducted in AFNI’s 3dClustSim using cluster-size thresholding based on Monte Carlo simulation. This creates multiple simulated null datasets from which a distribution of cluster sizes corresponding to a desired corrected p-value can be determined. An initial, uncorrected, statistical threshold of p<0.001 was chosen. Based on this threshold, the number of comparisons in our imaging volume and the smoothness of our imaging data, as measured by 3dFWHMx, a minimum cluster size of 85 voxels was required to have a corrected p≤ 0.05. Mean BOLD values from significant clusters were extracted using the Volume of Interest (VOI) tool in SPM8 for the cluster average for those clusters exhibiting a significant positive association with the dlPFC time series using the above threshold. These extracted values were then used in a statistical model using IBM SPSS Statistics 22 (Chicago, IL, USA).
Relationship between SCN2A-modulated dlPFC intrinsic coupling and “g”
A general linear model was utilized to assess the correlation between the extracted resting-state functional coupling values and “g” both entered as continuous variables. This model also included gender, age, diagnosis, and MDS components as covariates. A formal mediation model was tested using the PROCESS model 4 for SPSS (Hayes, 2012), with SCN2A rs10174400 genotype as the independent variable, dlPFC-dACC connectivity as the mediator, and “g” as the dependent variable. Again, gender, age, diagnosis, and MDS components were entered as covariates.
Replication Cohort 1
Participants
Participants in the Zucker Hillside Hospital (ZHH) cohort were recruited from the New York metropolitan area via advertisements, word of mouth, referrals, and study registries (Lencz et al., 2014). Participants had no history of a current or past DSM-IV Axis I major mood or psychotic disorder as assessed by structured diagnostic interview using the SCID-I (First, Spitzer, Gibbon, & Williams, 1995). Additional exclusion criteria included: (1) intellectual or learning disability; and (2) significant medical illness that could affect brain structure. Written informed consent according to the Institutional Review Board of the North Shore – Long Island Jewish Health System was obtained from all participants prior to neurocognitive testing and collection of blood (via venipuncture) for DNA extraction. All analyses were restricted to self-reported non-Hispanic Caucasian participants. Only participants with complete data were included in the present study, leading to a smaller sample than used in some previous reports (Lencz et al., 2014)
Genotyping
All participants were genotyped on the Illumina OmniExpress (San Diego, CA, USA) microarray platform for ~770 K SNPs. A standard GWAS quality control pipeline was applied to the dataset using the following parameters: SNP call rate >95%; sample call rate >90%; SNP Hardy–Weinberg equilibrium P > 10e-6. Possible occult population stratification was modeled using principal components analysis of genotypes in SVS 7.7.4 (GoldenHelix, Bozeman, MT, USA).
Cognitive Measures
Participants were recruited to serve as healthy comparisons for studies of patients with schizophrenia and other psychiatric disorders at ZHH. Thus, to target deficits most relevant to psychosis, the MATRICS Consensus Cognitive Battery (MCCB) was used as the cognitive assay (Kern et al., 2008; Nuechterlein et al., 2008). The MCCB evaluates seven domains of cognitive function including: 1) speed of processing using the Brief Assessment of Cognition in Schizophrenia (BACS), Trail Making Test part A, and Semantic Fluency (category: animals); 2) working memory using the spatial span task from the Wechsler Memory Scale - III (WMS-III) and a letter-number span task); 3) verbal learning using the Hopkins Verbal Learning Test - Revised (HVLT-R); 4) visual learning using the Brief Visuospatial Memory Test - Revised (BVMT-R); 5) reasoning and problem solving using the Neuropsychological Assessment Battery (NAB) Mazes subtest. Summary performance scores from each measure above were converted to z-scores. These standardized scores were entered into a PCA and the first principal component was used as the index of “g.”
The MCCB also generally tests attention/vigilance using the Continuous Performance Test – Identical Pairs version (CPT-IP); however, data were missing for >10% of participants and thus this measure was not included in the derivation of “g.” Additionally, the MCCB includes a measure of social cognition using the Mayer-Salovey-Caruso Emotional Intelligence Test (MSCEIT) that was not relevant for the present analyses.
Replication Cohort 2
Participants
Methods for this study have been detailed in a previous publication (Dickinson et al., 2014). In the present analysis, neurocognitive data was re-analyzed to derive “g” based on PCA for healthy participants with complete data, in order to keep methods consistent across cohorts and allow for the derivation of “g” factor scores similar to that in our discovery and first replication cohorts. Given that only participants with complete data were included, the present results include a smaller sample than used in some previous reports (Dickinson et al., 2014).
Overall Methods
SCN2A genotype association with “g”
The effect of SCN2A rs10174400 genotype on PCA-derived “g” factor scores was assessed using a general linear model for each individual cohort while covarying for effects of gender, age, and, when applicable, clinical diagnosis. While all analyses were restricted to non-Hispanic Caucasian participants, PCA or MDS derived ancestry components where utilized to account for possible occult genetic stratification within each cohort. Meta-analysis of the association between SCN2A rs10174400 genotype and “g” across the two replication cohorts as well as all three cohorts was performed using the Stouffer’s combined method.
Results
Cohort Demographics
Demographic information for each of the three cohorts is provided in Table 1. SCN2A rs10174400 allele frequencies did not deviate from Hardy-Weinberg equilibrium (p>0.05). Analyses in all three cohorts were limited to non-Hispanic Caucasian participants with complete data on all measures of interest including those used to derive “g” as well as SCN2A rs10174400 genotype. Additional participants were removed as genetic outliers. After trimming of genetic ancestry outliers, the final n=236 in the Discovery Cohort, n=149 in Replication Cohort 1 and n=119 in Replication Cohort 2. In our discovery cohort from the ongoing Duke Neurogenetics Study (DNS), 47 participants (19.9%) had a past or current DSM-IV defined Axis I diagnosis, the most common of which was substance abuse or dependence (13.5%). In Replication Cohort 1, 10 participants (6.7%) had a past or current DSM-IV diagnosis as determined by semi-structured interview, the most common of which was substance abuse or dependence (4.7%). Thus, all analyses in our discovery and replication 1 cohorts explicitly controlled for categorical presence of disorder in addition to age and gender. Participants from the replication 2 cohort were free of any past or current DSM-IV diagnosis as determined by structured clinical interview.
Table 1.
Demographic Characteristics of the Discovery and Replication Cohorts.
| Discovery Cohort | Replication Cohort 1 | Replication Cohort 2 | |
|---|---|---|---|
| N | 236 | 149 | 119 |
| Age (SD) | 19.72 (1.18) | 38.69 (18.1) | 31.46 (10.13) |
| Gender-Female | 50.8% | 54.4% | 56.2% |
| SCN2A rs10174400 | |||
| C/C | 110 | 56 | 42 |
| C/T | 99 | 73 | 64 |
| T/T | 27 | 20 | 13 |
Effect of SCN2A Genotype on “g”
A significant effect of genotype was found on “g” in our discovery cohort (p=0.021) after controlling for age, gender, diagnosis, and occult genetic stratification. Specifically, lower cognitive ability was observed in individuals with CC (b=−0.57, p=0.008) and CT (b=−0.57, p=0.009) genotypes compared to those with the TT genotype (Figure 1a, Table 2). Overall, SCN2A rs10174400 genotype accounted for approximately 2% (R2=0.021) of the inter-individual variability in “g” in our discovery cohort.
Figure 1. SCN2A rs10174400 Genotype Effects on Mean PCA-Derived “g” Factor Scores in Three Cohorts.

(A) In our discovery cohort, the TT genotype was associated with significantly higher “g” scores in comparison with the CC and CT genotypes (p=0.021). (B) A trend for higher “g” scores were observed in the TT genotype group in the first replication cohort (p=0.086). (C) A trend for higher “g” scores was found in the TT genotype group in the second replication cohort (p=0.051). Plots reflect the model adjusted mean after controlling for sex, age, diagnosis and genetic ancestry. The Y axes reflect least squares means. Error bars reflect standard errors.
Table 2.
Association of SCN2A rs10174400 (T allele) with general intelligence (g) in three independent cohorts of non-Hispanic Caucasians.
| Genotype | Discovery cohort
|
Replication 1
|
Replication 2
|
MetaP† | Meta_P | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | SE | P | P* | R2 | Beta | SE | P | P* | R2 | Beta | SE | P | P* | R2 | |||
| Unadjusted | |||||||||||||||||
| 0 | −0.612 | 0.213 | 0.004 | 0.0137 | 0.036 | −0.428 | 0.209 | 0.043 | 0.0581 | 0.025 | −0.459 | 0.300 | 0.128 | 0.1284 | 0.028 | 0.01573 | 0.00069 |
| 1 | −0.579 | 0.215 | 0.008 | −0.481 | 0.202 | 0.019 | −0.138 | 0.288 | 0.633 | ||||||||
| 2 | 0.000 | . | . | 0.000 | . | . | 0.000 | . | . | ||||||||
| Adjusted | |||||||||||||||||
| 0 | −0.567 | 0.213 | 0.008 | 0.0209 | 0.021 | −0.408 | 0.216 | 0.062 | 0.0855 | 0.022 | −0.541 | 0.325 | 0.099 | 0.0507 | 0.048 | 0.00939 | 0.00055 |
| 1 | −0.570 | 0.215 | 0.009 | −0.467 | 0.210 | 0.028 | −0.073 | 0.312 | 0.815 | ||||||||
| 2 | 0.000 | . | . | 0 | . | . | 0.000 | . | . | ||||||||
Note: Genotype, the number of minor T alleles; P, p value for testing for the mean difference in g of specifically genotype in comparison with TT genotype; P*: genotypic test for association with three genotypes; R2, R-square for SCN2A rs10174400 genotype; Adjusted: analysis adjusted for genetic ancestry using first four PCA and MDS derived from whole genome of SNPs; meta-P, combined p value of three cohorts using Stouffer’s method for meta-analysis. MetaP†, combined p values for the two independent replication cohorts.
The same pattern was found in our two independent replication cohorts after controlling for age, gender, diagnosis and genetic stratification (Replication cohort 1, p=0.086, Figure 1b; Replication cohort 2, p=0.051, Figure 1C). Similar to our Discovery Cohort, in Replication Cohort 1, lower cognitive ability was observed in individuals with CC (b=−0.41, p=0.062) and CT (b=−0.47, p=0.028) genotypes compared to those with the TT genotype (Table 2). In Replication Cohort 2 lower cognitive ability was also observed in individuals with CC (b=−0.54, p=0.010) and CT (b=−0.07, p=0.82) genotypes compared to those with the TT genotype (Table 2). The models explained 2.2% (R2=0.022) of the inter-individual variability in the first replication cohort and 4.8% (R2=0.048) in the second.
Meta-analysis across our three independent cohorts adjusting for genetic ancestry provided stronger evidence for the impact of SCN2A rs10174400 on “g” (p=0.0006; see Table 2 for additional details). Similar results were obtained from a meta-analysis of the two replication cohorts only (p=0.009; Table 2).
Effect of SCN2A Genotype on Prefrontal Physiology
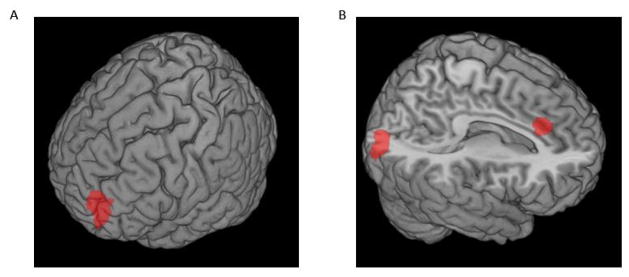
Analyses of resting-state fMRI data in our discovery cohort revealed significant modulatory effects of SCN2A genotype on intrinsic coupling of both right and left dlPFC (Figure 2). Specifically, coupling between the left dlPFC and dorsal anterior cingulate cortex (dACC; x=6, y=28, z=28; p<0.05 corrected, cluster size=94 voxels) as well as left dlPFC and middle occipital gyrus (MOG; x=20, y=−86, z=12; p<0.05 corrected, cluster size=197 voxels) was modulated by SCN2A genotype with higher coupling in minor T allele carriers. SCN2A genotype also modulated intrinsic coupling between the right dlPFC and middle frontal gyrus (MFG; x=−40, y=56, z=12; p<0.05 corrected, cluster size=383) again with greater coupling in minor T allele carriers. As just described, the T allele was also associated with better cognitive performance on neuropsychological testing.
Figure 2. SCN2A rs10174400 Genotype Modulates Intrinsic Coupling of Left and Right dlPFC.
(A) The minor T allele was associated with higher resting-state functional correlations between a right dlPFC seed and a cluster in the left middle frontal gyrus (MFG; x=−40, y=56, z=12; p<0.05 corrected, k=383). (B) The minor T allele was also associated with higher correlations between a left dlPFC seed and clusters in the dorsal anterior cingulate cortex (dACC; x=6, y=28, z=28; p<0.05 corrected, cluster size=94 voxels) and middle occipital gyrus (MOG; x=20, y=−86, z=12; p<0.05 corrected, cluster size=197 voxels).
Prefrontal Physiology Mediates Effects of SCN2A Genotype on “g”
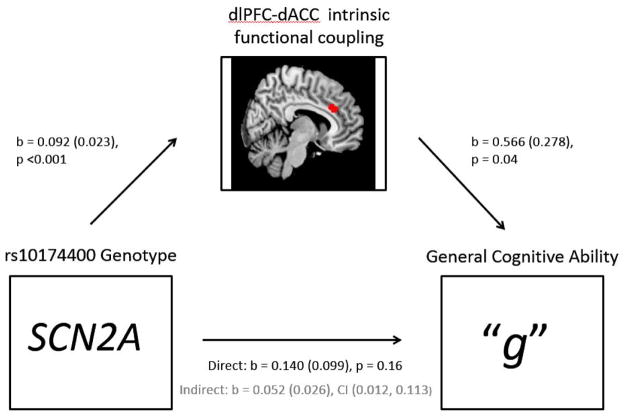
Analyses between general cognitive ability and the magnitude of SCN2A genotype-modulated instrinsic coupling between left dlPFC-dACC, left dlPFC-MOG, and right dlPFC-MFG, revealed a significant positive correlation only between “g” and coupling of left dlPFC-dACC (b=0.161, p=0.014, Adj. R2=0.025). This correlation was robust to the inclusion of age, gender, genetic stratification, and diagnosis as covariates, and survived a Bonferroni corrected threshold for multiple comparisons reflecting the three connectivity clusters included in the analyses (p<0.0167). Formal test of mediation (Figure 3) revealed that increased coupling between the left dlPFC and dACC mediated the association between the SCN2A rs10174400 minor T allele and higher “g” scores (direct effect: b=0.140 (0.099), p=0.16; indirect effect b=0.052 (0.026), CI (0.012, 0.113).
Figure 3. Mediation Model Demonstrating Effects of SCN2A on “g” Through Prefrontal Functional Coupling.
SCN2A rs10174400 minor T allele is associated with “g” in part through its effects on increased intrinsic functional coupling between the dlPFC and dACC (direct effect: b=0.140 (0.099), p=0.16; indirect effect b=0.052 (0.026), CI (0.012, 0.113). Note: the low p-value between SCN2A and dlPFC-dACC connectivity in the model is a result of the region being selected due to its modulation by SCN2A.
Discussion
Building from results of a recent genome-wide association study of cognition in patients with schizophrenia and earlier work on de novo mutations associated with intellectual disability and neurodevelopmental disorders (Dickinson et al., 2014; Hoischen et al., 2014; Sanders et al., 2012), we provide converging evidence in three independent samples that a common polymorphism in SCN2A accounts for significant inter-individual variability in general cognitive ability or “g.” Across the three independent samples of healthy participants SCN2A rs10174400 explained approximately 2–5% of the variance in “g” with higher scores in carriers of the minor T allele. Analyses of in vivo prefrontal cortex physiology assayed with resting-state fMRI in our discovery cohort suggest that this effect of SCN2A rs10174400 on “g” is mediated through relatively increased intrinsic coupling between regions of prefrontal cortex in minor T allele carriers.
SCN2A encodes for the α2 subunit (Nav1.2) of voltage-gated type II sodium channels, which are expressed throughout the central nervous system (Felts, Yokoyama, Dib-Hajj, Black, & Waxman, 1997; Whitaker et al., 2000), and may contribute to the development of cortical and hippocampal circuits (Eijkelkamp et al., 2012; Lai & Jan, 2006). Interestingly, the antiepileptic drug topiramate, which acts in part by inhibiting voltage-gated sodium channels, has been associated with mild to moderate cognitive impairment in some patients (Mula, 2012). This is consistent with the divergent effects of the SCN2A rs10174400 minor T allele on Nav1.2 mRNA expression in the dlPFC with relatively increased expression in healthy controls but decreased expression in patients with schizophrenia as well as being associated with relatively “inefficient” dlPFC activity during a working memory task in schizophrenia (Dickinson et al. 2014). Thus, in healthy participants relatively increased expression of Nav1.2 associated with the minor T allele of SCN2A rs10174400 may represent a molecular mechanism facilitating increased neural circuit function supporting general cognitive ability.
The results of our resting-state fMRI analyses suggest that SCN2A rs10174400 genotype effects on prefrontal cortex physiology in vivo may represent a systems level mechanism contributing to the pro-cognitive effects of the minor T allele. Specifically, relatively increased intrinsic functional coupling between the left dlPFC and dACC mediated the better cognitive performance that we observed in minor T allele carriers. In other words, a tighter correlation between left dlPFC and dACC was found in individuals with at least one copy of the minor T allele, which appears to have a “downstream” effect on cognitive performance in these individuals. The dlPFC and dACC are key nodes within multiple overlapping prefrontal circuits supporting a broad range of cognitive processes including attention, working memory, conflict detection, and behavioral control (Kondo, Osaka, & Osaka, 2004; Jung & Haier, 2007). We hypothesize that the observed increase in intrinsic functional coupling between these two prefrontal nodes may reflect a greater capacity for information processing in support of general cognitive functioning or it may reflect that individuals with the minor T allele tend to experience the fMRI environment in a more cognitively coherent or predictable manner. The prior report of relatively decreased and thus more efficient dlPFC activity during working memory in healthy carriers of the minor T allele is consistent with this hypothesis (Dickinson et al. 2014).
There was also evidence that SCN2A rs10174400 genotype modulated the intrinsic functional coupling between the dlPFC and middle occipital gyrus. However, this pattern of intrinsic coupling did not mediate the association between SCN2A genotype and general cognitive ability. We speculate, that this effect of SCN2A genotype may reflect the importance of voltage-gated type II sodium channels to processing of specific sensory inputs rather than higher-order information processing. Further research could explore SCN2A rs10174400 genotype associations with performance on tasks eliciting variability in visual processing and/or disorders characterized by visual deficits.
Our current work is not without limitations. In an effort to isolate the possible effects of SCN2A rs10174400 on “g” and reduce confounds associated with genetic background reflected in ancestry, we restricted our analyses to non-Hispanic Caucasian participants. While allowing for a more incisive examination of genotype effects on general cognitive ability, our strategy necessarily limits our ability to generalize the observed associations to other groups, which should be extended in future studies. This selection strategy also reduced the sample size of each cohort in our study. However, the meta-analytic results provide support that the association between SCN2A rs10174400 and “g” is significant when pooled across the three datasets. Also, the pattern of association in the three samples was the same, with minor alleles being associated with higher “g.” Furthermore, this association was robust to possible effects of age, gender, genetic ancestry, and psychopathology, and expressed across a broad range of “g.” Nevertheless, larger sample sizes are welcome and would further afford opportunities to examine the extent to which epistatic and epigenetic mechanisms as well as gene-environment interactions may contribute to the divergent effects of the minor T allele in healthy controls and both patients with schizophrenia and their unaffected siblings (Dickinson et al., 2014). Lastly, the molecular mechanisms through which SCN2A rs10174400 genotype ultimately shapes inter-individual variability in “g” remain unclear. However, our current results provide a tangible intermediate neural phenotype as a starting point for further study.
These limitations notwithstanding, our findings provide convergent evidence that a common polymorphism in SCN2A accounts for significant inter-individual variability in human general cognitive ability, possibly by modulating prefrontal cortex physiology. Our findings further highlight how focused candidate studies of genes initially identified through de novo mutations and genome-wide screens can advance our understanding of biological mechanisms that help shape individual differences in complex traits. In turn, the elucidation of such biological mechanisms can inform the search for novel therapeutic targets to improve cognitive deficits we often see in common neurologic and psychiatric disorders such as autism, schizophrenia, and epilepsy.
Acknowledgments
We would like to thank Yuliya S. Nikolova, Jacob Miller, Annchen Knodt, Spenser Radtke, Johnna Swartz and Jamie Hanson from the Laboratory of NeuroGenetics at Duke University.
Funding
MS is supported by an NSF Graduate Research Fellowship. The DNS is supported by Duke University as well as NIDA grants R01DA031579 & R01DA033369. The work conducted at Zucker Hillside Hospital was supported in part by an Advanced Center for Intervention and Services Research (P30 MH090590) grant and a Center for Intervention Development and Applied Research (P50 MH080173) from NIH. DD is supported by the Division of Intramural Research Programs, NIMH, NIH.
Footnotes
Conflict of Interest Disclosures
AKM is on the Scientific Advisory Board of Genomind, Inc. and a consultant for Takeda, Forum Pharmaceuticals.
References
- Ahmed CM, Ware DH, Lee SC, Patten CD, Ferrer-Montiel AV, Schinder AF, Evans GA. Primary structure, chromosomal localization, and functional expression of a voltage-gated sodium channel from human brain. Proceedings of the National Academy of Sciences. 1992;89(17):8220–8224. doi: 10.1073/pnas.89.17.8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbey AK, Colom R, Grafman J. Dorsolateral prefrontal contributions to human intelligence. Neuropsychologia. 2013;51(7):1361–9. doi: 10.1016/j.neuropsychologia.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deary I. Intelligence. Annual Review of Psychology. 2012;63:453–82. doi: 10.1146/annurev-psych-120710-100353. [DOI] [PubMed] [Google Scholar]
- Deary I, Penke L, Johnson W. The neuroscience of human intelligence differences. Nature Reviews Neuroscience. 2010;11(March):201–211. doi: 10.1038/nrn2793. [DOI] [PubMed] [Google Scholar]
- Dickinson D, Straub RE, Trampush JW, Gao Y, Feng N, Xie B, Weinberger DR. Differential Effects of Common Variants in SCN2A on General Cognitive Ability, Brain Physiology, and messenger RNA Expression in Schizophrenia Cases and Control Individuals 71(6), 647–656. JAMA Psychiatry. 2014;21:205. doi: 10.1001/jamapsychiatry.2014.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan J, Seitz RJ, Kolodny J, Bor D, Herzog H, Ahmed A, Emslie H. A Neural Basis for General Intelligence. Science. 2000;289(5478):457–460. doi: 10.1126/science.289.5478.457. [DOI] [PubMed] [Google Scholar]
- Eijkelkamp N, Linley JE, Baker MD, Minett MS, Cregg R, Werdehausen R, Wood JN. Neurological perspectives on voltage-gated sodium channels. Brain_: A Journal of Neurology. 2012;135(Pt 9):2585–612. doi: 10.1093/brain/aws225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson N, Macpherson JM, Tung JY, Hon LS, Naughton B, Saxonov S, Mountain J. Web-based, participant-driven studies yield novel genetic associations for common traits. PLoS Genetics. 2010;6(6):e1000993. doi: 10.1371/journal.pgen.1000993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felts PA, Yokoyama S, Dib-Hajj S, Black JA, Waxman SG. Sodium channel a-subunit mRNAs I, II, III, NaG, Na6 and hNE (PN1): Different expression patterns in developing rat nervous system. Molecular Brain Research. 1997;45:71–82. doi: 10.1016/S0169-328X(96)00241-0. [DOI] [PubMed] [Google Scholar]
- First M, Spitzer R, Gibbon M, Williams J. Structured Clinical Interview for DSM-IV Axis I Disorders–Non-Patient Edition. New York: New York Biometrics Research Department, New York State Psychiatric Institute; 1995. [Google Scholar]
- Hayes AF. PROCESS: A versatile computational tool for observed variable mediation, moderation, and conditional process modeling. 2012 White Paper. [Google Scholar]
- Heron SE, Scheffer IE, Grinton BE, Eyre H, Oliver KL, Bain S, Mulley JC. Familial neonatal seizures with intellectual disability caused by a microduplication of chromosome 2q24.3. Epilepsia. 2010;51(9):1865–9. doi: 10.1111/j.1528-1167.2010.02558.x. [DOI] [PubMed] [Google Scholar]
- Hoischen A, Krumm N, Eichler EE. Prioritization of neurodevelopmental disease genes by discovery of new mutations. Nature Neuroscience. 2014;17(6):764–72. doi: 10.1038/nn.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joëls M, Fernandez G, Roozendaal B. Stress and emotional memory: a matter of timing. Trends in Cognitive Sciences. 2011;15(6):280–8. doi: 10.1016/j.tics.2011.04.004. [DOI] [PubMed] [Google Scholar]
- Johnson W, Bouchard TJ, Krueger RF, McGue M, Gottesman II. Just one g: consistent results from three test batteries. Intelligence. 2004;32(1):95–107. doi: 10.1016/S0160-2896(03)00062-X. [DOI] [Google Scholar]
- Jung RE, Haier RJ. The Parieto-Frontal Integration Theory (P-FIT) of intelligence: converging neuroimaging evidence. The Behavioral and Brain Sciences. 2007;30(2):135–54. doi: 10.1017/S0140525X07001185. discussion 154–87. [DOI] [PubMed] [Google Scholar]
- Kamiya K, Kaneda M, Sugawara T, Mazaki E, Okamura N, Montal M, Yamakawa K. A nonsense mutation of the sodium channel gene SCN2A in a patient with intractable epilepsy and mental decline. The Journal of Neuroscience. 2004;24(11):2690–8. doi: 10.1523/JNEUROSCI.3089-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney J, Plummer N, Smith M, Kapur J, Cummins T, Waxman S, Meisler M. A gain-of-function mutation in the sodium channel gene Scn2a results in seizures and behavioral abnormalities. Neuroscience. 2001;102(2):307–317. doi: 10.1016/S0306-4522(00)00479-6. [DOI] [PubMed] [Google Scholar]
- Kern RS, Nuechterlein KH, Green MF, Baade LE, Fenton WS, Gold JM, Marder SR. The MATRICS Consensus Cognitive Battery, part 2: co-norming and standardization. The American Journal of Psychiatry. 2008;165(2):214–20. doi: 10.1176/appi.ajp.2007.07010043. [DOI] [PubMed] [Google Scholar]
- Kondo H, Osaka N, Osaka M. Cooperation of the anterior cingulate cortex and dorsolateral prefrontal cortex for attention shifting. NeuroImage. 2004;23(2):670–9. doi: 10.1016/j.neuroimage.2004.06.014. [DOI] [PubMed] [Google Scholar]
- Lai HC, Jan LY. The distribution and targeting of neuronal voltage-gated ion channels. Nature Reviews Neuroscience. 2006;7(7):548–62. doi: 10.1038/nrn1938. [DOI] [PubMed] [Google Scholar]
- Lencz T, Knowles E, Davies G, Guha S, Liewald DC, Starr JM, Malhotra aK. Molecular genetic evidence for overlap between general cognitive ability and risk for schizophrenia: a report from the Cognitive Genomics consorTium (COGENT) Molecular Psychiatry. 2014;19(2):168–74. doi: 10.1038/mp.2013.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litt M. Localization of a human brain sodium channel gene (SCN2A) to chromosome 2. Genomics. 1989;5(2):204–208. doi: 10.1016/0888-7543(89)90047-5. [DOI] [PubMed] [Google Scholar]
- Mula M. Topiramate and cognitive impairment: evidence and clinical implications. Therapeutic Advances in Drug Safety. 2012;3(6):279–89. doi: 10.1177/2042098612455357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuechterlein KH, Green MF, Kern RS, Baade LE, Barch DM, Cohen JD, Marder SR. The MATRICS Consensus Cognitive Battery, part 1: test selection, reliability, and validity. The American Journal of Psychiatry. 2008;165(2):203–13. doi: 10.1176/appi.ajp.2007.07010042. [DOI] [PubMed] [Google Scholar]
- Plomin R, Haworth CMa, Meaburn EL, Price TS, Davis OSP. Common DNA markers can account for more than half of the genetic influence on cognitive abilities. Psychological Science. 2013;24(4):562–8. doi: 10.1177/0956797612457952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MaR, Bender D, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ree MJ, Earles JA. The stability of g across different methods of estimation. Intelligence. 1991;15(3):271–278. doi: 10.1016/0160-2896(91)90036-D. [DOI] [Google Scholar]
- Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey aJ, State MW. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485(7397):237–41. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spearman C. “General Intelligence,” Objectively Determined and Measured. The American Journal of Psychology. 1904;15(2):201. doi: 10.2307/1412107. [DOI] [Google Scholar]
- Sugawara T, Tsurubuchi Y, Agarwala KL, Ito M, Fukuma G, Mazaki-Miyazaki E, Yamakawa K. A missense mutation of the Na+ channel alpha II subunit gene Na(v)1.2 in a patient with febrile and afebrile seizures causes channel dysfunction. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(11):6384–9. doi: 10.1073/pnas.111065098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung JY, Do CB, Hinds Da, Kiefer AK, Macpherson JM, Chowdry AB, Eriksson N. Efficient replication of over 180 genetic associations with self-reported medical data. PloS One. 2011;6(8):e23473. doi: 10.1371/journal.pone.0023473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker WR, Clare JJ, Powell AJ, Chen YH, Faull RL, Emson PC. Distribution of voltage-gated sodium channel alpha-subunit and beta-subunit mRNAs in human hippocampal formation, cortex, and cerebellum. The Journal of Comparative Neurology. 2000;422:123–139. doi: 10.1002/(SICI)1096-9861(20000619)422:1<123::AID-CNE8>3.0.CO;2-X. pii. [DOI] [PubMed] [Google Scholar]