Abstract
Nanomaterials are increasingly prevalent throughout industry, manufacturing, and biomedical research. The need for tools and techniques that aid in the identification, localization, and characterization of nanoscale materials in biological samples is on the rise. Currently available methods, such as electron microscopy, tend to be resource-intensive, making their use prohibitive for much of the research community. Enhanced darkfield microscopy complemented with a hyperspectral imaging system may provide a solution to this bottleneck by enabling rapid and less expensive characterization of nanoparticles in histological samples. This method allows for high-contrast nanoscale imaging as well as nanomaterial identification. For this technique, histological tissue samples are prepared as they would be for light-based microscopy. First, positive control samples are analyzed to generate the reference spectra that will enable the detection of a material of interest in the sample. Negative controls without the material of interest are also analyzed in order to improve specificity (reduce false positives). Samples can then be imaged and analyzed using methods and software for hyperspectral microscopy or matched against these reference spectra in order to provide maps of the location of materials of interest in a sample. The technique is particularly well-suited for materials with highly unique reflectance spectra, such as noble metals, but is also applicable to other materials, such as semi-metallic oxides. This technique provides information that is difficult to acquire from histological samples without the use of electron microscopy techniques, which may provide higher sensitivity and resolution, but are vastly more resource-intensive and time-consuming than light microscopy.
Keywords: Bioengineering, Issue 106, Darkfield microscopy, hyperspectral imaging, metal oxide nanoparticles, histology, spectral angle mapping, hyperspectral mapping, tissue
Introduction
As nanomaterials are increasingly used in a variety of industries and applications, there is a need for nanoscale imaging and characterization methods that are more rapid, affordable, and convenient than traditional modalities, such as electron microscopy. To visualize nanoparticle (NP) interactions with cells, tissues, and living systems, many optical techniques have been employed, including differential interference contrast (DIC) microscopy1 and evanescent field-based approaches, such as total internal reflection (TIR) or near-field scanning optical microscopy (NSOM)2,3. However, these are high-end analytical approaches, out of reach of most non-specialist labs4. Electron microscopy, including transmission electron microscopy (TEM), has also been used to study NP interaction with cells5,6,7,8. High angle annular darkfield (HAADF) scanning transmission electron microscopy has been used to study the interaction of NPs with viruses9. Confocal microscopy is another popular technique used to study NP-cell interactions10.
In recent years, darkfield-based hyperspectral imaging (HSI) techniques have been used as a promising analytical tool for studying NPs in biological matrices11. HSI systems generate a three-dimensional representation of spatial and spectral data, known as a hypercube or datacube12. The spatial map of spectral variation is used for material identification. Spectral profiles of known materials can be generated and used as reference libraries for comparison to unknown samples. One of the major advantages with HSI systems is its ability to combine imaging with spectroscopy, thereby establishing the location and distribution of unknown NPs in vivo or ex vivo as well as linking them to another reference particle of known, similar composition.
There are several advantages of using HSI systems over conventional imaging techniques: minimal sample preparation is required; sample preparation is typically non-destructive in nature; image acquisition and analysis is faster; the technique is cost-effective13; and the spatial distribution and analysis of compounds of mixed composition and/or in complex matrices is more easily accomplished14.
For nanomaterials research involving precious samples, one of the most important considerations is the availability of a non-destructive imaging method, which allows for the potential to repeatedly examine samples by one or more methods. Repeated or multiple analyses may be desired to develop comprehensive datasets that would not be available from a single method. To this regard, studying its optical properties is the safest way to analyze the sample. By using an enhanced darkfield microscope (EDFM) and HSI system to study the optical response of the sample – namely reflectance, but also absorbance and transmittance – feature identification and characterization can be performed15. Potential characterization endpoints include an assessment of relative size and shape of nanoparticles or agglomerates and distribution of nanoparticles within a sample.
In this paper, we describe mapping methods specifically for metal oxide nanoparticles in post-mortem tissue using a HSI system based on a pixel-spectral match algorithm referred to as a spectral angle mapper (SAM). We chose this particular application because it has the potential to complement current and future in vivo nanotoxicology research, wherein animal models are used to evaluate the health implications of exposure to engineered nanomaterials. Application of this method could also inform nanoscale drug delivery research that utilizes tissue or animal models. In particular, nanoparticle absorption, distribution, metabolism, and excretion throughout organs and tissues could be investigated with this system. A wide variety of applications are being investigated for use in biomedical research11.
This method could be utilized for assessment of different biological samples (such as various tissues types, bronchoalveolar lavage samples, and blood smears) that have been exposed to nanoparticles of a variety of elemental compositions16-19. Furthermore, this method is useful for studying nanoparticle biodistribution in vivo and in vitro, which is pertinent for nanoscale drug delivery studies11. Beyond biological samples, EDFM and HSI can be used to evaluate nanoparticles in environmental samples, such as wastewater20. Occupational exposure assessment can be facilitated by the use of this technique as well, since it may be used to evaluate the efficacy of personal protective equipment in preventing nanoparticle penetration. Furthermore, the research team is currently developing a similar EDFM and HSI protocol for the evaluation of filter media samples of nanoparticles collected from occupational exposure assessments. While preparation of these different sample types for EDFM and HSI may vary, it is important that they are prepared in such a way that they may be easily visualized by the optical system. Typically, the sample should be prepared as if it would be visualized via traditional brightfield microscopy. There are several hyperspectral imaging systems commercially available11.
Protocol
Animal protocols were approved by the Institutional Animal Care and Use Committee at the investigators’ collaborating institution, Stony Brook University. A list of specific materials and equipment used for this paper can be found in Table 1.
1. Sample Preparation of Tissues
Prepare animal tissues exposed to metal oxide nanoparticles for histological or immunohistochemical staining as per previously described methods21-23.
2. Imaging
- Initializing the Microscope NOTE: For this study, a research grade microscope was used, outfitted with a high-performance darkfield light source, motorized XY stage controller, 14-bit depth hyperspectral camera, and multiple objective lenses (10X air, 40X air, 100X oil immersion). The system used here has a 64 nm spatial resolution at 100X (oil immersion) magnification. The condenser used for this study has both Koehler and critical illumination features and focuses a highly collimated light at oblique angles on the sample.
- Plug in and turn on the light source, XY stage controller, optical camera (for darkfield imaging), hyperspectral camera, and computer system. Set the light source to 75% power; raise the stage to maximum height; connect the light guide to the condenser for enhanced darkfield imaging or to the collimator on the back of the microscope for reflected brightfield imaging. NOTE: By setting the light source to 75% power, there is sufficient, uniform illumination of all pixels within the entire field of view that enhances duller pixels while still allowing for contrast between dull and bright pixels.
- Raise the condenser to operating position. Apply 3-5 drops of type-A immersion oil on the condenser lens carefully, avoiding the formation of any bubbles. Wipe away the oil and reapply it, if bubbles should form.
- Position the slide on the stage. Slowly raise the condenser until the immersion oil makes contact with the slide. This will be noticeable through the rapidly brightening ring of illumination where the oil makes contact with the slide.
- Put the 10X objective in place. Focus and align the condenser by examining through the oculars.
- Move the stage up and down via the coarse objective focus knob until brightness is maximized.
- Move the condenser focus up and down via the condenser adjustment knob until maximum brightness is found. Attempt to create the brightest central spot possible in the field of view.
- Adjust the condenser alignment knobs as necessary to center the bright spot, if needed.
- Use the fine objective focus knob to bring the bright spot into focus. When changing objectives to obtain a different magnification, readjust the focus plane. When utilizing the 100X objective, apply a drop of immersion oil over the coverslip to enhance the image and avoid damage to the objective lens.
- Capturing Images
- Use stage controller to find a region of interest. This region will be determined by the needs of the experiment. A typical indicator will be high contrast with surrounding regions, as areas suffused with metal oxide nanoparticles often appear brighter than areas without them.
- Bring the region into focus using the fine objective focus knob, adjusting the condenser focus as necessary to equilibrate illumination. Attain a high-contrast, well-defined region within the field of view.
- Determine what images (for the optical camera) or datacubes (for the hyperspectral camera) will be captured, and in what sequence. Typically, optical images are obtained with a 10X air objective, 40X air objective, and 100X oil immersion objective and a corresponding HSI datacube with a 100X oil immersion objective.
- Open the optical imaging software. Click on “Settings” in the menu bar. Select the “Image Capture Button for capture event”. Select the stored image format (TIFF was used for this experiment); assign a filename; browse and select a stored image folder; keep default timelapse; click “OK”.
- Select the exposure settings that create the highest contrast image in the “Exposure” menu (for this study, a level of 0.0%, gain of 3.0 dB, and shutter of 35 ms were used).
- Capture the image by clicking the “Image” button in the menu bar. Capture several low-magnification darkfield images in addition to those at high magnification by changing the microscope objectives, in order to provide context. NOTE: Capture optical images at the same magnification as any datacubes that will be captured with the hyperspectral camera, as these optical images tend to have a larger field of view and better aesthetic appearance for later visual inspection. It is essential that any datacubes that will later be spectrally analyzed use consistent magnification, as it is possible that changing the objective will alter the transmission spectra of the microscope optics, changing the captured spectra, and thus reducing the accuracy of the spectral angle mapper (SAM). If one datacube is compared to another datacube obtained with a different objective, the SAM function may not work.
- Select the hyperspectral camera imaging detector by redirecting the light source knob on the microscope to the hyperspectral camera. If the light is directed towards the hyperspectral camera, no image will be shown in the optical camera software. Minimize but do not close the optical imaging software window, as one may need it as specified in step 2.4.4.
- Capturing Datacubes
- Open the hyperspectral imaging software for acquisition of HSI datacubes. Ensure the light guide is directed to the hyperspectral camera.
- Open “Hyperspectral Microscope” in the menu bar and select “HSI Microscope Controls”.
- Set the objective magnification and the save path. Make sure to name all images and files distinctively so no overwriting occurs. Change the area capture in settings by adjusting the field of view or number of lines (use 720 lines for this study), with a significant amount of additional time required for capturing larger areas. Finally, set the exposure time (0.25 sec for this study). Leave everything else to default, and click “Preview HSI” to view the image. Note: The intensity graph that appears shows the HSI datacube that will be recorded in the horizontal axis. The vertical axis represents the wavelengths of the spectra that will be captured in the HSI datacube. Placing the cursor at any position on the intensity graph corresponding to a spectral wavelength causes the intensities of all the points across the image, at that wavelength, to be shown.
- Focus based on this preview by adjusting the fine objective focus to sharpen the peaks in this image. If this is too difficult, the optical camera’s focal length is fairly similar, so load the optical imaging software and shift the slide bar to the optical camera, focus, and then return to the hyperspectral imaging software and return the slide bar to the hyperspectral camera.
- Adjust the intensity from this preview by adjusting source brightness, condenser focus, or by cancelling the preview to adjust the exposure time. The latter yields the best results, but increased exposure time can take longer to image. The aim is for the more significant peaks to be sufficiently large, but not exceeding the maximum intensity for the detector; here, the ideal range is between 1,000 and 16,000 units.
- Click “capture”. The microscope will ask to take a dark current image. Redirect the upper slide bar or aperture (carefully, so as not to disturb the alignment and focus of any of the other optics), and click “OK”. Restore the slide bar or aperture to the correct position and click “OK” again when prompt to image appears. Imaging can take up to 30 min, though times of about 5 min are more typical. Longer exposure times lead to longer imaging times. A progress indicator is present. Be careful not to physically disturb the scope before the progress indicator completes.
- Observe four new windows with the names: “Available bands list”, “#1zoom”, “#1scroll” and “#1 RGB band”. Maximize the “#1RGB band” window as this is the datacube for all future references.
- Right click the datacube and save it as a TIFF and click “OK”. If finished imaging; put away all samples; clean all oil-exposed surfaces with 70% isopropanol in water; raise stage to maximum height; lower condenser to minimum height and press into non-operational position; shut down or unplug the light source, stage controller, and cameras.
3. Creation of Reference Spectral Libraries
- Selection of Reference Spectra
- Select a positive control that is known to contain the material of interest contained in the same matrix as the experimental samples, since HSI spectral profiles are matrix-dependent.
- For this study, use porcine skin injected with high doses of metal oxide nanoparticles as positive controls for comparison to the experimental porcine skin tissue from a topical exposure study. The spectral profiles of the metal oxide nanoparticles in suspension were generated and examined and found to be an inappropriate positive control for the histological experimental samples due to the different matrices.
- Obtain a datacube from the positive control as described in step 2.4., using the hyperspectral camera.
- Be particularly cautious when setting the intensity, as this is the primary metric by which the internal particle filter will identify the materials. Any intensities above the saturation point of the detector (here, 16,000 units) will result in invalid data (refer to step 2.4.5.).
- Right click in the image window, and left click “Z-profile spectrum”. A pop-up Spectral Profile window will appear. Left click onto pixels of interest on the datacube, particularly the brightest ones or those that can be confidently identified as representing the material of interest. Observe the Spectral Profile window showing the associated spectrum. Take note particularly of their lowest and highest value and which wavelength corresponds to it.
- When surveying the positive control sample, survey by clicking the regions of interest that are very bright relative to the surrounding tissue, especially those with readily identifiable particles. These particles are most likely to be the materials of interest, especially in the case of metals.
- Use the “Particle Filter” tool under the “Particle Analysis” menu to identify particles present in the datacube. In the new pop-up window, observe the “Spectral Max Must Exceed”. This will be determined by observations in step 3.1.3. Set this value so it is higher than background pixels, but lower than the materials of interest. The “Valid Data Max” is the maximum intensity (here, 16,000 units).
- Leave other parameters to default, but exclude objects based on size by adjusting the “Size Threshold” box. Save this data either at this point, or after running the analysis. For this experiment, use the following parameters: Spectral Max Must Exceed: 5,000; Valid Data Max: 16,000; Size Threshold (pixels): 400. Once all parameters have been set, click “OK”.
- Observe the resulting graph with the details of particles detected within the indicated intensity threshold; this data can be exported. If the characteristic maximum reflectance wavelength of the material of interest is known, select those particles that have a similar “Max WL” value; otherwise, click “select all”. Then click “Export”; “To Spectral Library”. Choose a file name then click “OK”.
- Remove False-positive Spectra
- Select a sample that will serve as a negative control. Such a sample should have been prepared and treated in the same way as all experimental samples save that the absence of nanoparticles similar or identical to the material of interest is guaranteed. NOTE: It is important that the matrix of the negative control is the same as the matrix of the experimental samples, as HSI spectral profiles are matrix-dependent. For this study, we used porcine skin that was not exposed to metal oxide nanomaterials as negative controls.
- Obtain several datacubes from the negative control as described in step 2.4.7, using the hyperspectral camera. At least one is required, but more can be captured to increase selectivity (this is especially important in the event that some contaminant may be in the reference spectra).
- Use the hyperspectral imaging software to filter the spectral library collected in step 3.1 (positive control) against each datacube captured in step 3.2.2 (negative controls) serially, as instructed in the following steps: NOTE: Save the resulting, filtered spectral library to a separate file, and use as the reference spectral library for the material of interest.
- Click “Filter Spectral Library” under the Analysis menu, located on the main program toolbar. Click “Open”; “New File” and select the spectral library created in step 3.1. for the positive control as the Input file. Click “OK”.
- For external source, select “Image” under the Spectral Data box. Keep default settings under the Processing Parameters box. Select an Output Name For Filtered Library (this should be different than the original, or the raw spectral library collected will be lost). Click “OK”.
- Click “Open”; “New File” and select the first datacube captured in step 3.2.2 for the negative control, when prompted to select a source image. NOTE: The software will analyze the spectral library and remove each spectrum that matches any spectrum of the negative control datacube. Removing the background from the selection criteria thereby reduces potential for false-positives. A summary will be available when this is done (note that this summary is not saved automatically).
- If additional filtering is desired, repeat from step 3.2.3.1, except: step 3.2.3.2, select the last filtered spectral library created (resulting after step 3.2.3.3. completes); in step 3.2.3.3, select the next datacube from step 3.2.2. NOTE: Correcting and normalizing for the lamp spectrum may be necessary if samples scatter a considerable amount of light (e.g., carbon nanotubes) and/or if zero spectra remain when filtering a positive control spectral library against a negative control. These circumstances did not arise for our study and so this correction was not performed.
4. Image Analysis
- Spectral Angle Mapping
- Open the “Spectral Angle Mapper” (SAM) from the Spectral menu, Mapping Methods submenu, using the hyperspectral imaging software once all the experimental datacubes have been acquired following step 2.4. The spectral angle mapper compares spectra by geometrically analyzing the changes in intensity as a function of wavelength (i.e., it compares two spectra by normalizing their intensity and comparing the angles required to trace the graph of each spectra)24.
- With the datacube open, select the name of the experimental datacube in the pop-up window and click “OK”. If no file names are listed, click “open”; “new file”, choose the experimental datacube, then click “OK”.
- Observe a new pop-up window called Endmember Collection:SAM. Click “Import” in the menu bar, then select “from Spectral Library file”. A pop-up window named Spectral Library Input File will appear. Open the Spectral Library of reference that was created in step 3.2.3. and click “OK”. Observe a new pop-up window named “Input Spectral Library”.
- Click “Select All Items”; click “OK” right click on “Color” and select “Apply default colors to all.” Click “Select all,” followed by “Apply” and then choose the output file name and click “OK”. The Spectral Angle Mapper will take a few seconds to analyze and save the data.
- Open the datacube in the hyperspectral imaging software. Select “Classification” in the Overlay menu of the image window, then navigate to the file name and click “OK”. The user can now overlay any spectrum from the library to see where they map to in the image.
- Select the “Merge Classes” option from the Option menu in the “Interactive class tool” opened through the Overlay menu in the image window, if a unified color scheme is desired, such as for easier analysis by other software (as described in step 4.2.).
- Highlight all the classifications to combine (typically, everything except “unclassified” in the “classes to merge into base” list), and select a single spectrum from the “base class” list, then click “OK”. That color will now represent all selected spectra.
- Click on the color selected and all matching spectra will be shown in that color.
- To obtain a dichromatic image that will show the matching spectra over a black background, cIick on the “unclassified” colors box, which is black by default. Observe a dichromatic image. This step can be reversed by clicking again on the “unclassified” colors box.
- Right click to save the image as a TIFF, including any overlays currently active. NOTE: For future imaging processing, a dichromatic image will be used.
- Select “Class Distribution” from the Option menu in the “Interactive Class Tool” window to acquire the mapping statistics. This data represents how many pixels were unmapped (unclassified) and how many were identified as the material of interest. Note that the number of pixels does not correspond with the number of particles mapped. This information is not automatically recorded.
- Particle Analysis in Hyperspectral Mapped Images
- Open the images in NIH ImageJ software.
- Use the Image menu, Adjust submenu, “Threshold” function. Select parameters that differentiate particles of interest from all other materials. Use the following threshold parameters for this study: default threshold method, color red, color space HSB, checked dark background checkbox, checked pass checkboxes for hue, saturation and brightness. NOTE: This may vary significantly from case to case. When analyzing a mapped datacube, this can be simplified by unifying all relevant classes to a single color and overlaying that color with all unclassified colors before generating the image (steps 4.1.4 to 4.1.8).
- Use the Analyze menu, “Analyze particles” function, which will retrieve information for area, mean, minimum and maximum values. For this study, use the parameters here: size 0-infinity, circularity 0-1, show nothing, checked display results checkbox.
- For statistical analysis, export this data to another software program enabling statistical comparison with the number and size of particles located in similar control samples. We suggest using a spreadsheet for analysis of ImageJ data. Data displayed in the results table after step 4.2.4 should be copied to a spreadsheet. The MAX function can be used on the first (untitled) column to determine the number of particles, while the AVERAGE function can be used on the Area column to determine mean size of a particle. In the future, experimental validation will be pursued to investigate this function in determining mean size against an established standard for sizing (e.g., TEM).
Representative Results
Hyperspectral microscopy is useful for its ability to identify materials in a manner similar to spectrophotometry. As indicated in Figure 1, each material has several characteristic spectra and an overall shape of its reflectance, which is unique. Furthermore, Figure 1 illustrates the matrix-dependent nature of the spectral profiles: the spectral profiles for each of the three metal oxides in histological tissue samples (top panel) are different from the spectral profiles for each of the three metal oxides in aqueous suspension (bottom panel). By mapping the characteristic spectral profiles to unknown samples in the same matrix, the technique is useful for determining presence or absence of a material, and can also semi-quantitatively compare relative amounts of material in samples.
The reference spectral libraries created from positive control skin samples are appropriate for mapping to experimental skin samples. As seen in Figure 2, the reference spectral libraries map well to the corresponding positive controls and do not map to the corresponding negative controls.
The most significant advantage of the technique is its ability to detect low levels of materials of interest in samples, as well as to differentiate them from contaminants. For example, individual nanotubes can be observed in lungs during inhalation of doses as low as 40 µg in a 20-30 g rat25. Figure 3 demonstrates this in histological skin samples: while some particles are readily obvious in the darkfield optical image (column 2), others are detected using hyperspectral imaging (column 3) that might have been missed using brightfield microscopy (column 1) or other methods. The use of darkfield HSI also serves to improve specificity over simple brightfield microscopy. These images can then be subject to more quantitative forms of analysis, notably particle analysis via ImageJ.
Enhanced darkfield microscopy has several different applications, even in the absence of hyperspectral imaging. The first is its ability to generate high quality, high-contrast images. This is best demonstrated by Figure 3, in which particles of interest are readily identifiable in comparison to their brightfield counterparts. These images can also be subject to quantitative particle analysis, though in the absence of hyperspectral mapping, caution is necessary to avoid confusing a contaminant particle from a particle of interest.
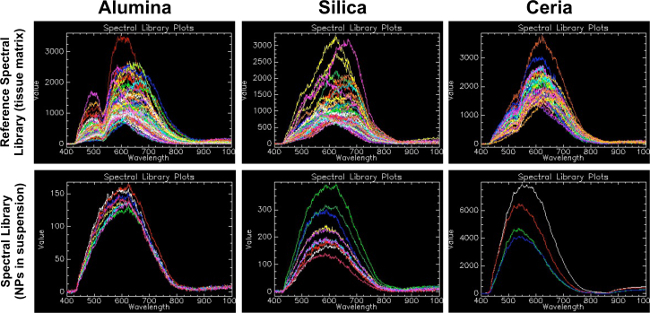 Figure 1. Reference spectral libraries (tissue matrix) compared to spectral libraries (nanoparticles in suspension). This figure demonstrates the importance of generating a reference spectral library from nanomaterials of interest in the same matrix as experimental samples. The first row shows the reference spectral library (RSL) for each material in this study (alumina, silica and ceria). Please click here to view a larger version of this figure.
Figure 1. Reference spectral libraries (tissue matrix) compared to spectral libraries (nanoparticles in suspension). This figure demonstrates the importance of generating a reference spectral library from nanomaterials of interest in the same matrix as experimental samples. The first row shows the reference spectral library (RSL) for each material in this study (alumina, silica and ceria). Please click here to view a larger version of this figure.
This RSL was created by the method described in this protocol, where positive control tissue samples were filtered against negative control tissue samples. The second row shows spectral libraries (SL) created from the nanoparticles of interest in liquid suspension on a glass slide. For alumina, a bimodal peak at wavelengths of 500 and 650 was found in the RSL, whereas a more curved and less distinctive peak was seen in the alumina NP suspension SL at around 600. For silica, a peak at a wavelength of around 650 was present in the RSL, whereas a more curved and less distinctive peak was seen in the silica NP suspension SL at around 600. For ceria, a bimodal peak was found at wavelengths of 520 and 620 in the RSL, whereas a less distinctive peak was seen in the ceria NP suspension SL at around 560. This suggests that the matrix in which the NPs are embedded creates a shift in spectra that can be considerable when selecting the controls to create the SL for as specific experiment.
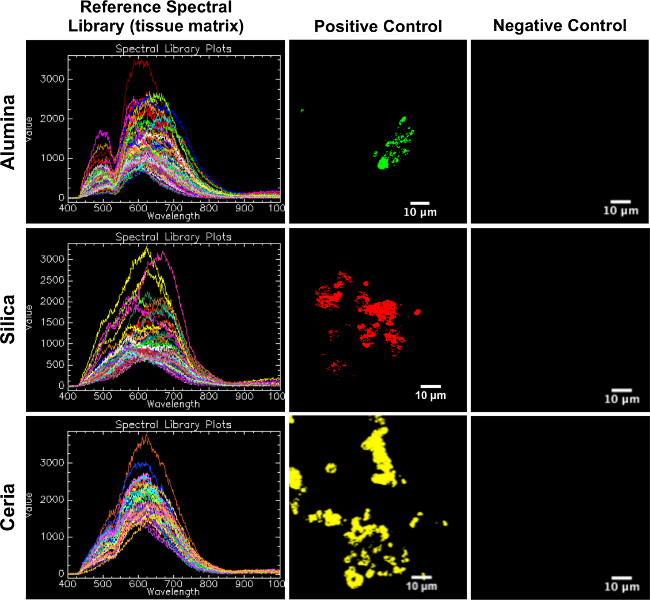 Figure 2. Reference spectral libraries (tissue matrix) mapped on positive control and negative control porcine skin tissue. This figure serves as confirmation of the RSLs as appropriate for mapping to experimental samples, in that each RSL (left column) maps well to its corresponding positive control (middle column) and does not map to its corresponding negative control (right column). Please click here to view a larger version of this figure.
Figure 2. Reference spectral libraries (tissue matrix) mapped on positive control and negative control porcine skin tissue. This figure serves as confirmation of the RSLs as appropriate for mapping to experimental samples, in that each RSL (left column) maps well to its corresponding positive control (middle column) and does not map to its corresponding negative control (right column). Please click here to view a larger version of this figure.
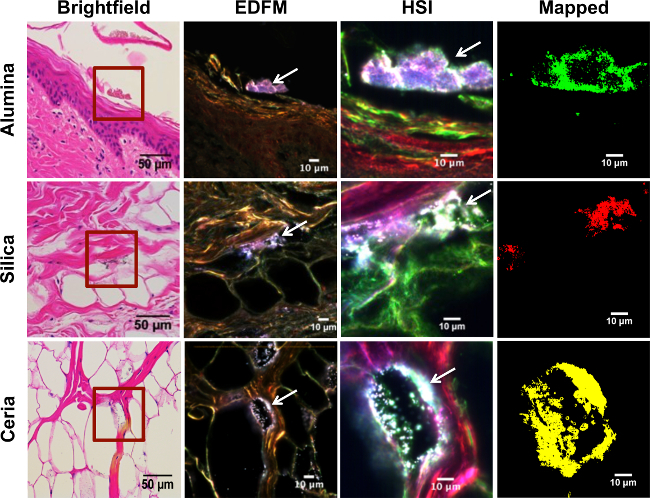 Figure 3. Hyperspectral mapping of experimental porcine skin tissue exposed to metal oxide nanoparticles. Rows from top to bottom correspond to different layers of the skin, from superficial to deepest layer: epidermis, dermis, and subcutaneous tissue, respectively. The first row shows the stratum corneum of the epidermis from a skin sample that was exposed to alumina NPs in suspension; the second row was exposed to silica NPs in suspension; and the third row to ceria NPs in suspension. Each column depicts the same area of the skin imaged with different techniques. The first column corresponds to brightfield microscopy (40X mag.; scale bar = 50 µm), where the area enclosed in a red square was magnified (100X mag; scale bar = 10 µm) with enhanced darkfield microscopy (EDFM) in the second column, where white arrows are pointing to the high contrast NPs . The third column was obtained with a hyperspectral camera (HSI), showing the same high contrast NPs (white arrows). EDFM and HSI images were taken at 100X magnification; scale bar = 10 µm. The fourth column shows the HSI image mapped against the respective RSL for each exposure group, where alumina NPs are shown in green, silica NPs in red, and ceria NPs in yellow, respectively. Please click here to view a larger version of this figure.
Figure 3. Hyperspectral mapping of experimental porcine skin tissue exposed to metal oxide nanoparticles. Rows from top to bottom correspond to different layers of the skin, from superficial to deepest layer: epidermis, dermis, and subcutaneous tissue, respectively. The first row shows the stratum corneum of the epidermis from a skin sample that was exposed to alumina NPs in suspension; the second row was exposed to silica NPs in suspension; and the third row to ceria NPs in suspension. Each column depicts the same area of the skin imaged with different techniques. The first column corresponds to brightfield microscopy (40X mag.; scale bar = 50 µm), where the area enclosed in a red square was magnified (100X mag; scale bar = 10 µm) with enhanced darkfield microscopy (EDFM) in the second column, where white arrows are pointing to the high contrast NPs . The third column was obtained with a hyperspectral camera (HSI), showing the same high contrast NPs (white arrows). EDFM and HSI images were taken at 100X magnification; scale bar = 10 µm. The fourth column shows the HSI image mapped against the respective RSL for each exposure group, where alumina NPs are shown in green, silica NPs in red, and ceria NPs in yellow, respectively. Please click here to view a larger version of this figure.
Discussion
For tissue samples that have undergone conventional histological staining, the identification and analysis of metal oxide nanoparticles may be achieved through a combination of EDFM, HSI mapping, and image analysis techniques. While there is flexibility in sample preparation for histology or immunohistochemistry (e.g., using fixed or frozen tissues; type of stain), it is important that samples are sectioned to a thickness of 5-10 µm for optimal visualization. Samples used here were formalin-fixed and paraffin-embedded prior to sectioning with a rotary microtome to 6 µm thickness, mounted on glass microscope slides, stained with hematoxylin and eosin and coverslipped. Porcine skin tissues from an ex vivo cutaneous penetration toxicology collaboration were used for this study. The tissues were exposed to metal oxide nanoparticles (alumina, silica, ceria) in aqueous suspensions. Detection of the region(s) of interest (high contrast elements) with EDFM is a critical first step that facilitates subsequent HSI mapping and analysis. Positive and negative control samples must be imaged and analyzed first in order to create a spectral library for reference. The collected spectra from the positive control are exported to a positive control spectral library. Then, all the spectra from the negative control images are subtracted from the positive control’s spectral library in order to improve specificity (reduce false positives). The resulting filtered spectral library is considered the RSL that serves for the analysis of materials of interest. All tissue samples undergo the same imaging process and are mapped against the RSL. The resulting image will contain only areas with elements of interest over a black background. This image could then be analyzed with ImageJ using its threshold and particle analysis functions to obtain the area of mapped particles per field of view. Numerical data obtained from ImageJ can be exported to a spreadsheet for further analysis.
It is important to consider that as biological samples are inherently different from one another, and staining methods may affect visualization through EDFM and HSI, the exposure settings should be determined in accordance to what produces the best high contrast image for a specific type of sample. Although reduction of false positives can be achieved through the filtration of spectral libraries, it is crucial to obtain reliable negative controls that have avoided contamination with the element of interest, as the spectra corresponding to this contamination could potentially be filtered from the positive control’s spectral library, increasing false negative rates. Also, the spectrum intensity range that is detectable with the hyperspectral imaging software cannot surpass the limit of the particular software (for this study, that is 16,000 units): areas with a high number of accumulated particles that produce spectral intensities above the intensity limit are left out of the spectral library, due to the risk of increasing the number of false negatives.
While the HSI system confers many advantages over traditional methods, there are some drawbacks and limitations to consider. One is that the large amount of optical data collected may require substantial computing power. Another is that HSI can be time-intensive, particularly at the beginning stages when reference spectral libraries are being created. Also, imaging time may require several minutes per image capture, making it slower than simple darkfield imaging; however, it is still faster than performing sampling preparation and visualization by electron microscopy. Additionally, complex systems may result in multiple characteristic spectra, which require the development of highly specialized controls and make the establishment of standardized, universal reference libraries difficult26. Finally, the technique results in lower resolution than probe- or electron-microscopy techniques, such as atomic force microscopy or transmission electron microscopy, which can resolve individual atoms. This technique’s resolution is limited due to its photonic nature, which prevents it from being a high-precision tool for measuring particle sizes at the nanometer level or for precisely locating materials at a sub-micron level. While the technique may be able to pinpoint particles within certain tissue compartments or cellular organelles greater than 1 µm (such as cell nuclei), smaller organelles or features are challenging to visualize accurately with this method. Also of note, given its spatial resolution, this method cannot differentiate between single nanoparticles and agglomerates11.
Other considerations include: certain materials (such as noble metals) have much higher reflectance and distinct spectral profiles, which can make them easier to analyze and spectrally map with this tool. Others, such as the semi-metallic oxides investigated in this study and carbon-based nanomaterials24, 27, may be more challenging due to their elemental composition, shape, and depending on the matrix. In two murine inhalation studies by Mercer et al., a similar system to the one employed in this study was used in order to locate carbon nanotubes in the lung and in secondary organs based on their remarkably high contrast with surrounding tissues. However, hyperspectral mapping was not demonstrated in either study, likely because the unique shape of carbon fibers was a sufficient trait for identification. Another consideration pertains specifically to tissues: since depositing nanoparticles of interest to specific organs through normal biophysical processes is unpredictable (and often itself the subject of study), determination of an applicable positive control can be difficult and requires consideration of how generation of the control might affect the state of a material of interest. For example, if a spectral library is created from pristine nanoparticles of interest, it may be difficult to map the library to those same nanoparticles in tissues or cells due to changes in the spectra resulting from alteration of the particle (e.g., due to change in pH, dissolution, agglomeration, protein binding) and the overall microenvironment or matrix. Finally, the technique is limited in its semi-quantitative nature: it can only be as quantitative as other two-dimensional microscopy techniques of similar resolution, which means it cannot be easily used to perform tasks such as characterizing total organ burden of a material.
Overall, EDFM and HSI provide several advantages over conventional nanomaterial imaging and characterization techniques, such as TEM, HAADF and DIC. EDFM/HSI allows for faster image acquisition and analysis, which saves time and cost in comparison to more intensive conventional techniques. Additionally, sample preparation for EDFM/HSI is typically both minimal and non-destructive, which saves time and also allows for more flexible analysis of a given sample since it may then be used for other techniques. Furthermore, HSI is versatile, allowing for analysis of nanoscale materials of many compositions and in a variety of matrices. The research team is working to refine the method described here for other materials and sample types, including an in-depth assessment of the specificity of the technology. A critical next step under investigation by the research team is validation of these techniques against traditional gold standards (e.g., Raman, TEM, SEM) for the materials and tissue types of interest.
Disclosures
The authors have nothing to disclose.
Acknowledgments
The authors thank Günter Oberdörster, DVM, PhD and Alison Elder, PhD (University of Rochester) and Mary Frame, PhD (Stony Brook University) for animal research collaborations resulting in tissue samples for analysis. Additionally, the authors thank: Christina Rotondi (Albany Medical College Histology Core); Rani Sellers, DVM, PhD and Barbara Cannella, PhD (Albert Einstein College of Medicine Histology and Comparative Pathology Facility); Leonardo Bezerra and Ahlam Abuawad (Brenner research team members); and Leslie Krauss, Byron Cheatham and Elyse Johnson (CytoViva). This work was supported in part by CDC-NIOSH grant OH-009990-01A1 and the NanoHealth and Safety Center, New York State, awarded to S.B.
References
- Sun W, et al. Endocytosis of a single mesoporous silica nanoparticle into a human lung cancer cell observed by differential interference contrast microscopy. Anal Bioanal Che. 2008;391(6):2119–2125. doi: 10.1007/s00216-008-2162-1. [DOI] [PubMed] [Google Scholar]
- Anselme K, et al. The interaction of cells and bacteria with surfaces structured at the nanometre scale. Acta Biomate. 2010;6(10):3824–3846. doi: 10.1016/j.actbio.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Anshup A, et al. Growth of gold nanoparticles in human cells. Langmui. 2005;21(25):11562–11567. doi: 10.1021/la0519249. [DOI] [PubMed] [Google Scholar]
- Weinkauf H, et al. Enhanced dark field microscopy for rapid artifact free detection of nanoparticle binding to Candida albicans cells and hyphae. Biotechnol. 2009;4(6):871–879. doi: 10.1002/biot.200800358. [DOI] [PubMed] [Google Scholar]
- Sondi I, et al. Silver nanoparticles as antimicrobial agent: a case study on E coli as a model for Gram negative bacteria. J Colloid Interface Sc. 2004;275(1):177–182. doi: 10.1016/j.jcis.2004.02.012. [DOI] [PubMed] [Google Scholar]
- Berry CC. Possible exploitation of magnetic nanoparticle cell interaction for biomedical applications. J Mater Chem. 2005;15(5):543–547. [Google Scholar]
- Chithrani BD, et al. Elucidating the mechanism of cellular uptake and removal of protein coated gold nanoparticles of different sizes and shapes. Nano Lett. 2007;7(6):1542–1550. doi: 10.1021/nl070363y. [DOI] [PubMed] [Google Scholar]
- Chithrani BD, et al. Determining the size and shape dependence of gold nanoparticle uptake into mammalian cells. Nano Lett. 2006;6(4):662–668. doi: 10.1021/nl052396o. [DOI] [PubMed] [Google Scholar]
- Elechiguerra JL, et al. Interaction of silver nanoparticles with HIV 1. J Nanobiotechnol. 2005;3(6):1–10. doi: 10.1186/1477-3155-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson C, et al. Unique cellular interaction of silver nanoparticles size dependent generation of reactive oxygen species. J Phys Chem. 2008;112(43):13608–13619. doi: 10.1021/jp712087m. [DOI] [PubMed] [Google Scholar]
- Roth GA, et al. Hyperspectral microscopy as an analytical tool for nanomaterials. WIREs Nanomed Nanobiotechnol In Press. 2015. [DOI] [PubMed]
- Williams P, et al. Maize kernel hardness classification by near infrared (NIR) hyperspectral imaging and multivariate data analysis. Anal Chim Act. 2009;653(2):121–130. doi: 10.1016/j.aca.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Ziph Schatzberg L. Hyperspectral imaging enables industrial applications. Industrial Photonic. 2014.
- Sun DW. Hyperspectral imaging for food quality analysis and contro. Elsevier; 2010. [Google Scholar]
- ElMasry G, et al. Hyperspectral imaging for nondestructive determination of some quality attributes for strawberry. J Food En. 2007;81(1):98–107. [Google Scholar]
- Mortimer M, et al. Potential of hyperspectral imaging microscopy for semi quantitative analysis of nanoparticle uptake by protozoa. Environ Sci Techno. 2014;48:8760–8767. doi: 10.1021/es500898j. [DOI] [PubMed] [Google Scholar]
- Sarlo K, et al. Tissue distribution of 20 nm 100 nm and 1000 nm fluorescent polystyrene latex nanospheres following acute systemic or acute and repeat airway exposure in the rat. Toxico. 2009;263:117–126. doi: 10.1016/j.tox.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Husain M, et al. Pulmonary instillation of low doses of titanium dioxide nanoparticles in mice leads to particle retention and gene expression changes in the absence of inflammation. Toxicol Appl Pharmacol. 2013;269:250–262. doi: 10.1016/j.taap.2013.03.018. [DOI] [PubMed] [Google Scholar]
- Meyer JN, et al. Intracellular uptake and associated toxicity of silver nanoparticles in Caenorhabditis elegan. Aquatic Toxicol. 2010;100:140–150. doi: 10.1016/j.aquatox.2010.07.016. [DOI] [PubMed] [Google Scholar]
- Badireddy AR, et al. characterization and abundance of engineered nanoparticles in complex waters by hyperspectral imagery with enhanced darkfield microscopy. Environ Sci Technol. 2012;46(18):10081–10088. doi: 10.1021/es204140s. [DOI] [PubMed] [Google Scholar]
- Dettmeyer RF. Staining techniques and microscopy Forensic histopathology fundamentals and perspectives. Berlin Heidelberg: Springer Verlag; 2011. 370 pp. vol 26 issue 453. [Google Scholar]
- Titford M. Progress in the development of microscopical techniques for diagnostic pathology. J Histotechnol. 2009;32(1):9–19. [Google Scholar]
- Kumar GL. Special stains and H&E. Carpinteria, California: Dako North America; 2014. [Google Scholar]
- Manolakis D, et al. Hyperspectral image processing for automatic target detection applications. Lincoln Laboratory Journal. 2003;14(1):79–116. [Google Scholar]
- Mercer RR, et al. Pulmonary fibrotic response to aspiration of multi walled carbon nanotubes. Part Fibre Toxicol. 2011;8(1):21. doi: 10.1186/1743-8977-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, et al. Hyperspectral reflectance and fluorescence imaging system for food quality and safety. T Am Soc Ag En. 2001;44(3):721–730. [Google Scholar]
- Mercer RR, et al. Extrapulmonary transport of MWCNT following inhalation exposure. Part Fibre Toxico. 2013;10(1):38. doi: 10.1186/1743-8977-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]


