Abstract
Objective assessment in human immunodeficiency virus (HIV)-related fatigue has been elusive because the biological mechanisms are not well characterized. We tried to identify low-abundance plasma proteins that correlate with self-reported fatigue intensity in people living with HIV. We used plasma samples from 32 patients with HIV with varying degrees of fatigue who were either treated with nucleoside reverse transcriptase inhibitors or treatment naïve. The plasma samples were enriched for low-abundance proteins and trypsinized. The peptides were analyzed using shotgun proteomics. Five targets correlated with severity of fatigue: apolipoprotein A-1 (ApoA1), apolipoprotein B (ApoB), histine-rich glycoprotein, alpha-1 B glycoprotein, and orosomucoid 2. These targets were selected based on total abundance and spectral count differences, and ApoA1 and ApoB were analyzed via Western blots to verify the mass spectrometry results. ApoA1 levels were higher in untreated patients, while ApoB results suggested a possible positive trend in treated patients. Further analysis is needed to identify additional low-abundance proteins and confirm already-identified proteins as potential fatigue biomarkers.
Keywords: antibodies, biomarkers, fatigue, HIV, mitochondria, proteins
Fatigue is a frequent symptom in patients with human immunodeficiency virus (HIV). Further, 55–65% of people living with HIV (PLWH) commonly report fatigue after initiating antiretroviral therapy (ART; Henderson, Safa, Easterbrook, & Hotopf, 2005; Pence et al., 2009; Voss, 2005). Negative outcomes of HIV-related fatigue (HRF) include reduced quality of life, social isolation, depression and anxiety, sleep disturbances, weakness, muscle aches, lack of concentration, and impaired memory function (Voss, 2005). Yet, despite its high prevalence, HRF is difficult to treat because its exact biological causes are undetermined and likely multifactorial.
Severity and frequency of HRF are measured using self-report scales. Yet, multiple investigators have recognized the subjectivity of using this type of measure for fatigue, with one patient describing mild fatigue as a state that another may describe as severe (Pence et al., 2009; Rodriguez, 2000). Pence and colleagues further demonstrated that self-reported severity of fatigue is very resistant to change over a period of 15 months. Trying to quantify fatigue using self-report techniques that do not account for individual tolerance makes it difficult to accurately measure fatigue and monitor changes in fatigue over time. However, neither of the two biological markers currently used to monitor the progression of HIV disease can be used to monitor HRF due to conflicting results in studies on the relationship between these measures and fatigue (Barroso, Pence, Salahuddin, Harmon, & Leserman, 2008; Yadav et al., 2011). The lack of biological measures that accurately represent the correlation between subjective perceptions of fatigue and the underlying biological function has made it very difficult to evaluate recommended behavioral, lifestyle, and pharmacological interventions in terms of their biological effects.
Previous studies have suggested that HRF may be a result of mitochondrial dysfunction and abnormalities in mitochondrial DNA (mtDNA; Côté et al., 2002). However, it is difficult to determine whether mitochondrial damage is solely the result of the HIV disease or is worsened by the onset of ART. Researchers have hypothesized that a combination of immune-mediated, organ-related, and ART-induced mitochondrial dysfunctions can lead to persistent or periodic tiredness and exhaustion that can last for more than 3 months (Payne et al., 2013). Studies on the effect of nucleoside reverse transcriptaseinhibitors (NRTIs) on fatigue have yielded contradictory results. While mitochondrial toxicity was associated with older NRTIs, the long-term effects of these medications on mitochondria are unclear. In a recent study, both treated and treatment-naive PLWH had significant decreases in adipose-tissue mtDNA content and citrate synthase function compared to the uninfected control group (100% mtDNA; Garrabou et al., 2011). The results did not indicate a significant difference between mtDNA content in adipose tissue of treatment-naive (62% remaining mtDNA) and treated PLWH (69% remaining mtDNA), suggesting that the mtDNA damage was independent of ART use (Garrabou et al., 2011; Voss, Dodd, Portillo, & Holzemer, 2006). This finding contradicts previous work that links mitochondrial dysfunction to ART. These studies suggested that the mtDNA polymerase % may be inhibited by NRTIs, leading to lower levels of mtDNA and compromising the ability of mitochondria to provide adenosine triphosphate (ATP) for cellular functions (Brinkman, ter Hofstede, Burger, Smeitink, & Koopmans, 1998; Lewis et al., 2006).
One current method to test mitochondrial damage is measurement of serum lactate levels. A blood sample is collected, and lactate levels are measured to determine the presence of hyperlactatemia risk factors: normal levels are ≤2.0 mmol/L and hyperlactatemic levels are >2.0 mmol/L (Boubaker et al., 2001; Tan, Walmsley, Shen, & Raboud, 2006). While higher serum lactate dehydrogenase levels have been associated with HIV disease progression, their relation to fatigue is still unknown (Butt, Michaels, & Kissinger, 2002). Serum lactate levels are not muscle- or organ-specific, and mitochondrial dysfunction may be due to other causes of mitochondrial toxicity such alcohol abuse (Tan et al., 2006). These factors make serum lactate levels an inaccurate measure of HRF.
A more precise method for identifying mitochondrial damage is the single twitch muscle fiber test, which examines the mitochondrial respiration in a single muscle fiber (Tonkonogi, Harris, & Sahlin, 1998). Elevated oxygen levels indicate mitochondrial damage and may therefore be associated with fatigue (Schuh, Jackson, Khairallah, Ward, & Spangenburg, 2011). This test, however, is costly and invasive, which severely limits its practicability to only a fraction of the HIV patients followed by a trained neurologist. There is thus a clear need for the development of new methods to measure mitochondrial damage and assess its relationship to fatigue.
Our future goals are to develop a biomarker that will elucidate the role of mitochondrial function in HRF and to develop a simpler and more accessible tool to further investigate the resulting model. In the present study, our goal was to generate future potential protein targets that are related to HRF.
Materials and Method
Plasma Samples
The plasma samples we used for the present study came from a natural history study of fatigue and mitochondrial dysfunction at the National Institutes of Health and were transferred to the University of Washington. The samples were categorized by fatigue scores, HIV status, and HIV treatment status. The self-report instrument that had been used to identify level of fatigue in the participants was the Piper Fatigue scale (PFS), one of the earliest of these instruments. First developed in 1989 by Barbara Piper and colleagues for use in breast cancer patients, the scale was shortened and revised in 1998. The PFS-Revised is a 22-item scale that measures four dimensions of fatigue: behavioral/severity, affective meaning, sensory, and cognitive/mood (Piper et al., 1998). It was developed based on the concept that, because fatigue is a multidimensional phenomenon, it must be measured in a multidimensional way. The total fatigue score is the mean of the average scores of the four subgroups, with a 0–10 (mild–severe fatigue) range (Dagnelie et al., 2006).
Participants from whom the samples had been collected were NRTI-treated (n = 5) and treatment-naive (n = 3) severely fatigued PLWH (fatigue score > 7), NRTI-treated (n = 5) and treatment-naive (n = 4) moderately fatigued PLWH (fatigue score 4–7), NRTI-treated (n = 5) and treatment-naive (n = 5) nonfatigued PLWH (fatigue score <4), and healthy control samples (n = 5; Table 1). Plasma was purified and stored at −80°C until analysis. The University of Washington institutional review board approved the study protocol.
Table 1.
Participants (n) by Treatment Group and Fatigue Score.
| Treatment group | Fatigue score
|
||
|---|---|---|---|
| 0–3 | 4–7 | 8–10 | |
| Antiretroviral treatment | 5 | 5 | 5 |
| No antiretroviral treatment | 3 | 4 | 5 |
| Healthy | 5 | ||
Note. Fatigue was measured with the revised Piper Fatigue scale. Total score ranges from 0 to 10, with higher scores indicating greater fatigue intensity.
Procedures
Removal of High-Abundance Proteins
A Multi Affinity Removal Spin Column (HU-7HC, Agilent Technologies, Santa Clara, CA) was used to remove seven high-abundance proteins from the plasma samples: IgG, IgA, transferrin, haptoglobin, antitrypsin, albumin, and fibrinogen. Plasma was diluted with Buffer A (Agilent Technologies) and filtered via centrifugation (Beckman Coulter Microfuge 22R centrifuge) for 30 s at 5,000 rpm using a 0.22-μm cellulose acetate spin filter (Agilent Technologies). Buffer A was used to adjust the pH of the filtrate to 6.9 and wash the Multi Affinity Removal Spin Column. Diluted sample of 200 μl was added to the column and centrifuged for 2 min at 1,000 rpm. Low-abundance proteins were collected in the runoff. Buffer A was added, and the column was centrifuged for an additional 2 min to collect any remaining low-abundance proteins. Runoffs collected after centrifugation for each participant were combined. Buffer B was slowly pushed through the column using a syringe to elude the high-abundance proteins. The retentate was collected for use in sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The column was re-equilibrated with Buffer A and the procedure was repeated for the remaining diluted sample. The runoff and elution were stored at −80°C.
Runoff, Elution, and Protein Concentration
Samples containing the low-abundance proteins were placed in a 5 K spin concentrator filter and centrifuged for 30 min at 4°C at 7,000 relative centrifugal force. The retentate was collected and stored. Elution samples with high-abundance proteins were pipetted into a Microcon sample reservoir (Millipore, Billerica, MA) and centrifuged in a Jouan GR 2022 centrifuge for 30 min at 14,000 rpm. The retentate was mixed with Buffer B and inverted before centrifuging for 5 min at 5,000 rpm. The filtrate was stored at −80°C. A standard protein curve was generated using a Pierce BCA protein assay kit (Thermo Fischer Scientific, Rockford, IL). Low-abundance protein samples were prepared according to kit specifications. A Milton Roy Spectronic 1001Plus was used to measure the absorbance of the samples at 562 nm.
Gel Electrophoresis
SDS-PAGE gels were performed for each patient. Original plasma samples, elution samples with high-abundance proteins, and filtered samples containing low-abundance proteins were prepared for gel electrophoresis by adding 4× loading buffer (NuPAGE, Life Technologies, Grand Island, NY) to 20 μl of sample. Samples were heated for 5 min at 90°C before loading onto a 4–12% Bis–Tris 1.0 mm, 10-well gel (NuPAGE). Gels were run using 1 × 3-(N-morpholino)pro-panesulfonic acid (MOPS) SDS running buffer (BioRad, Hercules, CA) for 1 hr at 180 V with a BioRad Power Pak 200 and stained with a BioRad silver stain kit.
Mass Spectrometry Preparation
Samples were pooled by treatment and fatigue group before adding cold 20% (v/v) trichloroacetic acid (TCA) to the low-abundance protein samples. Samples were placed on ice for 30 min and then centrifuged at 14,000 rpm at 4°C for 15 min using an Eppendorf centrifuge 5417R (Pasadena, TX). The pellet was washed with cold 20% (v/v) TCA and the supernatant was discarded. Protein pellets with a mass of 500 μg to 1 mg were broken down by trypsin digestion and resuspended in 100 μl of a 6 M urea in 50 mM ammonium bicarbonate solution. Of the 1.5 M Tris pH 8.8, 6.7 μl was added to adjust the pH before adding 2.5 μl of 200 mM tris(2-carboxyethyl)phosphine and incubating for 1 hr at 37°C. Following incubation, 20 μl of 200 mM iodoacetamide alkylation solution was added before vortexing for 1 hr at room temperature (21°C) in the dark. Of the 200-mM dithiothreitol (DTT), 20 μl was added before vortexing for another hour at room temperature (21°C). The urea was diluted by adding 800μl of 25 mM NH4HCO3 to each tube followed by 200 μl methanol (MeOH). Trypsin (Promega sequencing grade, San Luis Obispo, CA) was added to the sample in a protein-to-trypsin ratio of 50:1. The sample was incubated overnight at 37°C and then stored at −80°C. Samples were diluted using 5% acetonitrile (ACN) and 0.1% trifluoric acid (TFA). A MacroSpin C18 column was prepped using 1 ml of 100% MeOH and 2 ml of 0.1% TFA at a flow rate of 2–10 ml/min (Waters, Milford, MA). Sample was added to the column in 100-μl aliquots and allowed to pass through without assistance. The column was then washed with 3 ml of 0.1% TFA. The bound proteins were eluted using 1 ml of 80% ACN, 0.1% of TFA, and the flow through was collected. The eluted protein samples were reduced to no less than 10 μl using a speed vac for 3 hr. Proteins were resuspended in 110 μl of 5% ACN, 0.1% formic acid, and centrifuged for 2 min at 14,000 rpm at 4°C. From the top layer of the sample, 100 μl was collected.
Protein Identification and Database Search
Proteins were identified from peptide tandem mass spectra acquired during LC-mass spectrometry (MS)/MS experiments using collision-induced dissociation (CID). A protein sample was proteolyzed, and the resulting peptides were subjected to CID (during LC-MS/MS analysis) to generate peptide tandem mass spectra. Both the linear ion trap–Fourier Transform (LTQ-FT) and linear ion trap–Orbitrap (LTQ-OT) mass spectrometers had an LTQ where peptides were sequenced with a high-resolution, high-mass-accuracy detector (FT or the OT, where the peptides’ mass values are recorded). With either instrument, data were acquired in quadruplicate. Sets of precursor ion data (i.e., MS1) were used to derive peptide sequences from MS2 spectra. Software tools were used to calculate protein identifications from peptide information and changes in relative protein expression from these peptide data. Acquired peptide tandem mass spectra were matched to a sequence in databases (SEQUEST, Peptide and ProteinProphet within the context of the Trans-Proteomics Pipeline [TPP]) using two pieces of information. First, the parent mass of the peptide provided theoretical amino acid compositions, which, when compared to a sequence database, provided a list of potential peptide sequences within the measured peptide’s parent mass tolerance (~2–5 ppm). Second, from this list of theoretical peptide tandem mass spectra, a best fit to the observed peptide’s tandem mass spectrum was generated by a sliding cross-correlation score. The SEQUEST software was used to generate peptide sequence matches. Algorithms of peptide and ProteinProphet statistical routines were used to assign probability scores to the peptide sequence best fit and the likelihood that the parent protein was present. The web-based TPP was used to consolidate all of these software tools.
Relative Expression Analysis
Differences in relative expression of proteins were calculated using in-house software tools: either peptide spectral counting or peptide ion current area (PICA) algorithms. Peptide spectral counting uses tandem mass spectrometry data (MS2) to estimate, by statistical means, changes in relative abundance. The MS2-based methods estimate differences in relative protein expression by accounting for either the extent of protein sequence coverage or the number of tandem mass spectra generated. The more accurate PICA methods use precursor ion (MS1) data to calculate “area under the curve” for individual peptide response before generating protein expression levels from all peptides from a given protein. The PICA methods require reproducible C18 chromatography and are used for studies with large sample numbers.
Pathway Analysis of Marker Proteins
Identified (mitochondrial and nuclear-encoded) proteins were analyzed with Gene Ontology (http://www.geneontology.org/) for their possible cellular compartmentalization and molecular functions. For each identified protein, ProteinProphet probability score, protein annotation, tissue-type expression, and false discovery rate were calculated. Identified proteins were also analyzed for their pathway involvement by Teranode pathway analysis software through the Kyoto Encyclopedia of Genes and Genomes database. These types of data display and analyses allow visualization of the complex mitochondria and mitochondria-relevant nuclear protein systems.
Protein Verification Using Western Blots
Western blots were used to confirm the proteins identified by mass spectrometry. Initially, five antibodies were tested: apolipoprotein A-1 (ApoA1), apolipoprotein B (ApoB), histine-rich glycoprotein (HRG), orosomucoid 2 (ORM2), and alpha-1 B glycoprotein (A1BG). Antibodies were selected based on abundance and differences between treated and nontreated patients with mild, moderate, and severe fatigue. Participants’ unfiltered plasma samples were combined according to group. All ART 0–3 samples were combined, all ART 4–7 samples were combined, all ART 8–10 samples were combined, and so on, for a total of seven sample groups. Pooled samples were purified using the Affinity Column Removal procedures described previously.
Determination of Sample Concentration
A Pierce BCA micro-plate protein assay was used according to the kit recommendations to determine the protein concentration of each pooled sample. The absorbance of the samples was calculated at 562 nm. If the protein concentration was less than 1 μg/μl, TCA precipitation was used to concentrate the samples. Cold 20% (v/v) TCA was added, and samples were placed on ice for 30 min. Samples were centrifuged at 14,000 rpm for 15 min at 4°C in an Eppendorf 517R centrifuge. The supernatant was removed and the pellet was washed twice with cold acetone. For each wash, the sample was centrifuged for 10 min at 14,000 rpm at 4°C. After the final wash, the acetone was removed and the pellet was allowed to air dry. Pellets were resuspended in 0.5× phosphate-buffered saline (PBS). A Pierce BCA microplate protein assay was used to determine the new protein concentration. TCA precipitation was repeated until each pooled sample had a minimum concentration of 400 μg/μl. Sample aliquots were placed in a speed vac. Dry samples were resuspended in deionized water to a concentration of 2.0 μg/μl. For samples with a protein concentration between 1.0 and 2.0 μg/μl, a speed vac was used on the manual run setting. Dry samples were resuspended in deionized water to a concentration of 2.0 μg/μl and stored at −80°C.
Gel Electrophoresis and Staining
Two SDS-PAGE gels were generated for each antibody tested. Concentrated protein samples containing the low-abundance proteins were mixed with LDS sample buffer (NuPAGE). The samples were heated to 90°C for 5 min on an Eppendorf thermomixer. An XCell SureLock™ Mini Cell (Invitrogen) gel-running apparatus was setup with two NuPAGE 4–15% Bis–Tris gels (1.0 mm × 15 wells) and filled with 1 × MOPS SDS running buffer. Gels were run for 1 hr at 180 V. One gel was stained using Coomassie Blue staining solution (BioRad) for 1 hr. The remaining gel was used for membrane transfer to verify protein presence via Western blots.
Antibody Parameters in Western Blots
The transfer membrane (0.45-μm pore size, Invitrogen) was prepared by soaking in methanol for 40 s, then in deionized water for 10 min, and finally in transfer buffer for 10 min (5% 20× BioRad transfer buffer, 10% methanol). XCell II Blot Module (Invitrogen) was assembled and run for 1 hr at 30 V using an EC 1000-90 Thermo transfer device. The membrane was incubated in 5% milk, 0.05% Tween in PBS (PBS Tween) solution for 30 min. After blocking, the membrane was incubated with the primary antibody overnight: ApoA1 (mouse monoclonal from Cell Signaling Technology, Danvers, MA, at 10 μl/10 ml), ApoB (mouse monoclonal from R&D Systems, Minneapolis, MN, at 40 μl/10 ml), HRG (rabbit polyclonal from Epitomics, Burlingame, CA, at 125 μl/10 ml), and A1BG (rabbit polyclonal from Abcam, Cambridge, MA, at 125 μl/10 ml). Following incubation, the membrane was washed 4 times for 10 min in PBS Tween solution. The membrane was incubated with an anti-mouse Ig horseradish perioxidase antibody from sheep (GE Healthcare, Pittsburg, PA) for 30 min with an antibody-to-PBS Tween solution ratio of 1:5000. The membrane was washed 5 times for 10 min with PBS Tween before rinsing in 1 × PBS. An ECL plus kit (Amersham, Piscataway, NJ) was used to prepare imaging solution. Solution A was mixed with Solution B in a 40:1 ratio. The prepared solution was applied to the membrane immediately before photographing. Chemiluminescence was measured on an Alpha Inotech and used to calculate the relative abundance (in pixels) of proteins for each sample.
Results
Comparison of Elution, Plasma, and Filtrate Using Gel Electrophoresis
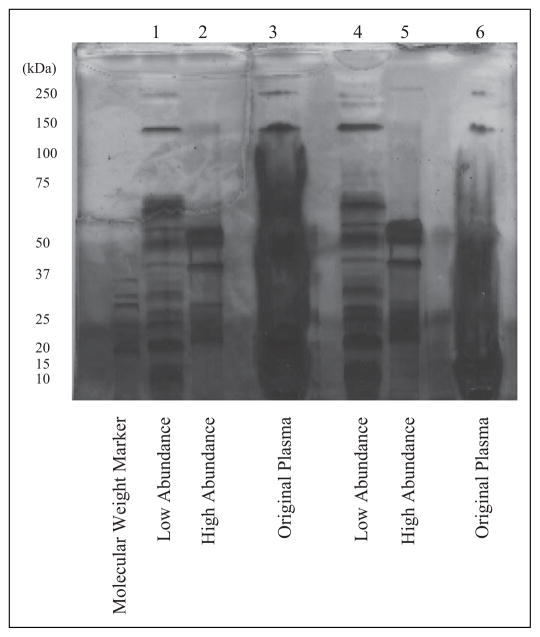
SDS-PAGE gels stained with a silver stain were used to compare the proteins present in the plasma, elution, and working samples (Figure 1). The low-abundance proteins (Lanes 1 and 4) had ~15 distinct bands of varying intensity, while the high-abundance proteins (Lanes 2 and 5) had large broad bands at 50 kDa, single bands at 37 kDa, and stained regions between 20 and 25 kDa. The consistency between the original plasma samples (Lanes 3 and 6) confirmed the presence of the IgG, IgA, antitrypsin, albumin, and haptoglobin in the high-abundance protein samples. A comparison of the whole plasma, high-abundance protein, and low-abundance protein lanes suggests that the column selectively bound high-abundance proteins of interest.
Figure 1.
SDS-Page results after purification with affinity removal column. SDS-PAGE gel stained with silver stain for two patients from the antiretroviral treatment group with fatigue scores of 4–7 (moderately fatigued). Protein of 23 μg was loaded onto the low-abundance protein lane for Participant 1 (Lanes 1–3) and 20 μg for Participant 2 (Lanes 4–6). The amount of protein loaded onto the high-abundance protein and original plasma lanes was not determined. Comparison of the bands in the high- and low-abundance protein lanes confirms that the high-abundance proteins from the original plasma samples were selectively bound by the multiaffinity removal column. SDS-PAGE = sodium dodecyl sulfate polyacrylamide gel electrophoresis.
Protein Identification Using Mass Spectrometry
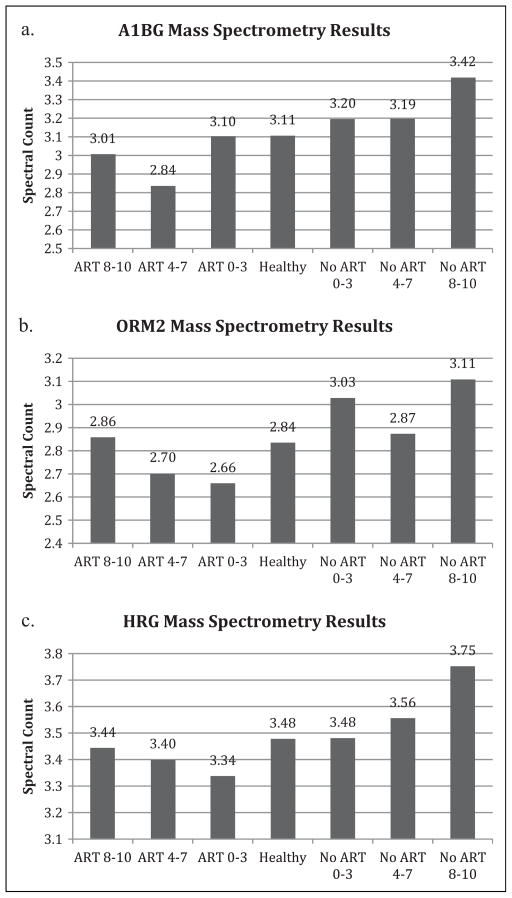
Mass spectrometry of the peptides detected the presence of 113 proteins. ApoB, ApoA1, HRG, ORM2, and A1BG (Figures 2a–c, 3d, 4d) with spectral counts (SCs) of 484, 391, 51, 32, and 41, respectively, were selected based on abundance as well as visible differences between fatigue severity groups within treatment groups and between treated and treatment-naive groups.
Figure 2.
Mass spectrometry results for the (a) A1BG, (b) ORM2, and (c) HRG proteins. Spectral counts from mass spectrometry showing the protein abundances for the seven different test groups shown in Table 1. A1BG was selected because of its high abundance, but the mass spectrometry results suggest no trends. ORM2 results suggest that protein concentration increased with increasing fatigue for the ART groups but does not indicate any trends for the no-ART groups. Results for HRG suggest that protein concentration increased with increasing fatigue for both ART and no-ART groups. Groups were categorized by both treatment and score on the Piper Fatigue scale–Revised, with scores of 8–10 indicating severely fatigued, 4–7 indicating moderately fatigued, and 0–3 indicating nonfatigued. A1BG = alpha-1 B glycoprotein; ART = antiretroviral therapy; HRG = histine-rich glycoprotein; ORM2 = orosomucoid 2.
Figure 3.
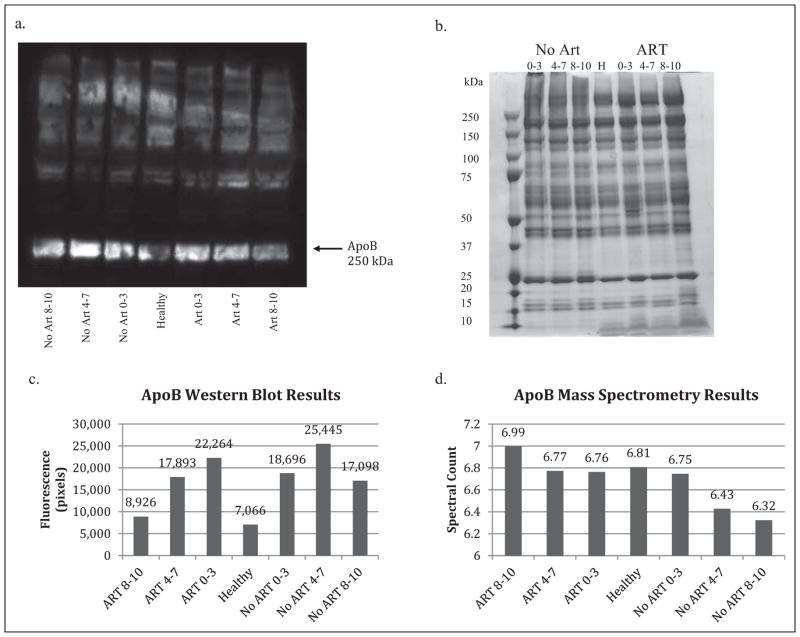
Western blot and mass spectrometry results for ApoB. (a) The immunofluorescent image from the Western blot procedure using the ApoB antibody shows a fluorescent band at 250 kDa, which is consistent with values in the literature for monomeric ApoB. Fluorescent intensity was used to determine relative protein concentrations in place of calculating protein amounts with a standard. ApoB was abundant in all seven test groups. (b) Gel electrophoresis using an SDS-PAGE gel stained with Coomassie Blue and samples identical to those used for the Western blot was used to confirm the presence of low-abundance proteins in the samples. The distinct bands indicate the presence of multiple proteins in these samples. (c) Fluorescence was calculated in pixels based on the Western blot image for ApoB using Alpha Inotech software in order to illustrate the relative protein concentrations in the seven test groups shown in Table 1. Western blot results were used to confirm the presence of proteins chosen from the mass spectrometry results. (d) Spectral counts from mass spectrometry showed differences in ApoB abundance among the test groups. The high spectral counts confirm the protein’s abundance. Mass spectrometry results suggest that protein concentration decreased with increasing fatigue for the no-ART groups and increased with increasing fatigue for the ART groups. Groups were categorized by both treatment and score on the Piper Fatigue scale–Revised, with scores of 8–10 indicating severely fatigued, 4–7 indicating moderately fatigued, and 0–3 indicating nonfatigued. ApoB = apolipoprotein B; ART = antiretroviral therapy; SDS-PAGE = sodium dodecyl sulfate polyacrylamide gel electrophoresis.
Figure 4.
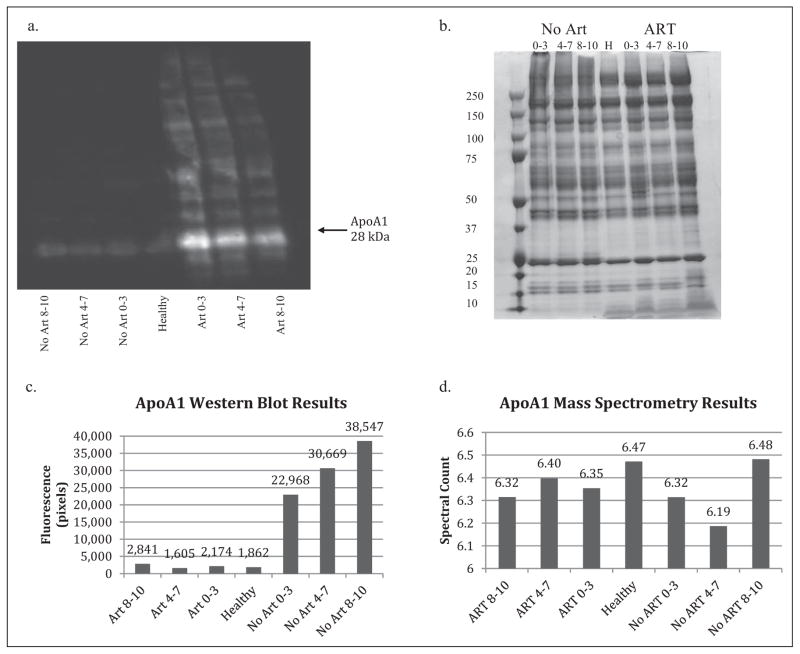
Western blot and mass spectrometry results for ApoA1. (a) Immunofluorescent image from the Western blot procedure for the ApoA1 antibody shows a lower band around 28 kDA, which is consistent with values in the literature for ApoA1. The fluorescence is significantly less in the no-ART and healthy groups but is still detectable as a faint band. (b) Gel electrophoresis using SDS-PAGE gel stained with Coomassie Blue and samples identical to those used for the Western blot was used to confirm the presence of low-abundance proteins in the samples, shown by the numerous distinct bands in the resulting image. (c) Fluorescence was calculated (in pixels) from the Western blot image for ApoA1 with Alpha Inotech software and in order to illustrate the relative protein concentrations in the seven test groups. Results for the no-ART groups suggest that protein concentration increases with increasing fatigue. No trend is suggested for the ART groups. (d) The mass spectrometry results do not indicate any trends among the seven test groups. The high spectral count confirms the protein’s abundance, suggesting its potential for use as a biomarker. Groups were categorized by both treatment and score on the Piper Fatigue scale–Revised, with scores of 8–10 indicating severely fatigued, 4–7 indicating moderately fatigued, and 0–3 indicating nonfatigued. ApoA1 = apolipoprotein A-1; ART = anti-retroviral therapy; SDS-PAGE = sodium dodecyl sulfate polyacrylamide gel electrophoresis.
ApoB and A1BG were selected because of SC differences between the moderately and severely fatigued treated and naive groups. Mass spectrometry for A1BG (Figure 2a) shows slightly higher SCs for the no-ART than the ART groups, while ApoB results indicate higher SCs for the ART groups. ApoA1 mass spectrometry results are fairly constant, with levels ranging from 6.19 to 6.48 SC. ORM2 mass spectrometry results suggest a positive correlation between fatigue severity and SC for the ART groups (Figure 2b). There does not appear to be a relationship between protein abundance and fatigue severity within the no-ART groups (Figure 2c). Mass spectrometry results for HRG suggest a positive trend between fatigue severity and SC for both the ART and no-ART groups. Results ranged from 3.34 SC for the nonfatigued ART group to 3.44 SC for the severely fatigued ART group and from 3.28 SC for the nonfatigued no-ART group to 3.75 SC for severely fatigued no-ART group. A1BG levels remained fairly constant among the different groups, with levels ranging from 2.84 to 3.42 SC (Figure 2a).
Trends in Protein Fluorescence From Western Blots
The stained gels were used to verify the presence of low-abundance proteins in the samples (Figures 3b and 4b). The ladder-like progression of the bands in the gels indicates the presence of multiple proteins in the samples. Additionally, the dark distinct bands suggest that the proteins were present in amounts adequate for detection by both gel electrophoresis and immunofluorescence. Western blots were performed for four of the five proteins of interest: ApoA1, ApoB, HRG, and A1BG. Antibodies were not available for ORM2 at the time of this report, and thus we could include no data. Only ApoA1 and ApoB yielded immunofluorescent results (Figures 3a and 4a). We used these results to confirm the presence of the proteins identified via mass spectrometry.
The ApoA1 fluorescent image shows a band around 28 kDa, which is consistent with values reported in the literature. There are bright distinguishable bands for the ART groups, but weakly fluorescent bands for the no-ART and healthy groups (Figure 4a). ApoB shows a strong fluorescent band around 250 kDa, which is consistent with values reported in the literature, for treated, treatment-naive, and control groups (Figure 3a).
Analysis of Western blot results was limited to differences in relative fluorescence since no standard curve was generated to relate pixels to protein amounts. We determined possible trends by comparing fluorescence, as we had loaded equal amounts of proteins onto each lane. Results for ApoB suggest a negative trend between fatigue severity and protein fluorescence for the treated groups, with the nonfatigued group having the highest level and the severely fatigued group the lowest (Figure 3c). No trend is suggested for the treatment-naive groups. Comparison of treated and treatment-naive groups shows increased protein fluorescence in the no-ART moderately and severely fatigued groups compared to the treated groups.
Western blot results for ApoA1 indicate a positive trend between fatigue severity and protein fluorescence for treatment-naive groups (Figure 4c), with the nonfatigued no-ART group having the lowest fluorescence and the severely fatigued the highest. No trend is suggested for the treated groups. Comparison between treatment groups indicates higher fluorescence levels for treatment-naive groups (>22,000 pixels) than for treated groups (<3,000 pixels).
We have not yet collected data for HRG and A1BG.
Discussion
This study was a primary step in a long-term research project aimed at identification of fatigue biomarkers. The identification of 113 low-abundance proteins provides an overview of protein expression in treated and treatment-naive PLWH. Because no previous work has been done concerning the identification of a fatigue biomarker for PLWH, a general sense of the relative abundance of the different proteins is helpful in identifying biomarker candidates. However, our assessment of protein abundance from Western blots was limited to visual differences in fluorescence rather than quantification of proteins present in the samples. To quantify the proteins tested, we would have had to generate a standard curve for each antibody. By loading known amounts of antibody onto a gel and using Western blot analysis to measure the fluorescence of each known sample, we could determine the relationship between protein concentration and relative fluorescence. From these data, it would be possible to use the relative fluorescence of each test group to calculate the amount of protein in the samples. This calculation would provide a more accurate representation of protein abundance than the relative fluorescence and would therefore be useful in determining differences in protein expression among the test groups.
Protein abundance is important to consider, not only when looking at differences between test groups but also when selecting proteins for the development of dipstick assays, which require detectable amounts of proteins to elicit a color change. Based on the data generated from our Western blots and mass spectrometry analyses and taking into consideration protein function, we identified three proteins, ApoB, ApoA1, and mitofilin, as targets for further study. ApoB has a high abundance, making it easy to detect, and our results also suggest downregulation of the protein as fatigue severity increases for treated groups. ApoA1 also has a high abundance, and Western blot results show large differences in relative fluorescence between treated and treatment-naive groups as well as upregulation of the protein with increasing fatigue severity for treatment-naive groups. Mitofilin, while not detected in the mass spectrometry results, is important in the fatty acid cycle and may play a role in generation of ATP in the mitochondria. Isolation and detection of mitofilin from plasma samples may provide insight into the relationship between mitochondrial dysfunction and fatigue. At this point, it is unclear how these markers may be impacted by factors such as antiretroviral treatments, duration of existing HIV infection, gender, and other demographic characteristics.
Reliable biomarker identification would allow health care professionals to quantify and track fatigue with the least amount of effort in diverse populations of PLWH. Biomarker measurement could aid in testing the efficacy of interventions and consequently improve quality of life. For example, the results of the present study suggest the potential for a new tool that could be similar to existing dipstick assays developed by the company MitoSciences (http://www.mitosciences.com/dip-stick_assays.html). These assays use antibodies on a nitrocellulose membrane to detect target proteins (Völkel et al., 2003). They take only 30–120 min to perform, which would greatly increase the ability of health care providers to monitor mitochondrial dysfunction and fatigue biomarker changes. The development of a similar test for fatigue in PLWH would, thus, have important implications for monitoring disease progression, preventing disabling side effects, and providing important information for the development of intervention methods.
Affinity Column Removal Assessment
Qualitative assessment of the silver-stained gel images suggests that the Multi Affinity Removal Spin Column eliminated the seven high-abundance proteins, IgG, IgA, transferrin, haptoglobin, antitrypsin, albumin, and fibrinogen, from the original plasma sample without removing low-abundance proteins. However, there are inherent problems with using a multiaffinity column. Low-abundance proteins with binding affinities similar to those of the high-abundance proteins may also bind to the column during this purification step. They may be undetectable in the high-abundance protein lane of the gel either because they are present in undetectable amounts or because they have molecular weights similar to those of the high-abundance proteins. Further analysis would need to be done, including identification of the proteins present in the elution, to eliminate this possibility.
Yadav and colleagues (2011) analyzed the specificity of affinity removal columns by identifying the proteins present in an eluted fraction of plasma depleted by affinity columns. The results of the study indicate that nontargeted proteins are removed during affinity removal and that the number of proteins removed increases with increasing numbers of target proteins. Several of the nontargeted proteins present in the elution were previously identified as disease biomarkers, including retinol-binding protein-4, a biomarker for renal dysfunction. However, while Yadav and colleagues found as many as 61 nontargeted proteins in eluted samples, comparable studies have detected the presence of only 3 (Yadav et al., 2011).
While the columns successfully remove high-abundance proteins, they are not able to selectively target these proteins without removing low-abundance proteins. This limitation is, in part, due to the interactions that occur between different plasma proteins. The nontargeted proteins may bind with the high-abundance proteins bound to the column, resulting in their removal from the filtrate along with those high-abundance proteins. The number of possible protein–protein interactions makes this phenomenon difficult to prevent, and it is one of the limitations of using a multiaffinity column for the isolation of low-abundance proteins from plasma samples.
Additionally, the nontargeted proteins may be present in both the elution and the filtrate. The amount of protein in the two samples would depend on the binding affinity between the high- and low-abundance proteins. Retention of low-abundance proteins in the elution presents a problem in the quantification of low-abundance proteins and, consequently, for the identification of biomarkers. In order to account for the possible presence of low-abundance proteins in the eluted sample, investigators would need to analyze both the filtrate and the elution. However, this process would be less efficient, more costly, and dependent on previous research that has identified low-abundance proteins of interest that can be identified and isolated from the elution.
Limitations
Mass spectrometry limitations present a problem in data analysis. Because some proteins are more easily detected via mass spectrometry than others, SCs may not accurately reflect differences in protein abundance. Additionally, the significance of differences between SCs cannot be determined without additional methods such as Western blots.
In the present study, we looked at differences between pooled patient samples but did not measure protein amounts in individual patients. While we saw some indication of upregulation and downregulation of proteins in different fatigue severity groups, we cannot determine whether these trends are consistent with individual trends. We also had a small sample size, which makes it difficult to determine whether or not the trends observed apply to larger populations.
Implications for Research and Clinical Practice
In the present study, we provide a basic overview of the low-abundance proteins present in plasma samples from PLWH, identifying which proteins should be considered in follow-up experiments. Our results suggest that ApoA1 and ApoB may serve as potential fatigue biomarkers for PLWH. Future studies should build on these results by considering individual protein levels within each of the seven groups to confirm the general patient trends and should include the generation of a standard curve to calculate actual protein concentrations as well as larger sample sizes. Following the measurement of protein levels in individuals, studies will be needed to explore the cellular effects of the proteins to increase our understanding of individual protein’s roles in fatigue. Such studies would involve reversibly inhibiting protein expression via RNA interference and observing the cellular response. After these analyses are complete, investigators should be able to identify an appropriate biomarker and develop a rapid protein detection system such as a dipstick assay for use in health care settings to monitor fatigue in PLWH.
Acknowledgments
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the NIH 5K22NR008672-02 K22 Career Transition Award, ITHS Technology Access Grant UL1RR025014, and the Robert Wood Johnson Nurse Faculty Scholar Program.
Footnotes
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- Barroso J, Pence BW, Salahuddin N, Harmon JL, Leserman J. Physiological correlates of HIV-related fatigue. Clinical Nursing Research. 2008;17:5–19. doi: 10.1177/1054773807311382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boubaker K, Flepp M, Sudre P, Furrer H, Haensel A, Hirschel B, Telenti A. Hyperlactatemia and antiretroviral therapy: The Swiss HIV cohort study. HIV/AIDS. 2001;33:1931–1937. doi: 10.1086/324353. [DOI] [PubMed] [Google Scholar]
- Brinkman K, ter Hofstede HJ, Burger DM, Smeitink JA, Koopmans PP. Adverse effects of reverse transcriptase inhibitors: Mitochondrial toxicity as common pathway. AIDS. 1998;12:1735–1744. doi: 10.1097/00002030-199814000-00004. [DOI] [PubMed] [Google Scholar]
- Butt AA, Michaels S, Kissinger P. The association of serum lactate dehydrogenase levels with selected opportunistic infections and HIV progression. International Journal of Infectious Diseases. 2002;6:178–181. doi: 10.1016/s1201-9712(02)90107-4. [DOI] [PubMed] [Google Scholar]
- Côté HC, Brumme ZL, Craib KJ, Alexander CS, Wynhoven B, Ting L, Montaner JS. Changes in mitochondrial DNA as a marker of nucleoside toxicity in HIV-infected patients. New England Journal of Medicine. 2002;346:811–820. doi: 10.1056/NEJMoa012035. [DOI] [PubMed] [Google Scholar]
- Dagnelie PC, Pijls-Johannesma MC, Pijpe A, Boumans BJ, Skrabanja AT, Lambin P, Kempen GI. Psychometric properties of the revised Piper Fatigue Scale in Dutch cancer patients were satisfactory. Journal of Clinical Epidemiology. 2006;59:642–649. doi: 10.1016/j.jclinepi.2005.09.015. [DOI] [PubMed] [Google Scholar]
- Garrabou G, López S, Morén C, Martínez E, Fontdevila J, Cardellach F, Miró Ó. Mitochondrial damage in adipose tissue of untreated HIV-infected patients. AIDS. 2011;25:165–170. doi: 10.1097/QAD.0b013e3283423219. [DOI] [PubMed] [Google Scholar]
- Henderson M, Safa F, Easterbrook P, Hotopf M. Fatigue among HIV-infected patients in the era of highly active antiretroviral therapy. HIV Medicine. 2005;6:347–352. doi: 10.1111/j.1468-1293.2005.00319.x. [DOI] [PubMed] [Google Scholar]
- Lewis W, Kohler JJ, Hosseini SH, Haase CP, Copeland WC, Bienstock RJ, Santoianni R. Antiretroviral nucleosides, deoxynucleotide carrier and mitochondrial DNA: Evidence supporting the DNA pol gamma hypothesis. AIDS. 2006;20:675–684. doi: 10.1097/01.aids.0000216367.23325.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne B, Hateley C, Ong E, Premchand N, Schmid M, Schwab U, Price D. HIV-associated fatigue in the era of highly active antiretroviral therapy: Novel biological mechanisms? HIV Medicine. 2013;14:247–251. doi: 10.1111/j.1468-1293.2012.01050.x. [DOI] [PubMed] [Google Scholar]
- Pence BW, Barroso J, Harmon JL, Leserman J, Salahuddin N, Hammill BG. Chronicity and remission of fatigue in patients with established HIV infection. Aids Patient Care and STDs. 2009;23:239–244. doi: 10.1089/apc.2008.0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper BF, Dibble SL, Dodd MJ, Weiss MC, Slaughter RE, Paul SM. The revised piper fatigue scale: Psychometric evaluation in women with breast cancer. Oncology Nursing Forum. 1998;25:677–684. [PubMed] [Google Scholar]
- Rodriguez T. The challenge of evaluating fatigue. Continuing Education. 2000;12:329–338. doi: 10.1111/j.1745-7599.2000.tb00315.x. [DOI] [PubMed] [Google Scholar]
- Schuh RA, Jackson KC, Khairallah RJ, Ward CW, Spangenburg EE. Measuring mitochondrial respiration in intact single muscle fibers. American Journal of Physiology. 2011;302:712–719. doi: 10.1152/ajpregu.00229.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D, Walmsley S, Shen S, Raboud J. Mild to moderate symptoms do not correlate with lactate levels in HIV-positive patients on nucleoside reverse transcriptase inhibitors. HIV Clinical Trials. 2006;7:107–115. doi: 10.1310/95VE-A6KJ-WRQ0-1F8Y. [DOI] [PubMed] [Google Scholar]
- Tonkonogi M, Harris B, Sahlin K. Mitochondrial oxidative function in human saponin-skinned muscle fibre: Effects of prolonged exercise. Journal of Physiology. 1998;510:279–286. doi: 10.1111/j.1469-7793.1998.279bz.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Völkel D, Zimmermann K, Breitwieser A, Pable S, Glatzel M, Scheiflinger F, Dorner F. Immunochemical detection of prion protein on dipsticks prepared with crystalline bacterial cell-surface layers. Transfusion. 2003;43:1677–1682. doi: 10.1111/j.0041-1132.2003.00567.x. [DOI] [PubMed] [Google Scholar]
- Voss J, Dodd M, Portillo C, Holzemer W. Theories of fatigue: Application in HIV/AIDS. Journal of the Association of Nurses in AIDS Care. 2006;17:37–50. doi: 10.1016/j.jana.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Voss JG. Predictors and correlates of fatigue in HIV/AIDS. Journal of Pain and Symptom Management. 2005;29:173–184. doi: 10.1016/j.jpainsymman.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Yadav A, Bhardwaj G, Basak T, Kumar D, Ahmad S, Priyadarshini R, Sengupta S. A systematic analysis of eluted fraction of plasma post immunoaffinity depletion: Implications in biomarker discovery. PLoS One. 2011;6:e24442. doi: 10.1371/journal.pone.0024442. [DOI] [PMC free article] [PubMed] [Google Scholar]