Abstract
Summary: Identification of driver mutations in human diseases is often limited by cohort size and availability of appropriate statistical models. We propose a method for the systematic discovery of genetic alterations that are causal determinants of disease, by prioritizing genes upstream of functional disease drivers, within regulatory networks inferred de novo from experimental data. Here we present the implementation of Driver-gene Inference by Genetical-Genomic Information Theory as an R-system package.
Availability and implementation: The diggit package is freely available under the GPL-2 license from Bioconductor (http://www.bioconductor.org).
Contact: ma2581@cumc.columbia.edu or ac2248@cumc.columbia.edu
1 Introduction
Identification of somatic mutations and germline variants associated to cancer and other complex diseases (driver mutations) is mostly performed on a statistical basis (Lawrence et al., 2013). Achieving appropriate statistical power, however, requires large effect sizes or large cohorts to control for errors arising from the large number of tested hypothesis (Califano et al., 2012). In addition, since statistical approaches do not provide mechanistic insight, many disease risk determinants, such as apolipoprotein E, were discovered long before they were mechanistically elucidated (Liu et al., 2013).
We have recently developed an approach that boosts the statistical power of GWAS by focusing the analysis to genes mechanistically linked to a specific phenotype or cell state, termed master regulators (MR, Aytes et al., 2014, Carro et al., 2010, Lefebvre et al., 2010). Driver-gene Inference by Genetical-Genomic Information Theory (DIGGIT) searches for genetic alterations associated with dysregulation of MR protein activity, reducing the number of hypothesis to test, while providing regulatory clues to help elucidate associated biological mechanisms (Chen et al., 2014). We have recently used DIGGIT to identify causal genetic determinants of the mesenchymal subtype of human glioblastoma (GBM, Chen et al., 2014). Here, we present the R-system implementation of DIGGIT, which is available as a software package from Bioconductor.
2 Approach
DIGGIT evaluates candidate alterations within a set of functional disease drivers and their upstream regulators (Chen et al., 2014). This is accomplished by a five-step process, requiring gene expression, matched genetic-variant profiles, specifically copy number variation data (CNV), and context-specific transcriptional (Basso et al., 2005) and post-translational (Wang et al., 2009) regulatory models.
The first step reduces the number of candidate genetic alterations, by selecting those whose ploidy is informative of gene expression as candidate functional CNVs (F-CNVs). This is assessed based on mutual information (MI) between copy number and expression. During the second step, the MRs for a specific phenotypic transition are inferred. The third step reduces the list of candidate genetic alterations by considering only the loci coding for MRs and their upstream post-translational modulators, as inferred by the MINDy algorithm. During the forth step, the statistical association between the functional genetic alterations, steps 2 and 3, and the activity of the MRs is inferred by MI analysis (activity Quantitative Trait Loci, aQTL). Finally, a conditional association analysis is performed to determine which, among multiple genes affected by the same amplified or deleted regions, are the most probable drivers of MR dysregulation.
3 Implementation
DIGGIT is implemented as an R-system package and it is available from Bioconductor. Input data and results are encapsulated in an S4 object of class diggit, requiring an expression dataset, CNV data, and an appropriate tissue lineage-matched regulatory network (interactome). Two alternative methods were implemented to compute the F-CNVs: MI and correlation analysis (Fig. 1A). MI is estimated using 1- and 2-dimensional Gaussian kernels with optimal bandwidth selection though a plug-in (hpi) approach, as implemented in the ks package (available from CRAN: http://cran.r-project.org). Finally, the statistical significance for the association is estimated by permutation analysis.
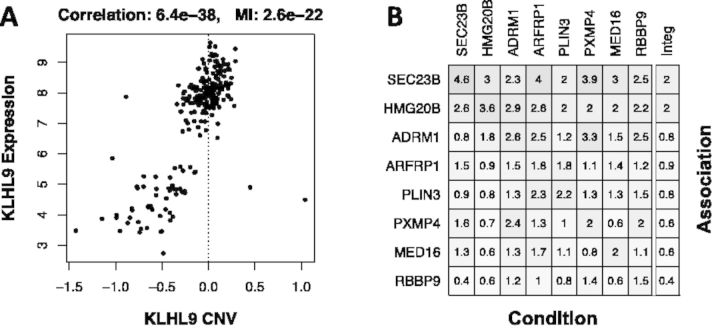
Fig. 1.
(A) Scatterplot for KLHL9 CNV vs. mRNA expression. Spearman correlation and MI p-values are indicated on top of the figure. (B) Heatmap showing the association (-log10(p)) between genes affected by genetic alterations (rows) and STAT3 inferred protein activity, while conditioning on each of the genetically altered genes (columns). The rightmost column indicates the weakest association for each gene
MR analysis is then performed with the msviper function implemented in the viper package (Bioconductor). Before aQTL analysis, the relative activity of the MRs for each individual sample is computed with the viper algorithm (viper package, Bioconductor). This step is critical, because MRs are usually dysregulated at the protein level, while rarely differentially expressed (Aytes et al., 2014; Carro et al., 2010; Lefebvre et al., 2010). Activity quantitative trail loci are then inferred by computing the statistical association between F-CNV and VIPER-inferred MR protein activity by MI or correlation analysis.
Finally, the conditional association analysis is performed by estimating the statistical association between samples harboring F-CNVs in a gene ‘a’ and the phenotype groups, after conditioning for the presence of CNVs in other genes ‘g’, one at a time, by Fisher’s exact test. Results for this analysis can be displayed as heatmaps (Fig. 1B). Selection of candidate genes for experimental validation and biochemical characterization can be performed based on the aQTL analysis p-values, either before or after correction by conditional association analysis.
4 Example application
We analyze 230 expression and CNV profiles for human GBM (The Cancer Genome Atlas, TCGA), distributed in the diggitdata package (Bioconductor), which also includes GBM-specific transcriptional and post-translational regulatory networks (Chen et al., 2014).
> library(diggit)
> data(gbm.expression, gbm.cnv, gbm.aracne, gbm.mindy,
+ package=″diggitdata″)
For the sake of speed, we restrict the analysis here to the first 1000 genes in the CNV profile, and infer the F-CNVs as follows:
> genes <- intersect(rownames(gbmExprs),
+ rownames(gbmCNV))[1:1000]
> gbmCNV <- gbmCNV[match(genes, rownames(gbmCNV)),]
> dobj <- diggitClass(expset=gbmExprs, cnv=gbmCNV,
+ regulon=gbmTFregulon, mindy=gbmMindy)
> set.seed(1)
> dobj <- fCNV(dobj, method=″mi″)
A scatterplot showing the association between CNV and expression for KLHL9 is shown in Figure 1A. Master regulators between the mesenchymal and proneural GBM subtypes can be inferred, and the top 20 most activated MRs displayed, with:
> set.seed(1)
> dobj <- marina(dobj, pheno=″subtype″, group1=″MES″,
+ group2=″PN″)
> sort(diggitMR(dobj), decreasing=TRUE)[1:20]
Then, aQTL analysis for two previously validated MRs (Carro et al., 2010), following by conditional association analysis for STAT3, can be performed as follows:
> set.seed(1)
> dobj <- aqtl(dobj, mr=c(″CEBPD″, ″STAT3″), method=″mi″)
> dobj <- conditional(dobj, pheno=″subtype″, group1=″MES″,
+ group2=″PN″, mr=″STAT3″, cnv=.15)
Conditional analysis results can be displayed as heatmap (Fig. 1B):
> plot(dobj, cluster=″2″)
Finally, results can be limited to MINDy-inferred post-translational modulators of the considered MRs, and summarized by:
> set.seed(1)
> dobj <- aqtl(dobj, mr=c(″CEBPD″, ″STAT3″), method=″mi″,
+ mindy=TRUE)
> dobj <- conditional(dobj, pheno=″subtype″, group1=″MES″,
+ group2=″PN″, mr=″STAT3″, cnv=.15)
> summary(dobj)
5 Discussion
Elucidating the causal genetic determinants of most complex diseases has proven more challenging than expected. Due to the large number of candidate loci, it is difficult to achieve enough statistical power to detect all but the most highly penetrant and frequent events. Furthermore, classic GWAS approaches are based on pure statistical associations, providing no mechanistic insights. DIGGIT aims to address both challenges by relying on context-specific models of cell regulation. It boosts the statistical power by focusing on the regulators mechanistically linked to the phenotype and on their upstream post-translational modulators (Chen et al., 2014). The algorithm relies on large (n > 100 samples) expression and genetic profiles and requires cell context-specific models of transcriptional and post-transcriptional regulation. Its R implementation, available as an R-system package from Bioconductor, has low computational requirements, running in most desktop workstations for an average (n ∼ 300 samples) dataset.
Funding
NCI CTD2 network 1RC2CA148308-01 (A.C.), National Centers for Biomedical Computing U54CA121852 (A.C., M.J.A.), R01 NS061776-05 (A.C.) and Kirschstein NRSA training grant T32GM082797 (J.C.C.).
Conflict of Interest: none declared.
References
- Aytes A., et al. (2014) Cross-species regulatory network analysis identifies a synergistic interaction between FOXM1 and CENPF that drives prostate cancer malignancy. Cancer Cell, 25, 638–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso K., et al. (2005) Reverse engineering of regulatory networks in human B cells. Nat. Genet., 37, 382–390. [DOI] [PubMed] [Google Scholar]
- Califano A., et al. (2012) Leveraging models of cell regulation and GWAS data in integrative network-based association studies. Nat. Genet., 44, 841–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carro M.S., et al. (2010) The transcriptional network for mesenchymal transformation of brain tumours. Nature, 463, 318–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.C., et al. (2014) Identification of causal genetic drivers of human disease through systems-level analysis of regulatory networks. Cell, 159, 402–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M.S., et al. (2013) Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature, 499, 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre C., et al. (2010) A human B-cell interactome identifies MYB and FOXM1 as master regulators of proliferation in germinal centers. Mol. Syst. Biol., 6, 377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.C., et al. (2013) Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat. Rev. Neurol., 9, 106–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., et al. (2009) Genome-wide identification of post-translational modulators of transcription factor activity in human B cells. Nat Biotechnol., 27, 829–839. [DOI] [PMC free article] [PubMed] [Google Scholar]