Abstract
Motivation: Accurate identification of genetic variants such as single-nucleotide polymorphisms (SNPs) or RNA editing sites from RNA-Seq reads is important, yet challenging, because it necessitates a very low false-positive rate in read mapping. Although many read aligners are available, no single aligner was specifically developed or tested as an effective tool for SNP and RNA editing prediction.
Results: We present RASER, an accurate read aligner with novel mapping schemes and index tree structure that aims to reduce false-positive mappings due to existence of highly similar regions. We demonstrate that RASER shows the best mapping accuracy compared with other popular algorithms and highest sensitivity in identifying multiply mapped reads. As a result, RASER displays superb efficacy in unbiased mapping of the alternative alleles of SNPs and in identification of RNA editing sites.
Availability and implementation: RASER is written in C++ and freely available for download at https://github.com/jaegyoonahn/RASER.
Contact: gxxiao@ucla.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
High-throughput RNA-sequencing (RNA-Seq) data are now indispensable in a wide range of biological and medical research areas, such as gene expression, gene regulation and functional studies of genetic abnormalities. As a result, accurate read alignment tools are highly desirable. Although many short read aligners exist, detection of single-nucleotide variants (SNVs) and small insertions or deletions (indels) harbored in the short RNA-Seq reads remains a very challenging task. Such applications require very low false-positive rate in read mapping to avoid calling spurious variants in the reads. In addition, different from studies of genetic variants using whole-genome sequencing data, a challenge specific to RNA-Seq is to accurately quantify the expression levels of alternative alleles of the variants based on RNA-Seq reads. For example, in studies of allele-specific expression (ASE) of single-nucleotide polymorphisms (SNPs), the expression levels of the two alleles of each SNP are quantified based on the number of mapped reads to determine whether a significant allelic bias (i.e. ASE) exists. A well-known issue is the mapping bias that favors reads harboring the reference allele (identical to the reference genome used for read alignment) of heterozygous SNPs (Degner et al., 2009; Heap et al., 2010; Pastinen, 2010). This problem is only partially alleviated if the reference genome is modified to encompass alternative SNP alleles that are known to exist in the specific dataset, making expensive whole-genome sequencing a necessity (Degner et al., 2009; Smith et al., 2013). In general, read alignment accuracy remains a fundamental limiting factor for quantification of ASEs.
A similar problem exists in studies of RNA editing. Recently, an increasing number of reports focused on identification of RNA editing sites using RNA-Seq data (reviewed in Lee et al., 2013). It is now well established that read mapping inaccuracy can lead to many false-positive predictions (Kleinman and Majewski, 2012; Lee et al., 2013; Lin et al., 2012; Pickrell et al., 2012). To increase editing prediction efficacy, strategies to combine multiple read aligners and apply a series of artifact filtering steps were utilized (Lee et al., 2013). Although many short read aligners are available (Engstrom et al., 2013) (http://en.wikipedia.org/wiki/List_of_sequence_alignment_software), no single aligner was specifically developed or tested as an effective tool for SNP and RNA editing prediction.
In our previous studies of ASE and RNA editing (Bahn et al., 2012; Li et al., 2012; Zhang and Xiao, 2015), we developed a read mapping pipeline that makes use of multiple read alignment tools and applies stringent requirements on the allowed mismatches of mapped reads. Specifically, it combines the alignment results of Bowtie (Langmead et al., 2009), BLAT (Kent, 2002) and TopHat (Trapnell et al., 2009) and applies two types of filters to retain reads that map uniquely with ≤n1 mismatches and do not mapped to any other genomic loci with ≤n2 mismatches (n1 < n2). We showed that this ‘double-filtering’ scheme is effective in reducing potential mapping bias or artifacts related to the presence of alternative sequence variants or homologous regions in the genome (Bahn et al., 2012; Lee et al., 2013; Li et al., 2012). However, this pipeline demands long CPU time and large hard drive space due to the usage of multiple read aligners and stringent mismatch filters.
Here, we present RASER (reads aligner for SNPs and editing sites of RNA) that is specifically designed for applications of RNA-Seq in studies involving SNPs, RNA editing or other types of SNVs. Since RASER is a standalone application, processing time and hard drive demand are greatly reduced compared with our previous read mapping strategy. A distinctive feature of RASER is its interrogation of a large number of mapping positions for each read using a novel tree structure that reduces ambiguity in mapping repeated sub-sequences, thus increasing mapping accuracy. The comprehensive search of many possible mapping positions of each read enables a comparative analysis of these alignments and elimination of ambiguous results likely due to existence of homologous regions in the genome. Built upon this strength, RASER further adopts a novel mismatch filtering scheme named ‘obviously best’ which aims to maximize mapping rate while maintaining high mapping accuracy.
Using both simulated and actual RNA-Seq datasets, we demonstrate that RASER shows the best mapping precision compared with other popular read mapping algorithms. The performance of RASER remains high for both short and long reads, which is also robust to mismatches and indels in the reads. Importantly, RASER shows superb efficacy in unbiased mapping of the alternative alleles of SNPs and in identification of RNA editing sites.
2 Methods
2.1 Building index
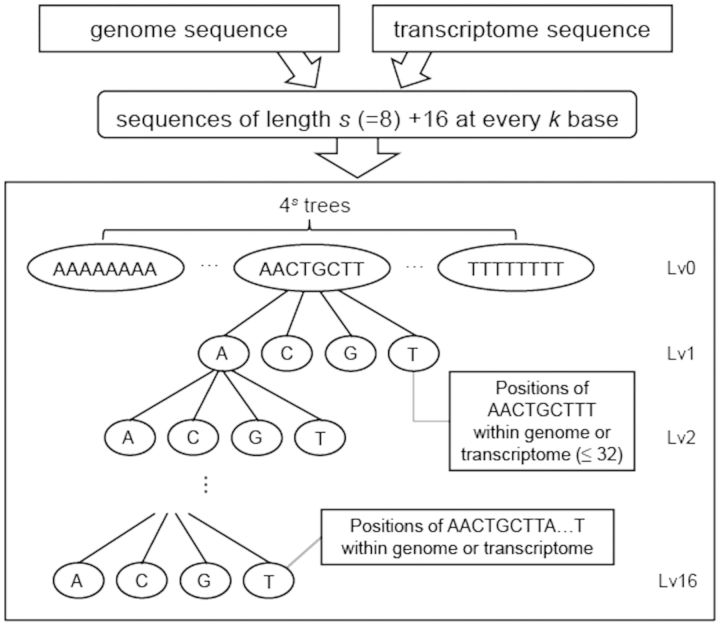
The index is a data structure that stores positions of specific nucleotide sequences within the reference and returns them during the read alignment search. For most DNA or RNA-Seq read aligners that require an index of the reference sequence, efficient storing and loading of the index are among the most important factors that facilitate accurate and fast alignment. One of the key features of the RASER index is that the length of indexed reference sub-sequence increases as it occurs more frequently in the reference, which reduces ambiguity in mapping of a read from the repeated regions. This feature enables reduction of processing time while maintaining sensitivity to resolve repetitive or homologous regions.
RASER index is composed of 4s trees as shown in Figure 1 where s is the predefined length of sequences corresponding to the root node (s = 8 in Fig. 1). The trees in the index have several properties. First, the root node of each tree is assigned with quaternary numbers to represent its associated sequence, where A = 0, C = 1, G = 2 and T = 3. We denote this quaternary number as qnum hereafter. A node with qnum N can have four child nodes whose qnums are (N x 4 + 0) ∼ (N x 4 + 3). For example, if qnum of a node is GAC ( = 2014), then its child nodes have qnums GACA ( = 20104), GACC ( = 20114), GACG ( = 20124) and GACT ( = 20134).
Fig. 1.
Index building. s + 16 bases at every k steps of genome and/or transcriptome reference sequences are indexed, where s and k are user parameters with default values 8 and 4, respectively. Nodes of level (lv) n (n > 0) store positions of sequences of length (s + n) within the reference. For example, the positions of ‘AACTGCTTT’ are stored in the lv1 node (which is a leaf node). The node is a leaf node if the number of stored positions is <32 or the maximum number of levels (16) is reached. See text for details
Second, the tree structure is intuitively defined in terms of levels. For example, the level of a root node is defined as 0, and its child node is defined as level 1 and so on. The maximum level of a tree is set to be 16. The level of a node can thus be calculated as (number of digit of qnum – s).
Third, a terminal or leaf node with qnum = N and level = v stores positions of all reference sequences whose first (v + s) nucleotides are calculated as N. A node is a terminal or leaf node if it stores≤32 reference positions or its level reaches the maximum v = 16. Nodes that do not meet the leaf node definition are split into four child nodes, according to qnums calculated from the first (v + s + 1) nucleotides of the sequences. For example, a node with qnum ‘GAC’ stores positions of ‘GAC…’ within the reference sequence if it is a leaf node. If the number of stored positions is >32 and the level is <16, positions of ‘GACA…’, ‘GACC…’, ‘GACG…’ and ‘GACT…’ are stored to the child nodes (if they are leaf nodes) whose qnums are ‘GACA’, ‘GACC’, ‘GACG’ and ‘GACT’, respectively.
According to the properties described above, the reference sequence of length (s + 16) at every k bases are indexed to the trees, where k is a user parameter whose default value is 4 (see below). The indexing process starts with trees whose qnum is equal to its first s bases and the tree branches are extended according to the rules defined above. The structure of the index and its building procedure are briefly illustrated in Figure 1.
The variable s is a user-defined parameter with a default value 8, which is recommended for the human genome. For genomes with smaller size, the value of s can be reduced. In general, a smaller s leads to smaller index sizes. However, if s is too small, alignment may become inefficient because smaller s leads to a decrease in the total number of nodes in the index tree and thus increases ambiguity in the mapping of reads originated from repetitive regions. The variable k is also a user parameter. Smaller k leads to larger index but can enable more accurate mapping. We recommend setting k to 4 for the human genome. It should be noted that the maximum level of the tree and the maximum number of positions stored in the nodes are fixed as 16 and 32, respectively. These variables are not user-defined parameters, because their optimal values are not sensitive to application-specific parameters such as the size of the reference genome or read length.
Using the above indexing scheme, two or more heterogeneous types of references, such as the genome and transcriptome, can be indexed together. The size of the index file is about 5.7 GB for hg19 only and 6 GB for hg19 and Encode transcriptome, using default user parameters. For all applications included in this article, an index of hg19 was used.
2.2 Read mapping
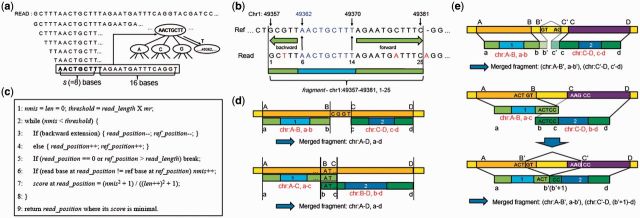
The first step to map a read is to apply a sliding window on the read sequence with a window size of (s + 16) and a step size of 1. Next, a root node is identified whose qnum is the same as the first s bases of the windowed (s + 16) read sequence. The tree branch of this root node that matches the ensuing nucleotides of the read sequence is then identified up to the leaf node. If assuming the level of the leaf node is n, positions stored in this leaf node correspond to reference sequences identical to the first (s + n) bases of the windowed read sequence. If a windowed read sequence contains ‘N’, it is ignored. This process is illustrated in Figure 2a. Note that if reference positions of all the windows of a read (pair) are obtained from leaf nodes (which may have far more than 32 reference positions), RASER terminates the mapping process and reports this read (pair) as unmapped. This ‘early termination’ procedure aims to reduce mapping time. Since such reads map to many positions in the reference, they will not be useful for applications that seek for unique or ‘obviously best’ mappings such as in the detection of SNPs or RNA editing sites. Thus, this procedure does not reduce the amount of final usable reads.
Fig. 2.
Read alignment. (a) Sliding windows are applied to a read with a window size s + 16 and step size 1. Genomic positions stored in the index are retrieved using the windowed sequence as illustrated. (b) An example to illustrate the extension process for a specific window with sequence ‘AACTGCTTT’ (blue) that was aligned to position 49362. The extension was carried out in both backward and forward directions. Mismatches and indels are labeled in red. (c) Pseudo code for the extension process in (b). (d) Merging of two fragments whose distance is less than the allowed number of insertions or deletions. Individual fragments are labeled in red. (e) Merging of two fragments whose distance is greater than the allowed number of insertions or deletions, which are examined as candidate spliced junction reads. Splice sites are determined using the GT-AG or GC-AG rules
Once the positions of a windowed read sequence are obtained, we extend these initial mappings as illustrated in Figure 2b. Such an extension is performed in both backward and forward directions. The pseudo-code for this process is given in Figure 2c. The variable mr (mismatch ratio) is a user-defined parameter with a default value of 0.08. The maximum total number of mismatches, insertions and deletions allowed in a mapped read is calculated as ‘read length x mr’. A score is calculated for each read position in the extension regions to represent the goodness of match of the extended read sequence (Fig. 2c). Mismatches (nmis) increase this score, but the score can still be small if the mapping size (= len) is large enough. In contrast, insertions or deletions are not considered in the extension step to avoid an inflation of the score due to possible existence of ensuing mismatches after indels. In the example shown in Figure 2b, read positions 6-14 were initially mapped to chr1:49 362–49 370 based on the index tree, which are marked in blue. Following backward and forward extension, read positions 1–25 was mapped to chr1:49 357–49 381. Hereafter, we denote this mapped sequence as a fragment.
After getting all the fragments of a read, the next step is to merge the fragments. If there are no insertions, deletions or clustered mismatches, many identical fragments are generated from the read. These duplicated fragments are discarded. For the remaining fragments, multiple scenarios may exist that call for different merging methods. Given two fragments ‘chr:A-B, a-b’ and ‘chr:C-D, c-d’ where A < D, the distance between them is C - B. If this distance is less than or equal to ‘read length x mr’, we assume that there are short deletions in the read relative to the reference and merge these fragments into a contiguous mapped region as in Figure 2d. Otherwise, we assume that the read spans an intron or large insertions in the reference and merge the fragments as illustrated in Figure 2e. For the latter case, RASER determines the best alignment using the known GT-AG or GC-AG splicing site sequences. If splice site signals are not found, the best alignment that results in the least number of mismatches and indels is chosen using the Smith–Waterman algorithm (Smith and Waterman, 1981). The maximum intron length is assumed to be 200 000 nt.
Fetching positions from the index is indeed an exact mapping process between windowed sequence of a read and positions within the reference, since this step does not allow mismatches or indels. However, given the sliding read window and the merging process of fragments, RASER is flexible enough to allow mismatches and indels.
The score of an alignment is defined as ‘the number of mismatches and indels/read length’. Only alignments with scores ≤ mr are reported (after read pairing if the sequencing data are paired end). Note that we apply the above mapping procedure to a read sequence and its reverse complement sequence separately.
2.3 Additional mapping schemes
The ‘double-filtering’ scheme (Bahn et al., 2012) is internally implemented in RASER and is an option that can be turn on or off by the user. As mentioned in Section 1, it is a mapping scheme where a read (pair) should be uniquely mapped with score less than or equal to mr and not mapped to anywhere in the reference with score more than df which is another user-defined parameter. Because a read (pair) with multiple aligned positions with less than ‘read length x mr’ mismatches is filtered out (failing the uniqueness requirement), it is not necessary to search for all the candidate mapping positions once two such alignments are reached, which leads to an accelerated alignment process.
RASER also implements an alternative new mapping scheme, namely ‘obviously best’, as an optional scheme to the user. By definition, this scheme finds the ‘obviously’ best mapping of a read (pair), whose mismatch score is less than or equal to mr and also less than scores of all other mappings by ob which is a user-defined parameter. Different from double filtering, this scheme necessitates a search of all possible candidate mappings with the larger mismatch ratio (mr + ob). This additional step somewhat slows down the alignment process but maximizes the mapping rate and minimizes false-positive mappings, as detailed in Section 3.
3 Results
3.1 Description of the experimental settings
We evaluated RASER using both simulated and actual RNA-Seq datasets. For simulated reads, we used BEER (Grant et al., 2011) to generate 1 million (M) paired-end reads of length 50, 100, 150 and 200 bases, respectively, each with three settings (thus, a total of 12 sets of simulated reads). The three settings (named SIM1, SIM2 and SIM3) represent three levels of sequencing errors: with a substitution rate of 0.001, 0.005 and 0.01 and an indel rate of 0.0005, 0.0025 and 0.005, respectively. Thus, SIM3 corresponds to the largest level of sequencing errors. The simulation used transcript annotations combining 10 annotation tracks including UCSC, RefSeq, RefSeq-Other, Ensembl, Vega, AceView, GenScan, GeneID, NSCAN and SGP (http://cbil.upenn.edu/BEERS/).
For real RNA-Seq data, we used three datasets referred to as GM12878, K562 and YH. The first two datasets were obtained by the ENCODE project that includes a total of 223 M and 213 M paired-end reads (2 × 76 nt) of the human lymphoblastoid and K562 cell line (polyA + cytoplasmic RNA), respectively (ENCODE Project Consortium, 2012). The third dataset YH was derived from lymphoblastoid cells of a Chinese individual (Peng et al., 2012) encompassing a total of 131 M paired-end (2 × 75 nt) and 30 M paired-end (2 × 100 nt) reads (polyA + RNA).
For both simulated and real datasets, we compared the performance of RASER with three state-of-the-art reads aligners, STAR 2.3.0 (Dobin et al., 2013), Tophat 2.0.9 (Kim et al., 2013) and GSNAP 2014-01-21 (Wu and Nacu, 2010). We also included NOVOALIGN (http://www.novocraft.com/products/novoalign/) in some analyses of simulated datasets. For all aligners, we used default parameters except that the maximum number of allowed mismatches was set to be 4 per 50 bases of read (e.g. mr = 0.05 for RASER). Specific to RASER, we used –d 0.09 for ‘double-filtering’ and –b 0.03 for the ‘obviously best’ mapping scheme. Unless otherwise noted, we used RASER with the ‘obviously best’ mapping scheme. For other aligners, unique mapping results were reported for all datasets. All read alignments were carried out against the human reference genome (hg19). Mapping results for all datasets are included in Supplementary Table S1.
3.2 Comparison of mapping performance
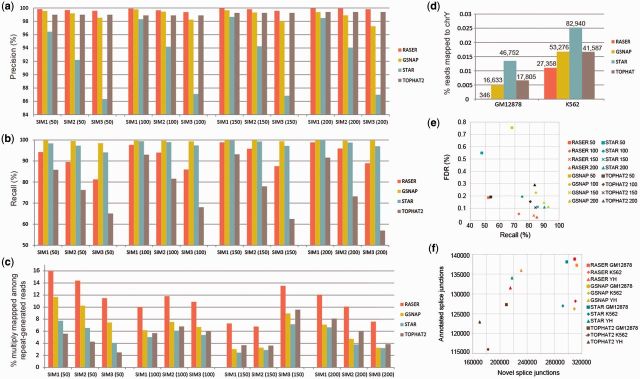
First, we compared mapping accuracy of different aligners using the 12 sets of simulated reads as described above (Fig. 3a). Mapping accuracy (i.e. precision) is defined as the percentage of correctly mapped among all mapped reads. A read is considered correctly mapped if more than 80% (Fig. 3a) or 50% (Supplementary Fig. S1a) of its nucleotides are mapped to their true positions. RASER, in its default mode (‘obviously best’ scheme, aka, OB), shows highest precision for all simulated datasets. The precision of RASER improves for longer reads, which is not observed for GSNAP or STAR. In addition, the precision of RASER is robust to sequencing errors, whereas that of GSNAP or STAR deteriorates as sequencing error increases. It should be noted that the precision of STAR is relatively low (Fig. 3a) in this analysis due to excessive soft clipping. However, its precision improves significantly if a lower % of mapped nucleotides per read is required to call a correct mapping (comparing Fig. 3a and Supplementary Fig. S1a). In general, Tophat2 is quite accurate and robust to sequencing errors, possibly at the cost of recall (see below).
Fig. 3.
Evaluation of mapping performance. (a) Comparison of precision (percentage of correctly mapped reads among all mapped reads) using simulated datasets of varying levels of sequencing errors (SIM1 < SIM2 < SIM3) and read length (in parenthesis of the x axis label). The mapping was deemed correct if more than 80% of the nucleotides in a read were correctly mapped to their original genomic loci. The ‘obviously best’ scheme was used for RASER and unique mapping was required for the other aligners. Same below. (b) Comparison of recall [percentage of mapped reads (unique or non-unique) among all reads] using the same simulated datasets as in (a). (c) Percentages of multiply mapped reads among all reads generated from repeats in the reference. (d) Percentage of reads mapped to chrY among all mapped reads of GM12878 and K562 datasets (derived from female cells). The numbers above each bar correspond to the number of mapped reads to chrY. (e) Identification of spliced junction reads. Simulated datasets as in (a) were analyzed. FDR is defined as (number of false-positive junction reads/total number of mapped junction reads). Recall is defined as (number of correctly mapped junction reads/total number of reads with junctions implanted in each simulated dataset). (f) The numbers of novel and annotated junctions detected by each aligner in real RNA-Seq dataset. Annotated splice junctions were defined as those that match perfectly or with less than 5 base difference to Genecode v19 annotation. Novel splice junctions were defined as those that were not included in the Gencode v19 annotation. For each junction, we required a minimum read coverage of 3 and a preference for canonical GT-AG or GC-AG splice sites
RASER out-performs the second best aligner in precision by 0.1 ∼ 0.5% which may appear to be small. However, incorrectly mapped reads are enriched with those that lead to false positive or inaccurate quantification of nucleotide variants (e.g. SNPs or RNA editing sites). The advantage of RASER in these aspects is demonstrated below.
In an alternative mode that only retains uniquely mapped reads (instead of OB), RASER again out-performs the others for most simulated datasets, especially those with larger sequencing errors or longer read length (Supplementary Fig. S1b and c). Since we implement three alternative mismatch filtering schemes [OB, unique (U), and double filtering (DF)], we compared the performance of RASER with these three schemes. The OB scheme shows superior mapping precision than the other two schemes (Supplementary Fig. S1d) but smaller recall (Supplementary Fig. S1e).
Another measure to evaluate the performance of aligners is recall, defined as the percentage of mapped reads (unique and non-unique) among all reads. In this comparison, GSNAP shows the highest recall values (Fig. 3b). Recall of RASER is lower compared with GSNAP or STAR but much higher than that of Tophat2. The lower recall of RASER is (at least partly) due to the early termination procedure described in Section 2 where reads expected to be mapped to many genomic positions are not reported.
NOVOALIGN is another often-used aligner. Since it does not directly map spliced junction reads, we excluded such reads in the simulated datasets to enable a fair comparison. NOVOALIGN shows relatively good recall but generally worse precision than other algorithms (Supplementary Fig. S1f–h). Given its suboptimal precision and the limitation in handling spliced junction reads, we did not include NOVOALIGN in further analyses.
A distinctive feature of RASER is manifested as the highest rate of multiple mappings (defined as the percentage of multiply mapped reads among all reads), which is often twice as high as those of other aligners (Supplementary Fig. S1i). In a related analysis, we compared the rate of multiply mapped reads among those reads that are originated from repeats in the reference genome (Fig. 3c). RASER again shows better performance than other aligners in capturing such non-unique reads. By design, RASER aims to identify as many mapping positions as possible for each read. This feature is important for reducing false-positive mappings when combined with the ‘double-filtering’ or ‘obviously best’ schemes, as it allows comparison of all or almost all possible mapped positions of a read. Since mapping of reads harboring SNPs or RNA editing sites is often complicated by the presence of highly similar regions across the genome, this feature of RASER enables improved performance in handling nucleotide variants in the reads, as demonstrated below.
To evaluate the performance of different aligners against real RNA-Seq data without ground truth, we examined the number of reads mapped to the Y chromosome (chrY) in RNA-Seq data derived from female cells: GM12878 and K562. Because of the absence of a Y chromosome in these cells, reads mapped to chrY should be considered as false-positive mappings. In this case, RASER (OB mode) out-performs the other aligners (unique mapping) considerably and yields the lowest number of false-positive mappings and lowest rate of false positives (defined as the ratio between the number of reads mapped to chrY and total number of mapped reads) (Fig. 3d). The high false-positive rates of the other aligners suggests that the Y chromosome harbors regions that have sequence similarity to other chromosomes in the genome, which is exacerbated by possible structural variations in the cancer cells (K562). The superior performance of RASER roots from its effectiveness in identifying and handling non-uniquely mapped reads when combined with the OB mismatch filtering scheme and its high precision in general.
Lastly, we evaluated the performance of RASER in identifying spliced junction reads resulted from RNA splicing. Two evaluation metrics were calculated: recall (defined as the number of correctly mapped junction reads/total number of reads with junctions implanted in each simulated dataset) and false discovery rate (FDR, defined as the number of false-positive junction reads/total number of mapped junction reads identified by an aligner). As shown in Figure 3e, RASER has the lowest FDR for simulated reads of all lengths, with comparable or lower recall than other aligners. Applied to real RNA-Seq datasets, RASER demonstrates the highest recall rates [defined as the number of perfectly or partially matched junctions/total number of junctions in Gencode (v19) annotation] for two of the three datasets (Supplementary Fig. S2). In addition, RASER also identifies the largest number of annotated spliced junctions in these datasets (Fig. 3f). Based on the above results for both simulated and real datasets, RASER is advantageous in identifying spliced junction reads.
3.3 Quantification of single-nucleotide expression in RNA-Seq
To evaluate the performance of RASER in quantifying SNVs in RNA-Seq reads, we implanted known SNPs (from the YH and GM12878 genomes) to the SIM1, SIM2 and SIM3 reads (read length being 50, 100, 150 and 200 nts). For each read that contains a known SNP, the probability for the SNP to have the reference or alternative allele was set to be 0.25, 0.5 and 0.75 (ratio defined as the number of reads containing the reference allele/total number of reads covering the SNP). Following read mapping, customized post-mapping filters were applied prior to calculation of allelic ratios, including removal of duplicated mappings, removal of reads where the mismatches (corresponding to SNPs) were (i) within 5 nt of read ends or (ii) with Sanger base quality less than 30, and removal of SNPs with less than 5 reads. We calculated allelic ratios for the remaining mismatches. Subsequently, we calculated for each SNP the absolute difference between its simulated allelic ratio and the observed allelic ratio from mapped reads, where allelic ratio is defined in the same way as described above (number of reads containing the reference allele/total number of reads covering the SNP). Thus, larger allelic ratio differences reflect increased mapping bias to the alternative alleles of the simulated SNP. At different thresholds of allelic ratio difference (0–0.2), we determined the FDR of SNP quantification as FP/(FP + TP), where FP and TP are numbers of false and true positives, respectively. At each threshold, a given SNP is defined as a true positive if its allelic ratio difference is less than the threshold; otherwise, this SNP is a false positive.
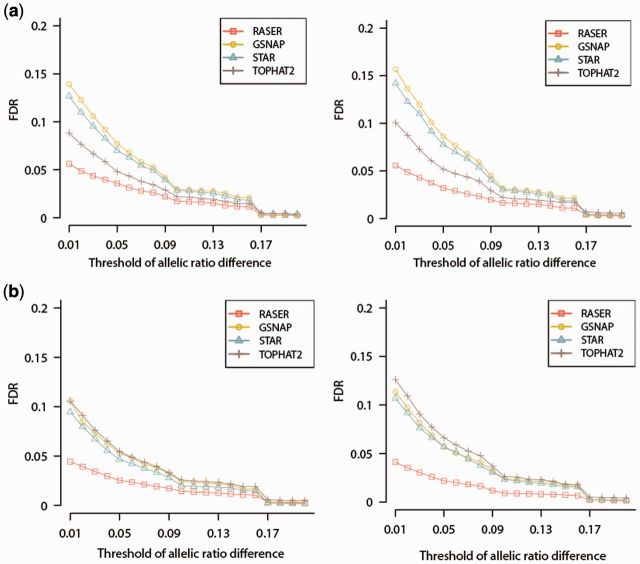
In this comparison, RASER offers the smallest FDR among all aligners tested for 55 cases among 72 cases (76%) (Fig. 4 and Supplementary Fig. S3). FDR of TOPHAT2 was smaller than RASER for 12 out of 18 cases of 50 nt read sets and GSNAP yielded smaller FDR than RASER for 5 out of the remaining 54 cases, but the differences were subtle. It is expected that aligners often have mapping bias favoring the reference allele. Consistent with this expectation, FDR was generally lower for 0.75 (higher proportion of the reference allele) compared with that for 0.25 (smaller proportion of the reference allele). More importantly, we observed that the difference in FDR between RASER and the second best aligner increases as allelic probability decreases, which confirms that RASER is less biased to reference allele. In addition, the allelic ratios resulted from RASER are not significantly different from simulated ratios for any dataset (P value > 0.05, Kolmogorov–Smirnov test) except two cases (SIM3 of Supplementary Fig. S4q and u), whereas other aligners often lead to significant deviations of allelic ratios from the ground truth (Supplementary Fig. S4).
Fig. 4.
FDR in the quantification of allelic ratios of SNPs expressed in RNA-Seq. SNPs were implanted in SIM1 data of (a) 100, (b) 150 bases in length. SNPs in the YH (left panels) or GM12878 samples (right panels), both of which had corresponding whole-genome sequencing data, were implanted into the simulated reads. The ‘obviously best’ scheme was used for RASER and unique mapping was reported from the other aligners. For a specific SNP, its allelic ratio is defined as (number of reads containing the reference allele/total number of reads covering the SNP). The allelic ratio difference of this SNP is defined as the absolute difference between its observed allelic ratio and the simulated allelic ratio. At different thresholds of allelic ratio difference (0–0.2, x axis), the FDR of SNP quantification is defined as FP/(FP + TP), where FP and TP are numbers of false and true positives, respectively. At each threshold, a given SNP is defined as a true positive if its allelic ratio difference is less than the threshold; otherwise, this SNP is a false positive
Applied to real RNA-Seq datasets, RASER also demonstrates superior performance in reducing mapping bias to alternative alleles of SNPs (Supplementary Fig. S5a and c). Using the two datasets derived from cells with whole-genome sequencing data (GM12878 and YH), we observed that the average allelic ratio of SNPs is closest to the expected 0.5 (ratio defined as number of reads containing the reference allele/total number of reads) for reads mapped by RASER. In contrast, the results from GSNAP, TOPHAT2 and STAR are more biased toward the reference allele (>0.5), which is a known issue where read mapping favors the reference allele of heterozygous SNPs since the reference genome was used.
The above analyses used the same customized post-mapping filters as described for simulated reads. As an alternative method for SNP filtering, we used GATK with default parameters (DePristo et al., 2011) to call SNPs based on the mapping results of each aligner in analyzing GM12878 and YH datasets. Supplementary Figure S5b and d show the distribution of allelic ratios of SNPs called by GATK in each dataset. RASER again shows the most unbiased results, which are similar to the distributions in Supplementary Figure S5a and c that used the SNP calling filters we implemented.
In addition, we evaluated the performance of the aligners in SNP calling resulted from GATK. We counted true-positive and false-positive SNPs and calculated FDR on SNP detection. True positives were defined as correctly detected SNPs by GATK when compared with known GM12878 or YH SNPs based on their respective genome sequencing data. False positives were defined as GATK-called SNPs that satisfied all three requirements: (i) the predicted SNP was not known GM12878 or YH SNPs; (ii) the predicted SNP was not listed as known human editing sites in the RADAR database (Ramaswami and Li, 2014) and (iii) the SNP was covered with more than 20 reads in total. Although these ‘false positives’ could indeed be true SNPs missed by genome sequencing or true RNA editing sites not in RADAR, we can reasonably assume that they proportionally represent the amount of wrong SNPs identified by each aligner.
For the YH data, RASER yielded the smallest FDR (Supplementary Fig. S5f). For GM12878, RASER demonstrated smaller FDR than GSNAP and STAR and slightly larger FDR than TOPHAT2 (Supplementary Fig S5e). These results can be explained by the fact that TOPHAT2 showed generally good performance for short (50 bp) reads and GM12878 reads (76 bp) are shorter than YH reads (100 bp). It should be noted that, although RASER has slightly lower recall rate in read mapping (Fig. 3b), its sensitivity in SNP calling is not lower than other aligners. As shown in Supplementary Figure S5e and f, the number of true-positive SNPs identified by RASER is often larger than those by other aligners.
3.4 Identification of RNA editing sites
Another application of single-nucleotide analysis of RNA-Seq data is to identify RNA editing sites, which is gaining broad attention in recent years (Lee et al., 2013). In previous studies, since read mapping is not perfect, a series of post-mapping filters were designed to remove likely false-positive editing sites resulted from possible mapping artifacts (Lee et al., 2013). We thus applied these artifact filters to the results of each aligner (see Supplementary Fig. S6 legend). RASER yielded the highest % AG values in all three datasets with or without these artifact filters (Supplementary Fig. S6).
The %AG value represents percentage of A-to-G mismatches among all predicted RNA–DNA differences. It is generally accepted that higher the %AG, the better the accuracy of predicted editing sites (Peng et al., 2012; Ramaswami et al., 2013). This is based on a prevailing assumption in the field that A-to-I editing (leading to A-to-G mismatches) should account for most of the observed editing sites.
Another metric to evaluate performance of aligners is to calculate an FDR defined as the percentage of originally identified editing sites that were removed by the artifact filters (Lee et al., 2013). By definition, these filters remove artifacts in the mapping results. Thus, the more sites that were removed by the filters, the more artifacts there were in the original results.
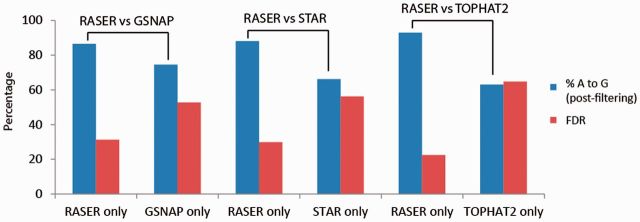
To focus on the differences of the aligners, we segregated the predicted editing sites into those that were only identified by RASER, only by the other aligner in comparison or by both RASER and the counterpart aligner. We calculated the %AG (post-filtering) and FDR for each subset of editing sites (Fig. 5 and Supplementary Fig. S7). The results showed that RASER out-performs the other aligners in terms of both %AG and FDR for all three datasets included in this study.
Fig. 5.
Identification of RNA editing sites in K562 data. The ‘obviously best’ scheme was used for RASER and unique mapping was reported from the other aligners. Editing sites that were only identified by RASER (‘RASER only’) or by the other aligner were analyzed. Percentage of A to G editing sites (blue) and FDR values (red) are shown. FDR is defined as the percentage of originally predicted editing sites that were removed by the artifact filters. These filters remove RNA–DNA differences sites that satisfy one of the following: (i) covered by reads with a strand bias, (ii) with 100% editing, (iii) with low-editing levels (<10% or <3 edited reads), (iv) close to splice sites (i.e. ≤4 nt from spliced junctions), (v) within simple repeats (defined by Repeatmasker), (vi) within homopolymer repeats (Repeatmasker) and (vii) overlapping known SNPs in public databases (dbSNP) (see Lee et al., 2013 for details)
It should be noted that editing sites common to RASER and another aligner generally had lower FDR than sites that were specific to one aligner (Supplementary Fig. S7c and d), except in the YH data where RASER-specific sites had the lowest FDR (Fig. S7e). This observation is consistent with the expectation that results common to multiple aligners should be highly accurate, which in one way confirms the validity of our method to evaluate accuracy (i.e. by FDR). It should also be noted that RASER normally identifies less editing sites than the other aligners (Supplementary Fig. S7c–e). However, because those sites that were unique to the other aligners had high FDR, RASER’s performance in accuracy outweighs its somewhat lower sensitivity because improved accuracy is highly desirable in editing analysis.
3.5 Evaluation of running time and memory
We compared running time of different aligners for 12 simulated datasets on a machine with Intel (R) Xeon (R) CPU E5-2680 v2 at 2.80 GHz × 40, and 256 GB of memory (Table 1). In general, running time of RASER is similar to that of TOPHAT2 (slightly faster than TOPHAT2 if the read length ≤ 100 bases and somewhat slower otherwise). RASER with the OB mapping scheme is slower than RASER without any mapping scheme or with the DF scheme. This is because OB mapping requires a search for all the possible candidate mappings with a large mismatch ratio (mr + ob). As expected, STAR shows extraordinarily short running time. GSNAP is relatively slow for short read length (50 nt), but the running time is improved for longer read length (100–200 nt). For memory, RASER required less than 50% of memory than GSNAP or STAR. TOPHAT2 is the most memory efficient but with increased memory requirement for longer reads. Supplementary Figure S8 shows a vignette-like example flow of RASER.
Table 1.
Runtime and memory benchmark results
| Read length | Algorithm | Runtime (s) |
Memory (GB) | ||
|---|---|---|---|---|---|
| SIM1 | SIM2 | SIM3 | |||
| 50 | RASER | 592.6 | 587.28 | 618.9 | 10.8 |
| RASER DF | 625.07 | 617.85 | 639.6 | 10.8 | |
| RASER OB | 706.51 | 698.05 | 721.27 | 10.8 | |
| GSNAP | 1869.37 | 1657.21 | 3310.93 | 24.8 | |
| STAR | 9 | 11 | 10 | 28.9 | |
| TOPHAT2 | 636 | 618 | 638 | 4.3 | |
| 100 | RASER | 724.93 | 748.41 | 768.43 | 10.8 |
| RASER DF | 854.06 | 899.75 | 919.89 | 10.8 | |
| RASER OB | 951.95 | 1013.68 | 1021.67 | 10.8 | |
| GSNAP | 549.58 | 366.21 | 482.04 | 24.8 | |
| STAR | 11 | 13 | 15 | 28.9 | |
| TOPHAT2 | 814 | 858 | 921 | 4.8 | |
| 150 | RASER | 1143.79 | 1241.51 | 1262.79 | 10.8 |
| RASER DF | 1502.06 | 1641.47 | 1557.84 | 10.8 | |
| RASER OB | 1644.49 | 1800.12 | 1687.85 | 10.8 | |
| GSNAP | 1015.73 | 848.68 | 1294.81 | 24.8 | |
| STAR | 23 | 25 | 32 | 28.9 | |
| TOPHAT2 | 1085 | 1189 | 1252 | 5.3 | |
| 200 | RASER | 1682.08 | 1789.76 | 1812.85 | 10.8 |
| RASER DF | 2278.6 | 2341.39 | 2411.74 | 10.8 | |
| RASER OB | 2542.26 | 2580.5 | 2657.54 | 10.8 | |
| GSNAP | 849.71 | 867.47 | 1773.6 | 24.8 | |
| STAR | 25 | 31 | 42 | 28.9 | |
| TOPHAT2 | 1361 | 1508 | 1651 | 5.9 | |
Comparison performed using 1 M reads, eight threads for each algorithm. There was no difference in memory usage for different sets of simulated datasets. RASER DF, RASER with double filtering; RASER OB, RASER with obviously best mapping.
4 Discussion
Accurate and unbiased identification and quantification of SNPs or RNA editing sites using RNA-Seq data proved to be challenging tasks (Degner et al., 2009; Heap et al., 2010; Lee et al., 2013; Pastinen, 2010). These challenges call for an in-depth analysis of the performance of short read aligners in handling SNVs and development of new aligners tailored for this purpose. Because of existence of sequence similarity across different regions of a genome, false-positive mappings may occur where reads originated from one region align to another region by mistake with a small number of mismatches. These mismatches are then exploited in subsequent analysis for studies of SNPs or RNA editing. Mapping errors may only affect a relatively small number of reads. However, since the total number of reads harboring true SNVs is expected to be small relative to all reads of a dataset, those incorrectly mapped reads often constitute a considerable portion of reads used to analyze for SNPs or RNA editing sites. Therefore, these applications call for very accurate read mapping.
To tackle this problem, we reason that if an aligner can find all or almost all mapping positions of a read, these results can then be examined strategically to exclude ambiguous mappings and reduce false-positive rate. RASER was designed according to this rationale. In contrast, existing aligners do not generally report all or almost all mapping positions of a read, thus impractical to be combined with the OB or DF schemes integrated into RASER. We showed that RASER can effectively identify repeat-generated reads as multiply mapped reads and its mapping precision is higher than other popular aligners. Although RASER outperforms other aligners in overall mapping precision by a small margin, this seemingly small improvement enabled considerably better results in the analyses of SNPs and RNA editing sites. This observation is again due to the sensitivity of such analyses to incorrectly mapped reads that contain apparent mismatches relative to the reference. In addition, RASER also demonstrates superior performance in identifying spliced junctions, a critical aspect of RNA-Seq read mapping.
Many aligners have been developed to map short reads generated by high-throughput sequencing experiments. It is important to note that different applications call for different aligners with specific properties. RASER is appropriate for applications where accurate mapping is highly desirable, especially if SNVs in the short reads are sought after. The improved mapping precision of RASER is largely resulted from its novel tree structure that reduces ambiguity in mapping of repetitive sequences and relatively complete search of all possible mappings. Despite its extensive search for alternative mapping positions, RASER shows comparable mapping speed to TOPHAT2 or GSNAP. Furthermore, RASER requires less memory than GSNAP and STAR, which is a desirable feature for aligners in parallel computing environments where memory usage is often an important issue.
Note that RASER benefits from removing ambiguously mapped reads for applications involving SNPs or RNA editing prediction. Removal of ambiguous reads may not be advisable for other applications, such as transcript reconstruction where ambiguous reads can be utilized (Pertea et al., 2015; Zickmann et al., 2014).
Supplementary Material
Acknowledgements
We thank members of the Xiao laboratory for helpful comments on this work. We thank the ENCODE consortium for generating the data and making their data available to the public.
Funding
This work was supported by the National Institute of Health [R01HG006264, U01HG007013 to X.X.] and the National Science Foundation [1262134 to X.X.].
Conflict of Interest: none declared.
References
- Bahn J.H., et al. (2012) Accurate identification of A-to-I RNA editing in human by transcriptome sequencing. Genome Res., 22, 142–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degner J.F., et al. (2009) Effect of read-mapping biases on detecting allele-specific expression from RNA-sequencing data. Bioinformatics, 25, 3207–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo M.A., et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet., 43, 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., et al. (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium (2012) An integrated encyclopedia of DNA elements in the human genome. Nature, 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrom P.G., et al. (2013) Systematic evaluation of spliced alignment programs for RNA-seq data. Nat. Methods, 10, 1185–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant G.R., et al. (2011) Comparative analysis of RNA-Seq alignment algorithms and the RNA-Seq unified mapper (RUM). Bioinformatics, 27, 2518–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heap G.A., et al. (2010) Genome-wide analysis of allelic expression imbalance in human primary cells by high-throughput transcriptome resequencing. Hum. Mol. Genet., 19, 122–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J. (2002) BLAT—the BLAST-like alignment tool. Genome Res., 12, 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., et al. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol., 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinman C.L., Majewski J. (2012) Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science, 335, 1302; author reply 1302. [DOI] [PubMed] [Google Scholar]
- Langmead B., et al. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol., 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., et al. (2013) Analysis and design of RNA sequencing experiments for identifying RNA editing and other single-nucleotide variants. RNA, 19, 725–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., et al. (2012) Identification of allele-specific alternative mRNA processing via transcriptome sequencing. Nucleic Acids Res., 40, e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W., et al. (2012) Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science, 335, 1302; author reply 1302. [DOI] [PubMed] [Google Scholar]
- Pastinen T. (2010) Genome-wide allele-specific analysis: insights into regulatory variation. Nat. Rev. Genet., 11, 533–538. [DOI] [PubMed] [Google Scholar]
- Peng Z., et al. (2012) Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat. Biotechnol., 30, 253–260. [DOI] [PubMed] [Google Scholar]
- Pertea M., et al. (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol., 33, 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell J.K., et al. (2012) Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”. Science, 335, 1302.; author reply 1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami G., Li J.B. (2014) RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res., 42, D109–D113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami G., et al. (2013) Identifying RNA editing sites using RNA sequencing data alone. Nat. Methods, 10, 128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R.M., et al. (2013) Whole transcriptome RNA-Seq allelic expression in human brain. BMC Genomics, 14, 571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T.F., Waterman M.S. (1981) Identification of common molecular subsequences. J. Mol. Biol., 147, 195–197. [DOI] [PubMed] [Google Scholar]
- Trapnell C., et al. (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics, 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T.D., Nacu S. (2010) Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics, 26, 873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Xiao X. (2015) Genome sequence-independent identification of RNA editing sites. Nat. Methods, 12, 347–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickmann F., et al. (2014) GIIRA—RNA-Seq driven gene finding incorporating ambiguous reads. Bioinformatics, 30, 606–613. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.