Abstract
Context:
To date, a clear understanding of dengue disease pathogenesis remains elusive. Some infected individuals display no symptoms while others develop severe life-threatening forms of the disease. It is widely believed that host genetic factors influence dengue severity.
Aims:
This study evaluates the relationship between certain polymorphisms and dengue severity in Sri Lankan patients.
Settings and Design:
Polymorphism studies are carried out on genes for; transporter associated with antigen presentation (TAP), promoter of tumor necrosis factor-α (TNF-α), and promoter of interleukin-10 (IL-10). In other populations, TAP1 (333), TAP2 (379), TNF-α (−308), and IL-10 (−1082, −819, −592) have been associated with dengue and a number of different diseases. Data have not been collected previously for these polymorphisms for dengue patients in Sri Lanka.
Materials and Methods:
The polymorphisms were typed by amplification refractory mutation system polymerase chain reaction in 107 dengue hemorrhagic fever (DHF) patients together with 62 healthy controls.
Statistical Analysis Used:
Pearson's Chi-square contingency table analysis with Yates′ correction.
Results:
Neither the TAP nor the IL-10 polymorphisms considered individually can define dengue disease outcome with regard to severity. However, the genotype combination, IL-10 (−592/−819/−1082) CCA/ATA was significantly associated with development of severe dengue in these patients, suggesting a risk factor to developing DHF. Also, identified is the genotype combination IL-10 (−592/−819/−1082) ATA/ATG which suggested a possibility for protection from DHF. The TNF-α (−308) GG genotype was also significantly associated with severe dengue, suggesting a significant risk factor.
Conclusions:
The results reported here are specific to the Sri Lankan population. Comparisons with previous reports imply that data may vary from population to population.
Keywords: Dengue, dengue hemorrhagic fever, interleukin-10, transporter associated with antigen presentation, tumor necrosis factor-α
INTRODUCTION
The dengue virus (DENV) belongs to the genus flavivirus. It has four antigenically related, but distinct, DENV serotypes of infection (DENV-1 to DENV-4) and can be transmitted to humans by Aedes mosquitoes that act as a vector and a reservoir of infection. DENV infection can cause a broad spectrum of clinical symptoms ranging from asymptomatic infections or a mild flu-like febrile illness (dengue fever [DF]) to the life-threatening severe forms of DF, which include dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS). Dengue is the most important mosquito-borne viral disease in tropical and subtropical regions, with an estimate of 50 million infections annually by the WHO and an estimate of 390 million infections/year by a study conducted recently using novel risk mapping techniques.[1,2] Less than 3% of these infections will develop into the more severe form of the infection.[3] Approximately, 90% of DHF/DSS cases occur in patients having a secondary infection with a heterologous dengue serotype.[3] This secondary infection carries a higher risk of plasma leakage, which if not appropriately supported clinically with fluid management, can lead to shock. However, a conclusive understanding of the mechanisms underlying why only some patients develop the more severe forms of the infection is lacking.[4]
While epidemiologic factors, ecological factors, and viral variations are factors involved in disease progression, it is believed that genetic heterogeneity of molecules involved in the host immune response to DENV infection contributes to disease progression. Host genetic factors may predispose some individuals to progress to the severe forms of the illness while others may protect individuals.[5,6,7,8] Genetic polymorphisms involved in the host immune mechanism can have a significant effect on disease progression due to the importance of the host immune response to the virus. Polymorphisms on transporter associated with antigen presentation (TAP), dendritic cell-specific intercellular adhesion molecule-3-Grabbing nonintegrin, interleukin-10 (IL-10), Vitamin D receptor, cytotoxic T-lymphocyte antigen 4, mannose-binding lectin, JAK1, tumor necrosis factor-α (TNF-α), MICB, and PLCE1 are some of the key molecules in which locus polymorphisms have been associated with DENV disease progression previously.[9,10,11,12,13,14] TAP is a key component involved in the transport of viral antigenic peptides for loading of major histocompatibility complex Class I molecules within the cell, which play a role in triggering an immunological response from the host. TAP is a key molecule involved in antigen presentation and is thought to help in DENV immune escape and DENV infection.[15,16,17] Polymorphisms within the gene for TAP have been implicated in cancer, autoimmune disorders and a number of viral infections including DENV itself.[18,19] Polymorphisms in the TAP gene region could influence the selection process that determines which antigen peptides contribute to the pathogenesis of dengue infection, which in turn may determine the severity of the infection. Polymorphisms of TAP1 and TAP2 genes have been typed, and associations have been uncovered for DENV in patients in Puducherry, India.[7,20]
The plasma leakage characteristic of severe dengue infections is suspected to be caused by malfunction of vascular endothelial cells induced by cytokines.[21] DF and DHF patients have been reported to have significant differences in cytokine levels, and the involvement of cytokines in DENV pathogenesis has been identified previously.[22,23] Plasma levels of cytokines such as TNF-α and IL-10 have been found to be significantly higher in DHF patients in comparison to DF patients.[24,25] Considering that the production of cytokines can be regulated by genetic polymorphisms and different genetic variants have been associated with DENV, we decided to study these particular polymorphic sites on TNF-α and IL-10 in relation to the Sri Lankan population.
The genetic variation on the promoter region of TNF-α at positions −308 results in two allelic forms. The presence of guanine (G) defines the common variant and the presence of adenine (A) defines the less common variant. TNF-α (−308) A allele is associated with increased gene transcription of TNF-α, in comparison to the common allele G.[26] Several studies have shown higher levels of TNF-α in patients with DHF and DF in comparison to controls. Some of these studies show plasma levels of TNF-α are higher in patients with DHF than patients with DF.[24] TNF-α (−308) A allele is a possible risk factor of DHF in a cuban and venezuelan population and the TNF-α (−308) GG genotype has been associated with protection from DHF.[8,27] However, studies conducted by Vejbaesya et al. (2009) in a Thai population and Garcνa-Trejo et al. in Mexico did not find an association between TNF-α (−308) polymorphisms and dengue disease severity.
Genetic variation on the IL-10 promoter regions −1082, −819 and −592 influence IL-10 production.[28] However, in previous studies, IL-10 promoter (−1082) A/G, (−819) C/T, (−592) C/A polymorphisms investigated individually did not show any significant association with dengue disease severity in Cuban and Venezuelan populations.[8,27] However, the IL-10 genotype combination IL-10 (−1082/−819/−592) ACC/ATA was reported to be a DHF risk factor in Cuban patients.[8]
Hence, the purpose of the present study was to evaluate the relationship between certain polymorphisms and the genetic susceptibility to the severe forms of DF in patients in Sri Lanka. The polymorphisms reported on here; TAP1 (333), TAP2 (379), TNF-α (−308), and IL-10 (−1082, −819, −592) have been previously associated with DENV disease pathogenesis as mentioned above and therefore chosen to be tested within the Sri Lankan population group investigated here. This information is important to determine if the host genetic background could play an important role in defining the clinical outcome of dengue infections in Sri Lankan patients.
MATERIALS AND METHODS
Sample collection
A total of 107 patients with suspected severe dengue infection, who were admitted to a Tertiary Care Hospital in Colombo, were recruited to the study following informed written consent. The study was approved by the Ethics Review Committee of the University of Sri Jayewardenepura, Sri Lanka. The severity of their dengue infection was classified based on the WHO 2011 guidelines.[25,29] Accordingly, all patients who were classified as having DHF either had clinical or laboratory evidence of fluid leakage. Based on the 2011 WHO diagnostic criteria, shock was defined as lowering of pulse pressure to 20 mmHg or less or the presence of signs of poor capillary perfusion (cold extremities, poor capillary refill or a rapid pulse rate). All clinical features such as the presence of fever, vomiting, diarrhea, myalgia, blood pressure, pulse rate and volume, general status of the patient, presence of any bleeding manifestations and presence of any possible fluid accumulation in the pleural cavity and abdomen were monitored several times a day from the time of admission to hospital, until they were discharged. Patients presenting with clinical symptoms of DHF and DSS were grouped together under the category of DHF based on the WHO 2011 criteria. Among 107 patients, 26 patients were classified as having DSS and 81 patients were classified as DHF. Laboratory investigations such as the full blood count were done several times a day and aspartate aminotransferase, alanine aminotransferase once during the critical phase, in all patients until they were discharged from hospital. Serum electrolytes, coagulation profiles, chest radiography and ultrasound scans of the abdomen were only done in selected patients who developed severe dengue due to resource limitations. Patients with DF were not recruited, as our aim was to determine the genetic polymorphisms that are associated with severe dengue or DHF, and therefore, we compared the genetic polymorphisms in patients with more severe forms of dengue, with those who had a past mild or asymptomatic dengue infection.
Sixty-two dengue seropositive individuals who had never been hospitalized due to a febrile illness were recruited as healthy controls following informed written consent. As they never had a febrile infection that warranted hospital admission and were found to have dengue specific IgG antibodies confirming seropositivity, they were considered to have had a mild/asymptomatic past dengue infection.
Serology
Acute dengue infection was confirmed by testing the serum samples that were collected after day 6 of illness with a commercial capture-IgM and IgG enzyme-linked immunosorbent assay (ELISA) (Panbio, Brisbane, Australia). The ELISA was performed and the results were interpreted according to the manufacturer's instructions. This ELISA assay has been validated as both sensitive and specific for primary and secondary DENV infections.[30,31] Patients who only had DENV specific IgM were classified as having a primary dengue infection while those who had a positive result for both IgM and IgG were classified as having a secondary dengue infection.
Dengue virus-specific real time-polymerase chain reaction
DENV RNA was extracted from serum using QIAamp viral RNA mini kit (Qiagen CA, USA). RNA was reverse transcribed and the polymerase chain reaction (PCR) was performed using primers and conditions as previously described.[32] When determining the serotype of the infecting DENV, positive controls for DEN1, DEN2, DEN3 and DEN4 were used in all experiments.
DNA extraction and genotyping
Human genomic DNA was extracted from whole blood using the QIAamp DNA Mini Kit (Qiagen CA, USA). Samples were stored at −20°C until further use. Polymorphisms on TAP, TNF-α and IL-10 were identified by amplification refractory mutation system (ARMS-PCR) methodology using primers designed previously.[33,34,35] The primers and ARMS-PCR conditions used are given as Tables S1 (804.6KB, tif) and S2 (1.3MB, tif) . All PCR reagents were from Promega Corporation, USA.
PCR primers and primer protocol
PCR amplification protocols
Polymorphic residues investigated were: Codon 333 on TAP1 (ATC to GTC, Ile to Val) and codon 379 on TAP2 (GTA to ATA, Val to Ile) gene regions; codon (−308) on the promoter region of TNF-α (A to G); and codons (−1082) (A to G), (−819) (C to T), and (−592) (A-C) on the promoter region of IL-10.
DNA regions selected were amplified and PCR products were separated by 2% agarose gel electrophoresis and stained with ethidium bromide. PCR fragments from randomly selected samples were sequenced to confirm the presence of the suspected polymorphisms.
Statistical analysis
Statistical analysis was performed using GraphPad Prism (GraphPad Software Inc. version 4.03) using the Pearson's Chi-square contingency table analysis with Yates′ correction. A P < 0.05 was considered statistically significant. An odds ratio (OR) >1 was interpreted as posing a risk for developing severe dengue or DHF and an OR <1 was interpreted as a possible implication of protection from developing severe dengue. This analysis was performed for the individual polymorphisms as well as allelic genotype combinations to investigate the hypothesis that certain alleles or genotype combinations may predispose individuals to progress to the severe forms of dengue or DHF.
RESULTS
According to the IgM, IgG, and DENV PCR results, the 107 patients studied were confirmed to have DHF. Of 107 patients, 26 patients had shock and 27 had clinical evidence of either a pleural effusion or ascites. Bleeding manifestations were seen in 43 (40.2%) patients. Of these 43 patients, 30 had mucosal or skin bleeding only (petechie and bleeding from the gums), while only 15 had significant bleeding from the gastrointestinal tract. 11 out of 15 patients who had significant gastrointestinal bleeding also had shock. Fifteen of 107 patients had primary DENV infections while 92 patients had secondary DENV infections. All patients recruited for the study were of Sri Lankan descent and nationality.
The ARMS-PCR fragments produced for genotyping were sequenced to confirm the accuracy of the system.
Transporter associated with antigen presentation polymorphisms
Table 1 shows the distribution of the TAP1 (333) and TAP2 (379) genotype variants and genotype frequencies. The genotype distribution for both polymorphisms did not defer significantly between severe dengue patients and controls. Genotype combinations of the two TAP polymorphic sites were also investigated [Table 2] without showing any statistically significant results.
Table 1.
Comparison of TAP, IL-10, and TNF-α single nucleotide polymorphisms between DHF patients and controls
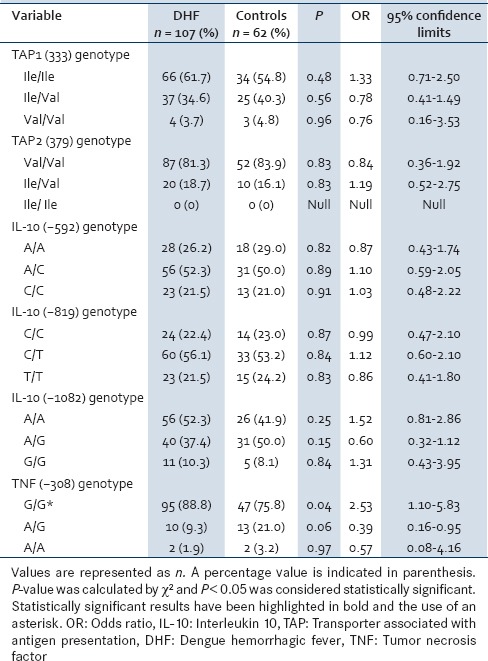
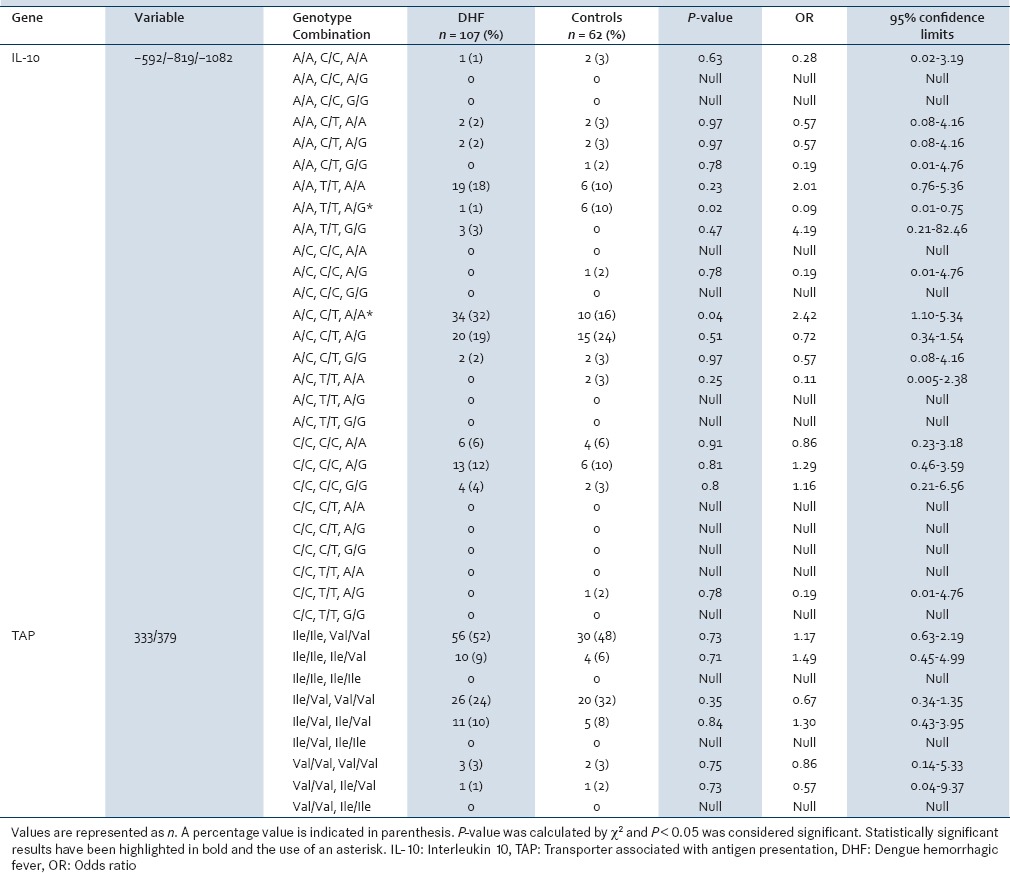
Table 2.
Comparison of IL-10 promoter and TAP genotype combinations between DHF patients and controls
Tumor necrosis factor-α polymorphisms
Table 1 shows the distribution of the TNF-α genotype frequencies for the −308 position for severe dengue patients and controls. Even though a higher frequency of genotype G/G was observed in both the severe dengue patients as well as the control group, there was a statistically significant association of the G/G genotype with severe dengue patients (P = 0.04, OR = 2.53, confidence interval [CI] = 95%).
Interleukin-10 polymorphisms
As shown in Table 1, IL-10 genotype data for positions −592, −819 and −1082 on the promoter region, considered individually did not show statistically significant results for an association with either of the conditions.
Genotype combinations of the polymorphic sites on IL-10 were then evaluated [Table 2] and contrary to the results of the individual positions, a certain IL-10 genotype combination, IL-10 (−592/−819/−1082) CCA/ATA was found to be significantly associated with the development of the severe form of DF (P = 0.04, OR = 2.42, CI = 95%). While the frequency of the genotype combination, ATA/ATA also implied an association with risk of severe dengue the P-value failed to reach significance.
The genotype combination IL-10 (−592/−819/−1082) ATA/ATG was found to have a statistically significant association with the control group with a P = 0.02 and an OR = 0.09 with a 95% CI.
DISCUSSION
Results obtained from the patients with DHF were compared with the healthy control group for all of the polymorphisms investigated. While serotyping was carried out for some of the samples due to sample and resource limitations it could not be completed. Only the DENV1 serotype was found, and this matches with other existing epidemiological data. However, these data were not used as it was beyond the scope of this study.
While results obtained for TAP1 and TAP2, polymorphisms within an Indian population demonstrated a significant association with disease progression.[7] The results obtained for the Sri Lankan population investigated here did not show any significant association with disease severity. Combinations of TAP genotypes were also investigated and compared with the control group but did not show statistically significant results. It has been seen from previous studies that associations for a number of polymorphisms change from population to population and do not remain a constant across all population groups worldwide.[5]
The analysis of the TNF-α −308 polymorphic site demonstrated an association between the G/G polymorphism and the development of severe dengue (P = 0.04, OR = 2.53, CI = 95%) suggesting that this polymorphism may be a risk factor for developing DHF among patients in Sri Lanka. This interpretation has been made cautiously keeping in mind that the P-value is just below the cut-off value of 0.05 and a study with a larger sample size will help to confirm these findings. This is the 1st time that the G/G genotype has been demonstrated as a risk factor in any population. The G/G genotype has been associated with protection from DHF in a population in Venezuela by Fernandez-mestre et al. Studies conducted by Vejbaesya et al. in Vietnam, Garcνa-Trejo et al. in Mexico and a recent study in India did not find any association between TNF-α −308 polymorphisms and dengue disease severity.[6,13,36] The A (−308) allele has been identified as the high expression allele of TNF-α, in comparison to the common allele G.[26] Studies have shown plasma levels of TNF-α are higher in patients with severe dengue.[24] The G allele is reported as the low expression allele and in the Sri Lankan patients studied here, appears to be predominant among patients with severe dengue, contrary to the theory that the presence of the high expression allele explains high TNF-α levels and increased vascular permeability in severe dengue patients.[8] However, what is not yet known is whether variation in the level of this cytokine is a primary cause for severe dengue or just reflects a secondary downstream effect of inflammation.
IL-10 is a major pleiotropic cytokine and high levels of IL-10 have been associated with DHF.[25] As seen in Table 1, IL-10 genotype data for positions (−592), (−819), and (−1082) on the promoter region imply that neither of these single polymorphisms considered individually can define the dengue disease outcome with regard to severity in Sri Lanka. Studying genotype combinations can provide a more accurate understanding of gene effects; therefore, we looked at genotype combinations of IL-10 as shown in Table 2. Our results show that the IL-10 genotype combination, IL-10 (−592/−819/−1082) CCA/ATA was found to be significantly associated with the development of DHF (P = 0.04, OR = 2.42, CI = 95%) suggesting a risk factor to developing severe dengue. Once again the interpretation has been made keeping in mind that the P-value is just below the cut-off value of 0.05. Interestingly, this result was seen among Cuban patients previously in a study conducted by Perez et al. in 2010. The IL-10 (−592/−819/−1082) CCA/ATA genotype combination has been previously related to low expression of IL-10.[37] IL-10 is an important anti-inflammatory cytokine and immune regulator. This result may imply that insufficient expression of IL-10 would result in inefficient immune regulation and resultant severity of disease. However, this does not explain the high levels of IL-10, also repeatedly reported in severe dengue patients previously.[25] The IL-10 genotype combination IL-10 (−592/−819/−1082) ATA/ATG was found to be associated with protection from severe dengue in Sri Lankan patients studied here. This association has not been reported previously in other populations. Other genotype combination associations seen within the Cuban population were not evident among the Sri Lankan patients investigated here. While certain other genetic variants appeared to be associated with risk of severe dengue, the P-values failed to reach significance. It is however, important to note that the IL-10 (−819) and (−592) polymorphisms are in linkage disequilibrium with each other.[37,38]
CONCLUSIONS
The IL-10 genotype combination, IL-10 (−592/−819/−1082) CCA/ATA was significantly associated with the development of DHF suggesting a risk factor to developing severe dengue, the IL-10 genotype combination IL-10 (−592/−819/−1082) ATA/ATG suggests a possibility of protection from DHF and the TNF-α (−308) GG genotype was found to be significantly associated with severe dengue suggesting a significant risk factor in Sri Lankan patients. Analysis of the results obtained here together with previous studies from different populations reveals that associations between host genetics and the clinical outcomes are complex. The results reported here are specific to the Sri Lankan population. Further studies with increased sample size and categorization of patients by infecting serotype may lead to a better understanding of the Sri Lankan situation. It is still unclear how many genes contribute to dengue susceptibility and the extent to which these genes interact with each other to cause severe disease. More data obtained from different population groups are crucial to a complete understanding of genetic associations to dengue infections. These results would help to understand the extent to which dengue pathogenesis could be genetically predicted and possibly help identify risk groups in populations where dengue is endemic.
Financial support and sponsorship
Supported through internal funds of The Centre for Dengue Research, University of Sri Jayewardenepura, Sri Lanka and Genetech Research Institute, Sri Lanka.
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
This work was supported by, The Centre for Dengue Research, University of Sri Jayewardenepura, Sri Lanka and Genetech Research Institute, Sri Lanka.
REFERENCES
- 1.World Health Organization. Impact of Dengue: World Health Organization. 2011. Apr 19, [Last accessed on 2015 Mar 15]. Available from: http://www.who.int/439csr/disease/dengue/impact/en/index.html .
- 2.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, et al. The global distribution and burden of dengue. Nature. 2013;496:504–7. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mathew A, Rothman AL. Understanding the contribution of cellular immunity to dengue disease pathogenesis. Immunol Rev. 2008;225:300–13. doi: 10.1111/j.1600-065X.2008.00678.x. [DOI] [PubMed] [Google Scholar]
- 4.Guzman MG, Halstead SB, Artsob H, Buchy P, Farrar J, Gubler DJ, et al. Dengue: A continuing global threat. Nat Rev Microbiol. 2010;8(12 Suppl):S7–16. doi: 10.1038/nrmicro2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fang X, Hu Z, Shang W, Zhu J, Xu C, Rao X. Genetic polymorphisms of molecules involved in host immune response to dengue virus infection. FEMS Immunol Med Microbiol. 2012;66:134–46. doi: 10.1111/j.1574-695X.2012.00995.x. [DOI] [PubMed] [Google Scholar]
- 6.García-Trejo AR, Falcón-Lezama JA, Juárez-Palma L, Granados J, Zúñiga-Ramos J, Rangel H, et al. Tumor necrosis factor alpha promoter polymorphisms in Mexican patients with dengue fever. Acta Trop. 2011;120:67–71. doi: 10.1016/j.actatropica.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Soundravally R, Hoti SL. Polymorphisms of the TAP1 and 2 gene may influence clinical outcome of primary dengue viral infection. Clin Immunol. 2008;67:618–25. doi: 10.1111/j.1365-3083.2008.02109.x. [DOI] [PubMed] [Google Scholar]
- 8.Perez AB, Sierra B, Garcia G, Aguirre E, Babel N, Alvarez M, et al. Tumor necrosis factor-alpha, transforming growth factor-β1, and interleukin-10 gene polymorphisms: Implication in protection or susceptibility to dengue hemorrhagic fever. Hum Immunol. 2010;71:1135–40. doi: 10.1016/j.humimm.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 9.Khor CC, Chau TN, Pang J, Davila S, Long HT, Ong RT, et al. Genome-wide association study identifies susceptibility loci for dengue shock syndrome at MICB and PLCE1. Nat Genet. 2011;43:1139–41. doi: 10.1038/ng.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whitehorn J, Chau TN, Nguyet NM, Kien DT, Quyen NT, Trung DT, et al. Genetic variants of MICB and PLCE1 and associations with non-severe dengue. PLoS One. 2013;8:e59067. doi: 10.1371/journal.pone.0059067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loke H, Bethell D, Phuong CX, Day N, White N, Farrar J, et al. Susceptibiilty to dengue hemorrhagic fever in Vietnam: Evidence of an association with variation in the vitamin d receptor and Fc gamma receptor IIa genes. Am J Trop Med Hyg. 2002;67:102–6. doi: 10.4269/ajtmh.2002.67.102. [DOI] [PubMed] [Google Scholar]
- 12.Wang L, Chen RF, Liu JW, Lee IK, Lee CP, Kuo HC, et al. DC-SIGN (CD209) Promoter -336 A/G polymorphism is associated with dengue hemorrhagic fever and correlated to DC-SIGN expression and immune augmentation. PLoS Negl Trop Dis. 2011;5:e934. doi: 10.1371/journal.pntd.0000934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vejbaesya S, Luangtrakool P, Luangtrakool K, Kalayanarooj S, Vaughn DW, Endy TP, et al. TNF and LTA gene, allele, and extended HLA haplotype associations with severe dengue virus infection in ethnic Thais. J Infect Dis. 2009;199:1442–8. doi: 10.1086/597422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lan NT, Hirayama K. Host genetic susceptibility to severe dengue infection. Trop Med Health. 2011;39(4 Suppl):73–81. doi: 10.2149/tmh.2011-S08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hershkovitz O, Zilka A, Bar-Ilan A, Abutbul S, Davidson A, Mazzon M, et al. Dengue virus replicon expressing the nonstructural proteins suffices to enhance membrane expression of HLA class I and inhibit lysis by human NK cells. J Virol. 2008;82:7666–76. doi: 10.1128/JVI.02274-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diamond MS. Evasion of innate and adaptive immunity by flaviviruses. Immunol Cell Biol. 2003;81:196–206. doi: 10.1046/j.1440-1711.2003.01157.x. [DOI] [PubMed] [Google Scholar]
- 17.Lobigs M, Blanden RV, Müllbacher A. Flavivirus-induced up-regulation of MHC class I antigens; implications for the induction of CD8+ T-cell-mediated autoimmunity. Immunol Rev. 1996;152:5–19. doi: 10.1111/j.1600-065X.1996.tb00908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sunder SR, Hanumanth SR, Gaddam S, Jonnalagada S, Valluri VL. Association of TAP 1 and 2 gene polymorphisms with human immunodeficiency virus-tuberculosis co-infection. Hum Immunol. 2011;72:908–11. doi: 10.1016/j.humimm.2011.07.304. [DOI] [PubMed] [Google Scholar]
- 19.Cerhan JR, Fredericksen ZS, Novak AJ, Ansell SM, Kay NE, Liebow M, et al. A two-stage evaluation of genetic variation in immune and inflammation genes with risk of non-Hodgkin lymphoma identifies new susceptibility locus in 6p21.3 region. Cancer Epidemiol Biomarkers Prev. 2012;21:1799–806. doi: 10.1158/1055-9965.EPI-12-0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soundravally R, Hoti SL. Immunopathogenesis of dengue hemorrhagic fever and shock syndrome: Role of TAP and HPA gene polymorphism. Hum Immunol. 2007;68:973–9. doi: 10.1016/j.humimm.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 21.Rothman AL. Cellular immunology of sequential dengue virus infection and its role in disease pathogenesis. Curr Top Microbiol Immunol. 2010;338:83–98. doi: 10.1007/978-3-642-02215-9_7. [DOI] [PubMed] [Google Scholar]
- 22.Mustafa AS, Elbishbishi EA, Agarwal R, Chaturvedi UC. Elevated levels of interleukin-13 and IL-18 in patients with dengue hemorrhagic fever. FEMS Immunol Med Microbiol. 2001;30:229–33. doi: 10.1111/j.1574-695X.2001.tb01575.x. [DOI] [PubMed] [Google Scholar]
- 23.Gunther VJ, Putnak R, Eckels KH, Mammen MP, Scherer JM, Lyons A, et al. A human challenge model for dengue infection reveals a possible protective role for sustained interferon gamma levels during the acute phase of illness. Vaccine. 2011;29:3895–3904. doi: 10.1016/j.vaccine.2011.03.038. [DOI] [PubMed] [Google Scholar]
- 24.Kurane I. Dengue hemorrhagic fever with special emphasis on immunopathogenesis. Comp Immunol Microbiol Infect Dis. 2007;30:329–40. doi: 10.1016/j.cimid.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 25.Malavige GN, Gomes L, Alles L, Chang T, Salimi M, Fernando S, et al. Serum IL-10 as a marker of severe dengue infection. BMC Infect Dis. 2013;13:341. doi: 10.1186/1471-2334-13-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson AG, Symons JA, McDowell TL, McDevitt HO, Duff GW. Effects of a polymorphism in the human tumor necrosis factor alpha promoter on transcriptional activation. Proc Natl Acad Sci U S A. 1997;94:3195–9. doi: 10.1073/pnas.94.7.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernández-Mestre MT, Gendzekhadze K, Rivas-Vetencourt P, Layrisse Z. TNF-alpha-308A allele, a possible severity risk factor of hemorrhagic manifestation in dengue fever patients. Tissue Antigens. 2004;64:469–72. doi: 10.1111/j.1399-0039.2004.00304.x. [DOI] [PubMed] [Google Scholar]
- 28.Boonnak K, Dambach KM, Donofrio GC, Tassaneetrithep B, Marovich MA. Cell type specificity and host genetic polymorphisms influence antibody-dependent enhancement of dengue virus infection. J Virol. 2011;85:1671–83. doi: 10.1128/JVI.00220-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.World Health Organization. World Health Organization, Regional Office for South-East Asia. 2011. Comprehensive Guidelines for Prevention and Control of Dengue and Dengue Haemorrhagic Fever. Revised and Expanded Edition. (SEARO Technical Publication Series No. 60). [Google Scholar]
- 30.Vaughn DW, Nisalak A, Solomon T, Kalayanarooj S, Nguyen MD, Kneen R, et al. Rapid serologic diagnosis of dengue virus infection using a commercial capture ELISA that distinguishes primary and secondary infections. Am J Trop Med Hyg. 1999;60:693–8. doi: 10.4269/ajtmh.1999.60.693. [DOI] [PubMed] [Google Scholar]
- 31.Sang CT, Cuzzubbo AJ, Devine PL. Evaluation of a commercial capture enzyme-linked immunosorbent assay for detection of immunoglobulin M and G antibodies produced during dengue infection. Clin Diagn Lab Immunol. 1998;5:7–10. doi: 10.1128/cdli.5.1.7-10.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam AV. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol. 1992;30:545–51. doi: 10.1128/jcm.30.3.545-551.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perrey C, Turner SJ, Pravica V, Howell WM, Hutchinson IV. ARMS-PCR methodologies to determine IL-10, TNF-alpha, TNF-beta and TGF-beta 1 gene polymorphisms. Transpl Immunol. 1999;7:127–8. doi: 10.1016/s0966-3274(99)80030-6. [DOI] [PubMed] [Google Scholar]
- 34.Powis SH, Tonks S, Mockridge I, Kelly AP, Bodmer JG, Trowsdale J. Alleles and haplotypes of the MHC-encoded ABC transporters TAP1 and TAP2. Immunogenetics. 1993;37:373–80. doi: 10.1007/BF00216802. [DOI] [PubMed] [Google Scholar]
- 35.Kingkeow D, McNicholl JM, Maneekarn N, Wongtrakul J, Taechareonkul S, Suriyanon V, et al. Frequencies of IL10 SNP genotypes by multiplex PCR-SSP and their association with viral load and CD4 counts in HIV-1-infected Thais. Asian Pac J Allergy Immunol. 2011;29:94–101. [PubMed] [Google Scholar]
- 36.Alagarasu K, Bachal RV, Tillu H, Mulay AP, Kakade MB, Shah PS, et al. Association of combinations of interleukin-10 and pro-inflammatory cytokine gene polymorphisms with dengue hemorrhagic fever. Cytokine. 2015;74:130–6. doi: 10.1016/j.cyto.2015.03.021. [DOI] [PubMed] [Google Scholar]
- 37.Turner DM, Williams DM, Sankaran D, Lazarus M, Sinnott PJ, Hutchinson IV. An investigation of polymorphism in the interleukin-10 gene promoter. Eur J Immunogenet. 1997;24:1–8. doi: 10.1111/j.1365-2370.1997.tb00001.x. [DOI] [PubMed] [Google Scholar]
- 38.Lazarus R, Klimecki WT, Palmer LJ, Kwiatkowski DJ, Silverman EK, Brown A, et al. Single-nucleotide polymorphisms in the interleukin-10 gene: Differences in frequencies, linkage disequilibrium patterns, and haplotypes in three United States ethnic groups. Genomics. 2002;80(Suppl 2):223–8. doi: 10.1006/geno.2002.6820. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PCR primers and primer protocol
PCR amplification protocols


