Summary
Mitochondrial‐derived peptides (MDP) are encoded by functional short open reading frames in the mitochondrial DNA (mtDNA). These include humanin, and the recently discovered mitochondrial open reading frame of the 12S rRNA‐c (MOTS‐c). Although more research is needed, we suggest that the m.1382A>C polymorphism located in the MOTS‐c encoding mtDNA, which is specific for the Northeast Asian population, may be among the putative biological mechanisms explaining the high longevity of Japanese people.
Keywords: aging, centenarians, longevity gene, longevity regulation, mitochondria, mitochondrial DNA, mitochondrial DNA abnormalities, molecular biology of aging
Background
The number of people aged ≥60 years is expected to almost triple by 2050, with the ‘oldest old’ group (>85 years) being the most rapidly expanding segment in Western societies (Waite, 2004). Among long‐lived individuals, those who reach exceptional longevity (EL, i.e., centenarians (≥100 years) and supercentenarians (SCs, ≥110 years)) are arguably the paradigm of successful aging (Andersen et al., 2012). Several genetic factors might contribute to EL, as suggested by the differences found in the frequency distribution of several genetic variants among centenarians compared with their ethnic‐matched referents of younger ages (Alexe et al., 2007; Ruiz et al., 2012; Garatachea et al., 2014). Factors related to inflammation (Basile et al., 2012), metabolism (Emanuele et al., 2014) or nutrition (Pareja‐Galeano et al., 2015), among others, can also influence the likelihood of reaching EL.
Japan has clearly the longest life expectancy in the world, as well as the highest number of SCs, as we recently reviewed (Santos‐Lozano et al., 2015). Thus, Japanese long‐lived people represent an interesting model to study the biology of EL, and to gain insight into the nature vs. nurture debate.
Mitochondrial haplogroups and EL
Mitochondrial DNA (mtDNA) can influence EL (He et al., 2014). The 16,569‐bp human mtDNA contains 13 genes that codify proteins involved in mitochondrial oxidative phosphorylation (OXPHOS), as well as 2 rRNA and 22 tRNA genes that are necessary for protein synthesis within mitochondria (Mercer et al., 2011). Mitochondria are one of the most important players to understand the aging process at the cellular level as they are both the main source and target of oxidative damage (Gomez‐Cabrera et al., 2012). Mitochondrial dysfunction is in fact a main hallmark of aging, which is partly caused by accumulation of mtDNA damage as we age (Lopez‐Otin et al., 2013). Thus, because mtDNA haplotypes or haplogroups (i.e., characteristic clusters of tightly linked mtDNA polymorphims that form continent‐specific genotypes) might influence individual susceptibility to mtDNA damage, they could also influence EL in a continent‐ or ethnic‐specific manner (Pinos et al., 2012). For instance, the association between mtDNA and EL is controversial in Spanish people, with Pinos et al. reporting no association between mtDNA haplogroups and EL (Pinos et al., 2012) but Dominguez‐Garrido and co‐workers finding that the Caucasian haplogroup J (which would be associated with lower mtDNA damage) might confer a higher chance to attain high longevity (85+years) compared with other haplogroups in Northern Spaniards (Domínguez‐Garrido et al., 2009). On the other hand, although mtDNA haplogroups D4b2b, D4a, and D5 are not associated with type 2 diabetes (Fuku et al., 2007), they are linked with EL in Japanese population (Alexe et al., 2007; Bilal et al., 2008). We also showed that the mtDNA m.1382A>C polymorphism, which is specific for the ancestor haplogroup D4b2, is associated with EL in the Japanese population (Alexe et al., 2007).
Mitochondrial‐derived peptide MOTS‐c
Mitochondrial‐derived peptides (MDP) are encoded by functional short open reading frames in the mtDNA. These include humanin, a 24‐amino acid peptide encoded in the 16S rRNA region with strong cytoprotective actions (Hashimoto et al., 2001) and the recently discovered mitochondrial open reading frame of the 12S rRNA‐c (MOTS‐c), which is a 16‐amino acid peptide that regulates insulin sensitivity and metabolic homeostasis (Lee et al., 2015). We have recently suggested that MOTS‐c might also be involved in the aging process (Alis et al., 2015).
The aforementioned m.1382A>C polymorphism is located in the MOTS‐c encoding mtDNA, that is a 51‐bp short open reading frame in the 12S rRNA region, from positions m.1343 to m.1393 (Table 1). The m.1382A>C variation causes a Lys14Gln replacement in the MOTS‐c peptide equivalent to nucleotide position 1382 of the mtDNA; this is likely to have functional consequences, as the physicochemical difference between the original and the altered aminoacid residues is relatively high, with a Grantham value of 53, that is, above the average value (=50) that differentiates radical from conservative single amino acid replacements (Grantham, 1974). This amino acid replacement is also predicted to have a functional effect with the PROVEAN (PROtein Variation Effect ANalyzer) tool (http://provean.jcvi.org), that is, yielding a score of −4.000, below the specifically predicted cutoff score (=−2.5) above which the variant would be ‘neutral’ (Choi et al., 2012). The m.1382A>C polymorphism is specific for the Northeast Asian population and may be among the putative biological mechanisms explaining the high longevity of Japanese people. Further, MOTS‐c is an important ‘mitokine’, with this term referring to mitochondrial‐derived signals that impact other cells in an endocrine‐like manner (Fig. 1).
Table 1.
The m.1382A>C polymorphism in the mtDNA region 12S rRNA (highlig‐hted in bold) causes Lys14Gln replacement in the mitochondrial‐derived peptide MOTS‐c
| Nucleotide positiona | Nucleotide sequence | Aminoacid (3‐letter code) | Aminoacid position |
|---|---|---|---|
| 1343 | atg | Met | 1 |
| 1346 | agg | Arg | 2 |
| 1349 | tgg | Trp | 3 |
| 1352 | caa | Gln | 4 |
| 1355 | gaa | Glu | 5 |
| 1358 | atg | Met | 6 |
| 1361 | ggc | Gly | 7 |
| 1364 | tac | Tyr | 8 |
| 1367 | att | Ile | 9 |
| 1370 | ttc | Phe | 10 |
| 1373 | tac | Tyr | 11 |
| 1376 | ccc | Pro | 12 |
| 1379 | aga | Arg | 13 |
| 1382 | [A>C]aa | Lys>Gln | 14 |
| 1385 | cta | Leu | 15 |
| 1388 | cga | Arg | 16 |
| 1391 | tag | Stop |
Position number based on the entire mtDNA sequence.
Figure 1.
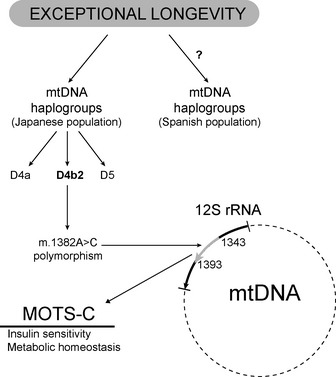
Putative biological link between the novel mitochondrial‐derived peptide MOTS‐c and exceptional longevity through the m.1382A>C mtDNA polymorphism. See text for abbreviations.
Conclusions
We suggest a biological link between MOTS‐c and extended lifespan through the putative endocrine action of this mitokine. Further mechanistic research is needed to determine the functional significance of the m.1382A>C polymorphism and the potential influence of MOTS‐c in the human aging process.
Funding
This work was supported in part by grants from the programs Grant‐in‐Aid for Scientific Research (B) (15H03081 to N.F.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan. Research in the field by A.L. is supported by Fondo de Investigaciones Sanitarias (FIS, grant#PI12/00914) and Fondos FEDER.
Author′s contributions
All authors have: (i) made substantial contributions to conception and design (NF, HP‐G, HZ, RA, AL, NH, YA); (ii) drafted the article (NF, HP‐G, HZ, RA) or revised it for critically for important intellectual content (AL, NH, YA); and (iii) gave final approval of the version to be published (NF, AL, NH, YA).
Conflict of interest
None declared.
References
- Alexe G, Fuku N, Bilal E, Ueno H, Nishigaki Y, Fujita Y, Ito M, Arai Y, Hirose N, Bhanot G, Tanaka M (2007) Enrichment of longevity phenotype in mtDNA haplogroups D4b2b, D4a, and D5 in the Japanese population. Hum. Genet. 121, 347–356. [DOI] [PubMed] [Google Scholar]
- Alis R, Lucia A, Blesa JR, Sanchis‐Gomar F (2015) The role of mitochondrial derived peptides (MDPs) in metabolism. J. Cell. Physiol. [DOI] [PubMed] [Google Scholar]
- Andersen SL, Sebastiani P, Dworkis DA, Feldman L, Perls TT (2012) Health span approximates life span among many supercentenarians: compression of morbidity at the approximate limit of life span. J. Gerontol. A Biol. Sci. Med. Sci. 67, 395–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basile G, Paffumi I, D'Angelo AG, Figliomeni P, Cucinotta MD, Pace E, Ferraro M, Saitta S, Lasco A, Gangemi S (2012) Healthy centenarians show high levels of circulating interleukin‐22 (IL‐22). Arch. Gerontol. Geriatr. 54, 459–461. [DOI] [PubMed] [Google Scholar]
- Bilal E, Rabadan R, Alexe G, Fuku N, Ueno H, Nishigaki Y, Fujita Y, Ito M, Arai Y, Hirose N, Ruckenstein A, Bhanot G, Tanaka M (2008) Mitochondrial DNA haplogroup D4a is a marker for extreme longevity in Japan. PLoS ONE 3, e2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Sims GE, Murphy S, Miller JR, Chan AP (2012) Predicting the functional effect of amino acid substitutions and indels. PLoS ONE 7, e46688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez‐Garrido E, Martínez‐Redondo D, Martín‐Ruiz C, Gómez‐Durán A, Ruiz‐Pesini E, Madero P, Tamparillas M, Montoya J, von Zglinicki T, Díez‐Sánchez C, López‐Pérez MJ (2009) Association of mitochondrial haplogroup J and mtDNA oxidative damage in two different North Spain elderly populations. Biogerontology 10, 435–442. [DOI] [PubMed] [Google Scholar]
- Emanuele E, Minoretti P, Pareja‐Galeano H, Sanchis‐Gomar F, Garatachea N, Lucia A (2014) Serum irisin levels, precocious myocardial infarction, and healthy exceptional longevity. Am. J. Med. 127, 888–890. [DOI] [PubMed] [Google Scholar]
- Fuku N, Park KS, Yamada Y, Nishigaki Y, Cho YM, Matsuo H, Segawa T, Watanabe S, Kato K, Yokoi K, Nozawa Y, Lee HK, Tanaka M (2007) Mitochondrial haplogroup N9a confers resistance against type 2 diabetes in Asians. Am. J. Hum. Genet. 80, 407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garatachea N, Fuku N, He ZH, Tian Y, Arai Y, Abe Y, Murakami H, Miyachi M, Yvert T, Venturini L, Santiago C, Santos‐Lozano A, Rodriguez G, Ricevuti G, Pareja‐Galeano H, Sanchis‐Gomar F, Emanuele E, Hirose N, Lucia A (2014) PTK2 rs7460 and rs7843014 polymorphisms and exceptional longevity: a functional replication study. Rejuvenation Res. 17, 430–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez‐Cabrera MC, Sanchis‐Gomar F, Garcia‐Valles R, Pareja‐Galeano H, Gambini J, Borras C, Vina J (2012) Mitochondria as sources and targets of damage in cellular aging. Clin. Chem. Lab. Med. 50, 1287–1295. [DOI] [PubMed] [Google Scholar]
- Grantham R (1974) Amino acid difference formula to help explain protein evolution. Science 185, 862–864. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y, Niikura T, Tajima H, Yasukawa T, Sudo H, Ito Y, Kita Y, Kawasumi M, Kouyama K, Doyu M, Sobue G, Koide T, Tsuji S, Lang J, Kurokawa K, Nishimoto I (2001) A rescue factor abolishing neuronal cell death by a wide spectrum of familial Alzheimer's disease genes and Abeta. Proc. Natl Acad. Sci. USA 98, 6336–6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YH, Lu X, Wu H, Cai WW, Yang LQ, Xu LY, Sun HP, Kong QP (2014) Mitochondrial DNA content contributes to healthy aging in Chinese: a study from nonagenarians and centenarians. Neurobiol. Aging 35, 1779 e1771‐1774. [DOI] [PubMed] [Google Scholar]
- Lee C, Zeng J, Drew BG, Sallam T, Martin‐Montalvo A, Wan J, Kim SJ, Mehta H, Hevener AL, de Cabo R, Cohen P (2015) The mitochondrial‐derived peptide MOTS‐c promotes metabolic homeostasis and reduces obesity and insulin resistance. Cell Metab. 21, 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez‐Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G (2013) The hallmarks of aging. Cell 153, 1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AM, Haugen E, Bracken CP, Rackham O, Stamatoyannopoulos JA, Filipovska A, Mattick JS (2011) The human mitochondrial transcriptome. Cell 146, 645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pareja‐Galeano H, Sanchis‐Gomar F, Santos‐Lozano A, Garatachea N, Fiuza‐Luces C, Lucia A, Emanuele E (2015) Serum eicosapentaenoic acid to arachidonic acid ratio is associated with cardio‐healthy exceptional longevity. Int. J. Cardiol. 184, 655–656. [DOI] [PubMed] [Google Scholar]
- Pinos T, Nogales‐Gadea G, Ruiz JR, Rodriguez‐Romo G, Santiago‐Dorrego C, Fiuza‐Luces C, Gomez‐Gallego F, Cano‐Nieto A, Garatachea N, Moran M, Angel Martin M, Arenas J, Andreu AL, Lucia A (2012) Are mitochondrial haplogroups associated with extreme longevity? A study on a Spanish cohort. Age (Dordr). 34, 227–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz JR, Fiuza‐Luces C, Buxens A, Cano‐Nieto A, Gomez‐Gallego F, Santiago C, Rodriguez‐Romo G, Garatachea N, Lao JI, Moran M, Lucia A (2012) Are centenarians genetically predisposed to lower disease risk? Age (Dordr). 34, 1269–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos‐Lozano A, Sanchis‐Gomar F, Pareja‐Galeano H, Fiuza‐Luces C, Emanuele E, Lucia A, Garatachea N (2015) Where are supercentenarians located? A worldwide demographic study Rejuvenation Res. 18, 14–19. [DOI] [PubMed] [Google Scholar]
- Waite LJ (2004) The demographic faces of the elderly. Popul. Dev. Rev. 30, 3–16. [PMC free article] [PubMed] [Google Scholar]


