Abstract
This study shows that the prenylated C‐terminus of RPGR can bind to PDE6δ with high affinity, suggesting two distinct binding sites of the RPGR/PDE6δ complex. The serine residue at the −3 position relative to the prenylated cysteine seems to play a key role in defining the selectivity of PDE6δ towards ciliary prenylated cargo.
Subject Categories: Membrane & Intracellular Transport, Molecular Biology of Disease, Structural Biology
Lee and Seo propose in their article 1 that RPGR binds to PDE6δ not with the N‐terminal RCC1‐like propeller domain but solely with the C‐terminus. They show, using an immunoprecipitation experiment, that FLAG‐tagged fragments missing the C‐terminal CaaX motif of RPGR fail to co‐immunoprecipitate together with myc‐tagged PDE6δ. We have previously shown that the N‐terminal 400 residues of RPGR form a stable RCC1‐like propeller domain and that this protein forms a complex with PDE6δ. This can be demonstrated by (untagged) pull‐down and gel permeation chromatography experiments. Additionally, the equilibrium dissociation constant was determined to be 500 nM by fluorescence polarization which also agrees with previous results published by Linari et al in which they report the affinity of RPGR (aa 1–392), using surface plasmon resonance, to PDE6δ to be 100 nM 2, 3. Finally, we have solved the structure of the complex PDE6δ–RPGR (aa 8–368) by X‐ray crystallography and verified the interaction interface by mutational analysis 3. Since immunoprecipitation experiments reflect dissociation kinetics rates rather than equilibrium dissociation, which are in turn very much dependent on many aspects of the experiments (see discussion below), we feel confident about the results by Wätzlich et al 3.
Lee and Seo then show that RPGR interacts with the C‐terminal end of RPGR which contains a CaaX motif. This finding may not be too surprising since we and others have previously shown that PDE6δ is a general prenyl‐binding protein that is required to shuttle lipidated proteins between membranes and that the binding is released by Arl2/3•GTP 4, 5, 6, 7. Our earlier structural studies revealed molecular details of the binding of farnesylated peptides/proteins to PDE6δ, showing how the farnesyl moiety is inserted into the hydrophobic cavity of PDE6δ and that in addition to the farnesyl group, only the last three residues of the farnesylated cargo make contacts with the binding pocket of PDE6δ 5. Zhang et al have shown that geranylgeranylated proteins such as rhodopsin kinase (GRK7) which carries a CaaX motif signifying geranylgeranylation, as does the CTIL motif of RPGR, bind to PDE6δ. Also localization of geranylgeranylated ciliary proteins such as the alpha' subunit of cone phosphodiesterase 6 (PDE6α') is dependent on PDE6δ 6, 7. However, so far no structural information exists that would explain how the PDE6δ hydrophobic pocket is able to accommodate an additional five carbon atoms (prenyl group) of a geranylgeranylated peptide. In order to investigate this structural plasticity, we solved the crystal structure of PDE6δ in complex with a geranylgeranylated and carboxy‐methylated peptide derived from the C‐terminus of PDE6α' at resolution of 2.1 Å. This peptide resembles the RPGR C‐terminal sequence.
Solid phase synthesis of the PDE6α' peptide (DDKKSKT‐C(gerger)‐OMe) was carried out on an Activotec P‐11 peptide synthesizer, on H‐Cys(Trt)‐ClTrt resin, using N‐Fmoc amino acids and HCTU as the coupling reagent (Merck Chemicals). The N‐terminal Asp was incorporated as Boc‐Asp(OtBu)‐OH. Methylation was as previously described 8. Following chain assembly and methylation, the peptidyl resin was cleaved, purified, geranylgeranylated, and purified as previously described 9.
The geranylgeranylated peptide from PDE6α' was dissolved in DMSO and mixed with PDE6δ at 1:1 molar ratio with final concentration of 500 μM. Crystals appeared in the PEGs II suite from Qiagen in conditions containing 0.1 M HEPES (pH 7.5), 0.2 M Li2SO4, 25% PEG4000, and 0.1 M NaOAc. X‐ray data were collected using the beamline X10SA at the Swiss Light Source. The data were processed using the XDS program, and the structure was solved by molecular replacement using the program Molrep and PDE6δ from the PDE6δ‐farnesylated RheB complex (PDB code: 3T5G) as a search model. Model building and structure refinement were carried out using WinCoot and Refmac5 programs, respectively. The final refinement and data collection statistics are summarized in Table 1.
Table 1.
Data collection and refinement statistics (molecular replacement)
| Gerger‐PDE6α'‐peptide•PDE6δ | |
|---|---|
| Data collection | |
| Space group | C222 1 |
| Cell dimensions | |
| a, b, c (Å) | 77.71, 81.43, 118.53 |
| α, β, γ (°) | 90.00, 90.00, 90.00 |
| Resolution (Å) | 29.63–2.10 |
| R sym or R merge | 5.0 (16.3) |
| I / σI | 23.46 (10.24) |
| Completeness (%) | 99.6 (100.0) |
| Redundancy | 5.29 (5.35) |
| Refinement | |
| Resolution (Å) | 2.10 |
| No. reflections | 21,194 |
| R work / R free | 20.05/26.56 |
| No. atoms | |
| Protein | 2,425 |
| Ligand/ion | 118 |
| Water | 146 |
| B‐factors | |
| Protein | 31.85 |
| Ligand/ion | 37.58 |
| Water | 41.47 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.018 |
| Bond angles (°) | 1.925 |
Numbers in parentheses represent the highest resolution bin.
The overall structure shows insertion of the geranylgeranyl moiety into the hydrophobic cavity of PDE6δ (Fig 1A, middle panel). Superimposition of the geranylgeranylated peptide with that of the farnesylated RheB (PDB code: 3T5G) shows that the prenyl groups and the first three residues proximal to the carboxy terminus superimpose well and that the extra prenyl group of the geranylgeranyl moiety inserts deeper into the cavity of PDE6δ (Fig 1A, middle panel). The deeper insertion is facilitated by structural rearrangement of the residues Phe133 and Leu17, which undergo a side chain shift enabling the insertion of the extra 5 carbon atoms (Fig 1A, left panel). Interestingly, the side chain of the serine at the −3 position relative to the geranylgeranylated cysteine in the PDE6α' peptide forms a hydrogen bond with the side chain of glutamic acid (Glu88) from PDE6δ (Fig 1A, right panel). In comparison, the side chain of lysine at the −3 position in RheB points away from the binding pocket of PDE6δ. Thus, we expected that this difference in the contact pattern of the side chains, in addition to the increased hydrophobic contact by the additional prenyl group of the geranylgeranyl moiety, might lead to a higher affinity of the geranylgeranylated PDE6α' peptide relative to the farnesylated RheB peptide.
Figure 1. PDE6δ interacts with C‐terminal geranylgeranylated peptides with high affinity.
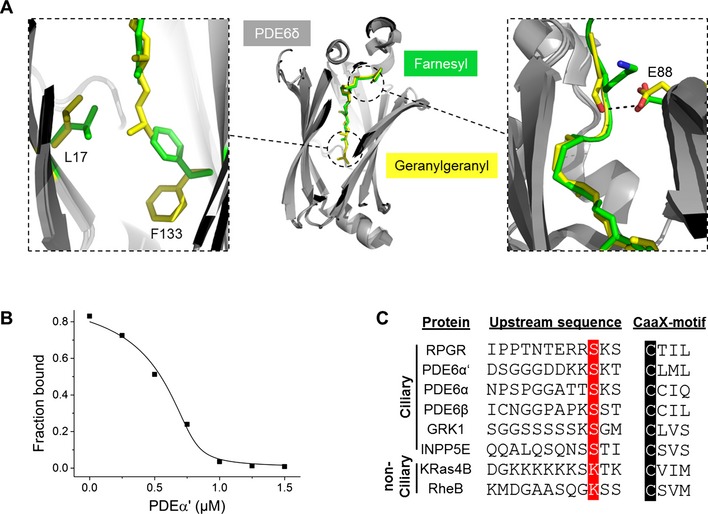
(A) Middle panel: crystal structure of PDE6δ (gray) in complex with geranylgeranylated and carboxy‐methylated PDE6α' peptide (yellow) superimposed with the structure of PDE6δ in complex with RheB (PDB code: 3T5G) showing the overlap with the farnesyl group (green). Left panel: side chain shift of Phe133 and Leu17 from PDE6δ enables the deeper insertion of the additional prenyl group of the geranylgeranyl moiety. Right panel: difference in the contact pattern of the residue at the −3 position relative to the prenylated cysteine. The PDB accession code for Gerger‐PDE6α'‐peptide in complex with PDE6δ is 5E8F. (B) Titrations of a preformed complex between 0.2 μM FITC‐labeled RheB peptide and 0.8 μM PDE6δ with increasing concentrations of geranylgeranylated PDE6α' peptide. Titration data were fitted with a competition model derived from the law of mass action as previously described 17. (C) Sequence alignment of C‐terminal part of PDE6δ ciliary and non‐ciliary interacting partners. The prenylated cysteine is highlighted in black; the serine at the −3 position upstream of the cysteine is highlighted in red.
To investigate this, we determined the binding affinity by titrating increasing concentrations of the geranylgeranylated PDE6α' peptide to a preformed complex between fluorescently labeled RheB peptide and PDE6δ in a fluorescence polarization‐based experiment. As shown in Fig 1B, the geranylgeranylated PDE6α' peptide binds to PDE6δ with an affinity in the low nanomolar range (K d = 2.3 ± 0.9 nM), which is almost 150 times higher than that of the farnesylated RheB peptide previously reported 5, 10. This difference in the affinity reveals the importance of the additional hydrophobic contact of the geranylgeranyl moiety as compared to the farnesyl moiety as well as the importance of the serine residue at the −3 position relative to the prenylated cysteine. This observations support the mutational analysis by Lee and Seo. Interestingly, the residue at the −3 position seems to be conserved in several PDE6δ ciliary interacting partners (Fig 1C), suggesting a major difference in the binding affinity with PDE6δ between ciliary (high affinity) and non‐ciliary (low affinity) cargos. PDE6δ is a shuttle factor of prenylated proteins between different membrane compartments, which undergoes continuous rounds of cargo loading and release 11. Thus, one would expect to have equilibrium of cargo‐loaded and cargo‐free PDE6δ in the cell. Moreover, the cellular content of PDE6δ is higher than that of the common low‐affinity cytoplasmic cargo molecules such as RheB and KRas 4. Finally, the ratio of cilium/cytoplasmic size is extremely small (~1:10,000); thus, one would assume that the total amount of cargo destined for the cilium is much lower than cytoplasmic cargo. In conclusion, we would expect that both low‐ and high‐affinity cargos can be shuttled by PDE6δ.
Up to date all structures of prenylated peptides or proteins in complex with PDE6δ including the new structure presented here in this study show that only the last three residues upstream of the prenylated cysteine are located inside the binding pocket of PDE6δ 5. Moreover, the last three residues of RPGR are almost identical with that of PDE6α' with only the substitution of threonine by serine at the −1 position (Fig 1C). Taking these two aspects into account, we would expect that the affinity of the C‐terminal part of RPGR would also fall into the low nanomolar range.
Thus, we fully support the notion by Lee and Seo that the geranylgeranylated C‐terminus of RPGR is fully capable of interacting with PDE6δ. Nevertheless, based on our and others' biochemical data in addition to our structural data, we do not exclude that the RPGR N‐terminus contacts PDE6δ. Furthermore, the structure of PDE6δ bound to the RPGR propeller shows the prenyl‐binding site to be in the open conformation and thus able to bind cargo 3. The estimated low nanomolar affinity of the C‐terminal end as compared to the 500 nM affinity of the N‐terminus could well explain the findings of the immunoprecipitation experiments by Lee and Seo. Whether PDE6δ binds the N‐ and C‐terminal ends simultaneously cannot be inferred from the present experiments, but there are precedents of other prenyl‐binding proteins such as RhoGDI or RabGDI which have a second binding site remote from the lipid‐binding site of their cargo 12, 13.
Our new data, in agreement with Lee and Seo, suggest that RPGR is also a cargo of PDE6δ. Nevertheless, the N‐terminal RCC1 domain stabilizes the open conformation of PDE6δ, as shown by our previous structure 3. Thus, in addition to being transported by PDE6δ, RPGR can act as a docking scaffold for other prenylated cargo complexed to PDE6δ. Additionally, the higher membrane binding affinity of the geranylgeranyl moiety as compared with the farnesyl moiety 14 might result in a slow off‐rate of the geranylgeranylated RPGR from membranes, retarding its extraction and shuttling by PDE6δ. One way to facilitate extraction from membranes would be a two‐step binding where PDE6δ binds first to the N‐terminus of RPGR followed by the binding to the geranylgeranylated C‐terminus; a similar scenario is observed by RabGDIs 15. Taken together, the interaction mode of PDE6δ with RPGR has the potential to mimic that of other prenyl‐binding proteins such as RhoGDI or RabGDI by having two distinct binding sites.
Additionally, the serine residue at the −3 position relative to the prenylated cysteine might play a role in increasing the binding affinity of PDE6δ with prenylated ciliary cargo. It is also possible that the presence of high concentration of Arl3•GTP in the ciliary compartment releases the prenylated C‐terminus. The release of the prenylated C‐terminus of RPGR by Arl3•GTP would be the first step to detach RPGR from its complex with PDE6δ followed by the specific interaction of RPGR with RPGRIP1 at the transition zone resulting in the complete dissociation of RPGR–PDE6δ complex. This assumption is supported by the fact that a ternary complex among RPGR, PDE6δ, and RPGRIP1 is structurally and biochemically not feasible 16.
Acknowledgements
The funding was supported by the Sonderforschungsbereich‐DFG (SFB 642). We would like to thank the staff of the beamline X10SA at the Swiss Light Source for their support and A. Porfetye, Dr. A. Richters, D. Bier and J. John for help with the data collection.
Reply to: J‐J Lee & S Seo
References
- 1. Lee JJ, Seo S (2015) EMBO Rep 16: 1581–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Linari M, Ueffing M, Manson F et al (1999) Proc Natl Acad Sci USA 96: 1315–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Watzlich D, Vetter I, Gotthardt K et al (2013) EMBO Rep 14: 465–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chandra A, Grecco HE, Pisupati V et al (2012) Nat Cell Biol 14: 148–158 [DOI] [PubMed] [Google Scholar]
- 5. Ismail SA, Chen YX, Rusinova A et al (2011) Nat Chem Biol 7: 942–949 [DOI] [PubMed] [Google Scholar]
- 6. Zhang H, Constantine R, Frederick JM et al (2012) Vision Res 75: 19–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang H, Liu XH, Zhang K et al (2004) J Biol Chem 279: 407–413 [DOI] [PubMed] [Google Scholar]
- 8. O'Reilly N, Charbin A, Lopez‐Serra L et al (2012) Yeast 29: 233–240 [DOI] [PubMed] [Google Scholar]
- 9. Riou P, Kjaer S, Garg R et al (2013) Cell 153: 640–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen YX, Koch S, Uhlenbrock K et al (2010) Angew Chem 49: 6090–6095 [DOI] [PubMed] [Google Scholar]
- 11. Schmick M, Vartak N, Papke B et al (2014) Cell 157: 459–471 [DOI] [PubMed] [Google Scholar]
- 12. Hoffman GR, Nassar N, Cerione RA (2000) Cell 100: 345–356 [DOI] [PubMed] [Google Scholar]
- 13. Rak A, Pylypenko O, Durek T et al (2003) Science 302: 646–650 [DOI] [PubMed] [Google Scholar]
- 14. Bhatnagar RS, Gordon JI (1997) Trends Cell Biol 7: 14–20 [DOI] [PubMed] [Google Scholar]
- 15. Pylypenko O, Rak A, Durek T et al (2006) EMBO J 25: 13–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Remans K, Burger M, Vetter IR et al (2014) Cell Rep 8: 1–9 [DOI] [PubMed] [Google Scholar]
- 17. Zimmermann G, Papke B, Ismail S et al (2013) Nature 497: 638–642 [DOI] [PubMed] [Google Scholar]


