Abstract
Contrary to the prior studies, PDE6D was found to bind more preferentially to the C‐terminal prenyl moiety than the N‐terminal region of RPGR in mammalian cells. This RPGR–PDE6D interaction is also dependent on the amino acid adjacent to the prenylation motif, providing an explanation as to how the specificity of PDE6D and prenylated protein interaction is determined.

Subject Categories: Membrane & Intracellular Transport, Molecular Biology of Disease
Ciliary dysfunction underlies multiple human genetic diseases, and mechanisms of protein trafficking to cilia are an area of active investigation. PDE6D (also known as PDEδ and PrBP) is a prenyl‐binding protein involved in ciliary targeting of prenylated proteins 1, 2, 3, 4. Previous studies uncovered that PDE6D interacts with RPGR 5, 6, mutations of which cause retinitis pigmentosa 7, 8, 9, 10. More recently, the structure of the PDE6D and RPGR (the N‐terminal half) complex was determined and RPGR was proposed as a scaffold protein that recruits cargo‐loaded PDE6D to the ciliary base 6. Here, we show an alternative mode of interaction between RPGR and PDE6D.
Two main isoforms of RPGR (RPGRex1–19 and RPGRORF15) are expressed in vertebrates 8, 11. Of these, RPGRORF15 appears to be expressed specifically in photoreceptor cells, while RPGRex1–19 is expressed ubiquitously. The RPGRex1–19 isoform (hereafter RPGR) has the CaaX prenylation motif and is geranylgeranylated at its C‐terminus. The interaction between RPGR and PDE6D was initially discovered by a yeast two‐hybrid screen and further verified by an in vitro GST pull‐down assay 5, 6. In these studies, PDE6D was found to interact with the RCC1‐like domain (RLD) within the N‐terminal half of RPGR. While we were conducting our research based on these studies, we found that the interaction between RPGR and PDE6D in mammalian cells is very different from what was observed in the initial studies, which were conducted in yeast and in vitro using purified proteins. Therefore, we set out to investigate the RPGR–PDE6D interaction in more detail in mammalian cells using co‐immunoprecipitation (co‐IP) assays.
Various RPGR mutant variants with N‐terminal FLAG tags were transfected with Myc‐tagged PDE6D into HEK293T cells, and lysates were subjected to IP with anti‐FLAG antibodies. As shown in Fig 1A (lane 2), the full‐length RPGR bound to PDE6D. However, contrary to the previous studies 5, 6, we were not able to detect any interaction between the N‐terminal region of RPGR and PDE6D (lanes 3 and 4). Deletion of the CaaX prenylation motif (aa 812–815) was sufficient to disrupt the PDE6D–RPGR interaction (lane 5), indicating that prenylation is essential for PDE6D binding in mammalian cells. Furthermore, a strong interaction was detected with the C‐terminal half of RPGR (lane 6). We further narrowed down the PDE6D interaction domain using GFP‐tagged RPGR‐mutant variants (Fig 1B). In this experiment, we found that the C‐terminal 9 residues (aa 807–815), which contain the CaaX motif, were sufficient to bind to PDE6D (lane 4). Substitution of the Cys residue (aa 812) within the CaaX motif to Ser completely abolished the RPGR–PDE6D interaction (lane 9). These data demonstrate that in mammalian cells, PDE6D binds to the C‐terminus of RPGR through its prenylation site.
Figure 1. PDE6D binds to the C‐terminus of RPGR through the prenylation site.
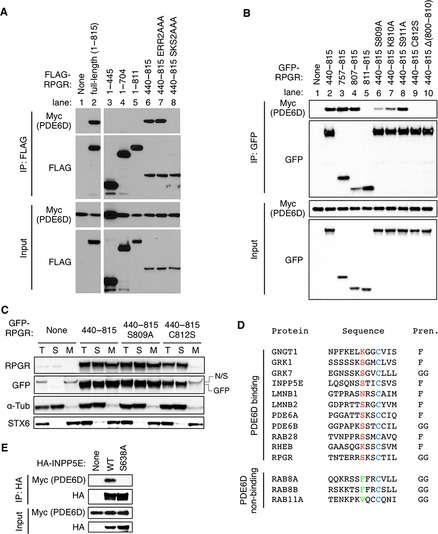
(A) PDE6D binds to the C‐terminal portion of RPGR in mammalian cells. Myc‐PDE6D and FLAG‐tagged RPGR mutant variants were transfected in HEK293T cells, and lysates were subjected to immunoprecipitation (IP) using anti‐FLAG antibodies. Numbers indicate the amino acids included in the constructs. ERR residues are at aa 806–808 and SKS at aa 809–811. (B) PDE6D binds to the C‐terminus of RPGR through the prenylation site. Various RPGR mutants (with a GFP tag) were transfected with Myc‐PDE6D in HEK293T cells, and lysates were subjected to co‐IP using anti‐GFP antibodies. (C) Distribution of RPGR mutant variants between soluble (S) and membrane (M) fractions. Indicated RPGR mutant variants were transfected in HEK293T cells, and cellular homogenates were subjected to ultracentrifugation at 127,000 g for 1 h after removing nuclei and unbroken cells. Fractions were subjected to SDS‐PAGE and immunoblotting with anti‐RPGR and anti‐GFP antibodies. α‐tubulin and STX6 were used as markers of soluble and membrane fractions, respectively. T: total cellular homogenate, N/S: non‐specific. (D) C‐terminal amino acid sequences of PDE6D‐binding and non‐binding prenylated proteins. Amino acid sequences of human proteins are shown. The Cys residue in the CaaX prenylation motif is in blue. Amino acids at the −3 position of known PDE6D‐binding and non‐binding prenylated proteins are shown in red and green, respectively. Types of prenylation (Pren.) are shown on the right: F, farnesylation; GG, geranylgeranylation. (E) The Ser residue at the −3 position in INPP5E is essential for PDE6D binding. Wild‐type (WT) and S638A mutant INPP5E were transfected with Myc‐PDE6D, and lysates were subjected to IP using anti‐HA antibodies.
While we were mapping the PDE6D interaction domain, we noticed that amino acids adjacent to the prenylation site in RPGR contribute to PDE6D binding. For example, deletion of 11 amino acids (Δ(800–810)) or substitution of three amino acids (SKS to AAA at 809–811) near the CaaX motif abrogated the RPGR–PDE6D interaction (Fig 1A lane 8; 1B lane 10). In particular, substitution of the Ser residue (aa 809) at the −3 position from the CaaX motif to Ala greatly reduced the ability of RPGR to bind PDE6D. The inability of the S809A mutant in PDE6D binding is unlikely due to impaired prenylation because its partitioning between the soluble and the membrane fractions was not significantly different from that of wild type, while the C812S mutant, which cannot be prenylated, was exclusively found in the soluble fraction (Fig 1C). When we compared the amino acid sequences of verified PDE6D‐binding and non‐binding prenylated proteins 1, 3 (and references therein), Ser was found to be the most preferred amino acid at the −3 position among PDE6D‐binding prenylated proteins (Fig 1D). Polar or positively charged residues (Asn and Lys) were also found. In contrast, hydrophobic residues were found in RAB8A, RAB8B, and RAB11A, which are geranylgeranylated but do not bind to PDE6D 3. To test whether the Ser residue at the −3 position plays an important role in PDE6D binding in other proteins, we mutated the Ser residue at the −3 position in INPP5E, a prenylated protein that binds to PDE6D and localizes to cilia 3, 4, to a hydrophobic amino acid Ala and examined its interaction with PDE6D. Indeed, substitution of the Ser to Ala abolished the interaction of INPP5E with PDE6D (Fig 1E). These findings indicate that amino acids at the −3 position in cargos play a critical role in PDE6D binding.
In summary, we found that in mammalian cells, PDE6D binds more preferentially to the C‐terminal prenylation site than the N‐terminal RLD in RPGR. Although we were not able to detect the interaction between PDE6D and the RLD of RPGR in our co‐IP assays, PDE6D may bind to the RLD with a relatively low affinity or in certain physiological conditions. Our data suggest that RPGR could be a cargo of PDE6D in addition to acting as a scaffold protein as previously proposed 6. Finally, our work indicates that the amino acids at the −3 position from the CaaX prenylation motif play a critical role in cargo–PDE6D interactions. This finding provides an explanation as to why certain prenylated proteins bind to PDE6D while others do not.
Comment on: D Wätzlich et al (May 2013)
See reply: EK Fansa et al
References
- 1. Baehr W (2014) Invest Ophthalmol Vis Sci 55: 8653–8666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhang H, Li S, Doan T et al (2007) Proc Natl Acad Sci USA 104: 8857–8862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Humbert MC, Weihbrecht K, Searby CC et al (2012) Proc Natl Acad Sci USA 109: 19691–19696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Thomas S, Wright KJ, Le Corre S et al (2014) Hum Mutat 35: 137–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Linari M, Ueffing M, Manson F et al (1999) Proc Natl Acad Sci USA 96: 1315–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Watzlich D, Vetter I, Gotthardt K et al (2013) EMBO Rep 14: 465–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Meindl A, Dry K, Herrmann K et al (1996) Nat Genet 13: 35–42 [DOI] [PubMed] [Google Scholar]
- 8. Kirschner R, Rosenberg T, Schultz‐Heienbrok R et al (1999) Hum Mol Genet 8: 1571–1578 [DOI] [PubMed] [Google Scholar]
- 9. Vervoort R, Lennon A, Bird AC et al (2000) Nat Genet 25: 462–466 [DOI] [PubMed] [Google Scholar]
- 10. Breuer DK, Yashar BM, Filippova E et al (2002) Am J Hum Genet 70: 1545–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hong DH, Li T (2002) Invest Ophthalmol Vis Sci 43: 3373–3382 [PubMed] [Google Scholar]


