Summary
We report on the X-ray structure of three 100-amino acid mbt repeats in h-l(3)mbt, a polycomb group protein involved in transcriptional repression, whose gene is located in a region of chromosome 20 associated with hematopoietic malignancies. Interdigitation between the extended arms and cores of the mbt repeats results in a three-leaved propeller-like architecture, containing a central cavity. We have identified one ligand binding pocket per mbt repeat, which accommodates either the morphilino ring of MES or the proline ring of the C-terminal peptide segment, within a cavity lined by aromatic amino acids. Strikingly, phenotypic alterations resulting from point mutations or deletions in the mbt repeats of the related Drosophila SCM protein are clustered in and around the ligand binding pocket.
Introduction
Deletion of the long arm of chromosome 20 (20q) is the second most common primary structural abnormality seen in hematological malignancies and is found in polycythemia vera, chronic myelogenous leukemia (CML) blast crisis (Mori et al., 1997), and, to a lesser extent, in acute myelogenous leukemia (AML) and myelodysplastic syndromes (MDS) (Kurtin et al., 1996). Inactivation of a putative suppressor gene(s) on 20q, which disrupts normal hematopoietic stem cell regulation, has been proposed to explain the pathogenesis of hematologic malignancies that contain 20q deletions (White et al., 1994). However, recently, it has been proposed that haploinsufficiency of an essential gene product could also underlie the pathogenesis of these disorders (Asimakopoulos and Green, 1996). Several chromosome 20 deletion mapping studies with primary patient material have identified common deleted regions (CDR) of 2.6 Mb in MPD patients and 2.7 Mb in MDS and AML patients, with a 1.7 Mb region of overlap (Bench et al., 2000). We used classical tumor suppressor gene investigative methods and a large number of myeloid leukemia cell lines (MacGrogan et al., 2001) to identify homozygous deletions or rearrangements within the CDR. We identified two cell lines with translocations within the CDR (MacGrogan et al., 2001) and performed exon trapping using three P1 artificial chromosome (PAC) clones that encompassed the translocation breakpoint, allowing us to isolate exons that encode portions of a human gene highly homologous to the Drosophila l(3)mbt (lethal malignant brain tumor) gene (Koga et al., 1999). The Drosophila l(3)mbt gene is a tumor suppressor gene, and it encodes a member of the PcG family of transcriptional regulators (Gateff et al., 1993; Wismar et al., 1995). Recessive or temperature-sensitive mutations of the Drosophila l(3)mbt gene are associated with malignant transformation of adult optic neuroblasts and ganglion mother cells in the larval brain (Wismar et al., 1995).
The human l(3)mbt is an ~110 kDa protein and, like the Drosophila l(3)mbt protein (Wismar et al., 1995), contains three readily identifiable structural domains: three central malignant brain tumor (mbt) repeats that are each ~100 amino acid residues in length, an ~20-amino acid stretch containing an unusual C2HC zinc finger, and a C-terminal α-helical SPM dimerization domain that is 60 amino acid residues in length (Bornemann et al., 1996; Peterson et al., 1997). Recent studies have shown that human l(3)mbt is a transcriptional repressor and that its activity is largely dependent on the presence of a region containing three mbt repeats (Boccuni et al., 2003). Overexpression of h-l(3)mbt leads to defective chromosome segregation and multinucleated cell formation, suggesting a role in normal cell division (Koga et al., 1999).
The mbt modules are found in several other PcG proteins and range from two to four repeats. Sex comb on midleg (SCM) protein from Drosophila and its human SCMH1 and SCML2 counterparts have been found to contain two mbt repeats (Bornemann et al., 1996), whereas the human l(3)mbt-like a/b proteins, murine Sfmbt protein, and KIAA1617 and RU1 proteins contain four mbt repeats (Usui et al., 2000; Wismar, 2001).
The described proteins that contain mbt repeats are presumed to belong to the PcG family of proteins, which have been grouped together because of their putative role in maintaining transcriptional repression of homeotic genes during development (Bunker and Kingston, 1994; Muller, 1995; Pirrotta, 1997a, 1997b; Poux et al., 2001; Simon, 1995) Drosophila that lack both maternal and zygotic PcG genes die with severe homeotic transformations (Breen and Duncan, 1986); they do not exhibit proper maintenance of homeotic genes as development progresses (Lewis, 1978; Simon et al., 1992; Struhl and Akam, 1985; Wedeen et al., 1986). Further, three hypomorphic Drosophila SCM mutations that map within the mbt repeats produce partial synthetic lethality in the embryo (Bornemann et al., 1998), suggesting that these mbt repeats play an important role in the function of PcG protein complexes.
To gain molecular insights into l(3)mbt and its putative role in PcG complexes and tumor suppression, we have determined the crystal structures of three mbt repeats from the human l(3)mbt homolog at resolutions up to 1.55 Å. These structures not only reveal the global architecture of the three mbt repeats, but also identify aromatic amino acid-lined binding pockets within each mbt repeat for the morphilino ring of MES and/or the proline ring of the C-terminal peptide segment. Strikingly, the available point mutations and deletions of the related Drosophila SCM protein map within and in the vicinity of the ligand/peptide binding pockets.
Results
Crystallization and X-Ray Structure Determination
A cDNA construct encoding the region of the h-l(3)mbt protein from residue 197 to 527 (henceforth designated 3mbt), which encompasses all three mbt repeats (Figure 1), was expressed in bacteria and purified to homogeneity. The initial crystals were trigonal (P32 space group), with three 3mbt molecules in the asymmetric unit. The structure was solved at 1.9 Å resolution by the multiple wavelength anomalous diffraction (MAD) technique on crystals of the selenomethionine-labeled protein (designated seleno-met) (Table 1). The seleno-met structure was refined to an R factor of 20.6 (Rfree = 23.8) with all data collected up to 1.85 Å resolution (Table 1). A second 3mbt crystal in the trigonal P32 space group (designated native I) was crystallized with GTP-preincubated 3mbt protein. The native I and seleno-met crystals are isomorphous, and both diffracted to a resolution of 1.85 Å. The native I structure was refined to an R factor of 21.1 (Rfree = 24.5). In both seleno-met and native I crystals, 312 amino acids (residues 206–518) per monomer are well defined in the electron density maps.
Figure 1. Sequence Alignments of Proteins Containing mbt Repeats.
(A) Multiple sequence alignment of the N-terminal arms of 3mbt repeats from h-l(3)mbt and related family members using CLUSTALW. Secondary structure elements defined by crystal structure of 3mbt repeats are shown above the alignment. α helices, wavy magenta lines; 310 helices, wavy orange lines; β strands, arrows; loops, straight lines. The helices are numbered consecutively from the N terminus. Helical structure αA, featured with a dashed line, only exists in the second and third repeats, while αD exists only in the first repeat. The loop with a dashed line only exists in the second and third repeats. Conserved hydrophobic, aromatic, positively charged, negatively charged, and polar residues are marked with yellow, green, red, blue, and orange. Accession numbers (SWISS-PORT, Genbank, or PIR) are as follows: H. sapiens (brain) l(3)mbt, Q9Y468; H. sapiens (brain) KIAA1798, Q96JM7; H. sapiens (testis), Q9Y4Q9; D. melanogaster, Q9VB52; M. fascicularis, Q95LL9; C. elegans (R06c7.7), Q21769; C. elegans (Y48GIA.6), Q9N3Q7; D. melanogaster SCM, Q9VHA0.
(B) Multiple sequence alignment of the β subunit core of three mbt repeats from h-l(3)mbt and related family members, with defined secondary structure elements as indicated in (A). The β2 region featured with a dashed line only exists in first repeat, while β6 only exists in the first and second repeats. The loop featured with a dashed line only exists in third repeat. Structure-based sequence alignment of the Tudor domain with the mbt β subunit core is shown at the bottom. Mutated residues in SCM and residues of the Tudor domain involved in binding Sm proteins and the GAR domain are boxed. Conserved residues that form the putative drug/peptide binding pocket are highlighted with asterisks, those involved in hydrogen bond formation (Figure 3D) are indicated by closed circles, and those involved in the hydrophobic pentad motif (Figure 3E) are indicated by open circles. The accession number for H. sapiens SMN1-Tudor is Q16637.
Table 1.
Crystallographic Analysis
| MAD Data Statistics for Seleno-Met | ||||
|---|---|---|---|---|
| λ4 (Low Energy) | λ3 (Inflection) | λ2(Peak) | λ1(Remote) | |
| Space group | P32 | |||
| Cell Parameters | a = b = 105.74 Å, c = 90.52 Å | |||
| Wavelength (Å) | 1.06334 | 0.97935 | 0.97914 | 0.95668 |
| Resolution range (Å) | 50.0–1.90 | |||
| Unique reflections (F > 0) | 89,576 | 88,659 | 89,360 | 88,565 |
| Redundancy | 3.1 | 10.48 | 11.6 | 7.8 |
| Completeness (%) | 100 | 100 | 99.9 | 100 |
| Rmergea(%) | 6.1 | 8.5 | 8.1 | 9.5 |
| Phasing powerb(Friedel) | 1.33 (1.48) | − (1.22) | 0.52 (1.43) | 0.95 (1.19) |
| Rcullisc(Friedel) | 0.73 (0.69) | − (0.74) | 0.94 (0.72) | 0.82 (0.78) |
| Mean FOMd | 0.62/0.91 | |||
| Refinement Statistics | ||||
| Crystal | Seleno-Met | Native I | Native II | |
| Space group | P32 | P32 | P21212 | |
| Cell parameters | a = b = 105.74 Å, c = 90.52 Å | a = b = 105.255 Å, c = 90.486 Å | a = 86.68 Å, b = 93.403 Å, c = 58.578 Å | |
| Beamline | APS-14ID | APS-19BM | APS-19BM | |
| Wavelength (Å) | 1.06334 | 1.03300 | 1.03300 | |
| Resolution range (Å) (last shell) | 20–1.85 (1.90–1.85) | 20–1.85 (1.90–1.85) | 20–1.55 (1.59–1.55) | |
| Unique reflections (last shell) | 88,637 (4,088) | 112,768 (8,263) | 65,305 (4,700) | |
| Redundancy | 3.1 | 3.1 | 6.7 | |
| Completeness (%) (last shell) | 97.0 (94.2) | 99.9 (100) | 98.8 (98.2) | |
| Rmergea% (last shell) | 5.9 (55.0) | 5.5 (47.8) | 6.7 (57.2) | |
| R factor, Rfreee(%) (last shell) | 20.6 (50.4), 23.8 (53.0) | 21.1 (26.1), 24.5 (33.5) | 18.9 (22.6), 21.9 (24.8) | |
| Number of residues in refinement | 3 × 312 = 936 amino acids, 540 H2O, 14 SO4, 8 MES | 3 × 312 = 936 amino acids, 492 H2O, 9 SO4, 6 MES | 323 amino acids, 528 H2O, 4 SO4, 4 MES | |
| Rmsd bonds (Å), angles (°) | 0.017, 1.58 | 0.017, 1.82 | 0.017, 1.64 | |
| Average B factor (Å2) | 31.2 | 24.2 | 16.9 | |
Rmerge = ΣhklΣi|I(hkl)i − <I(hkl)>|/ΣhklΣi<I(hkl)i> for i measurements of the intensity I of a reflection hkl.
Phasing power is root mean square of (|Fh|/E), where |Fh| is the heavy atom structure factor amplitude and E is the residual lack of closure error; resolution range 27.5–2.5 Å.
RCullis is the mean residual lack of closure error divided by the mean anomalous difference; resolution range 27.5–2.5 Å.
Prior to and after density modification with phase extension to 1.9 Å.
R factor = Σ‖F(obs) | − |F(calc)‖/Σ|F(obs)|; Rfree is the same, calculated with 5% of data withheld from refinement.
We also obtained orthorhombic (P21212 space group) crystals containing one 3mbt molecule in the asymmetric unit (designated native II), using GTP-preincubated conditions. The native II crystals diffracted to 1.55 Å, the highest resolution of all studied 3mbt crystals. The structure was solved by molecular replacement and refined to an R factor of 18.9 (Rfree = 21.9). In native II crystals, the polypeptide chain could be traced for 323 amino acids (residues 204–527), including nine additional C-terminal amino acids (residues 519–527) that were disordered in the trigonal crystals. The final refinement statistics for all three structures (seleno-met, native I, and native II) are given in Table 1. The 3mbt structures found in the seleno-met, native I, and native II crystals are similar. The average root mean square deviations (rmsd’s) for main chain atoms (residues 206–518) is less than 0.75 Å between any two superimposed final models selected from a total of seven crystallographically independent 3mbt structures. Sulfate anions and 2(N-morpholino-ethane sulfonic acid) (MES) ligands were also bound to 3mbt, in addition to water molecules.
3mbt Architecture
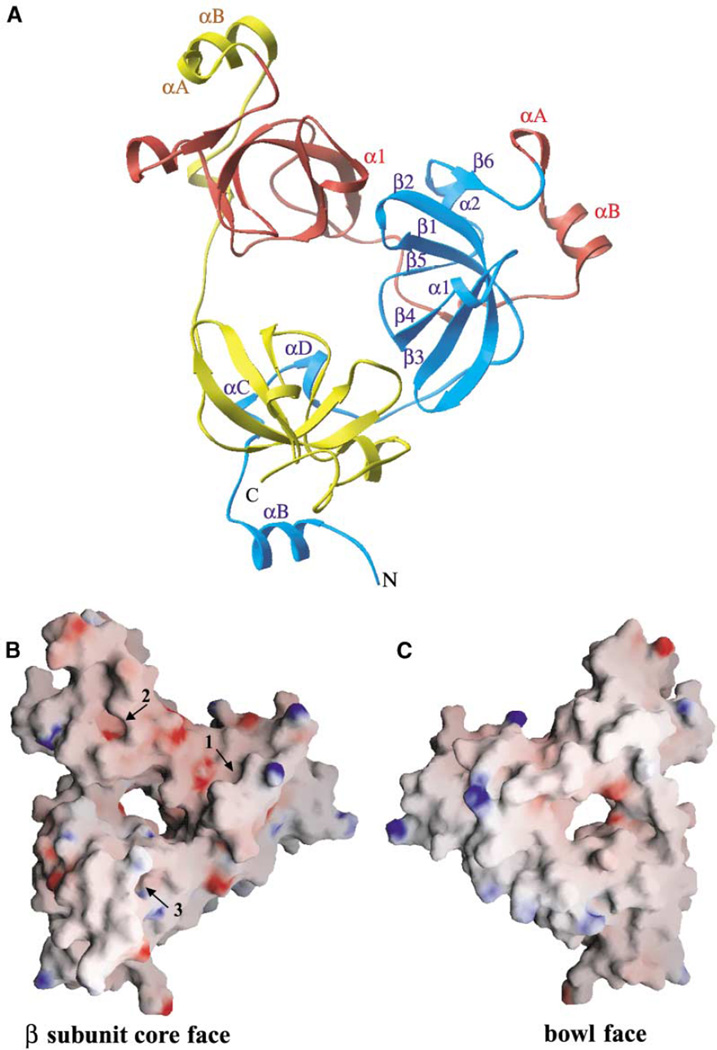
The overall architecture of 3mbt is shown in Figure 2A. The orthorhombic crystal structure is used for representation because of its highest (1.55 Å) resolution. The three mbt repeats are colored cyan, red, and yellow from the N terminus to the C terminus. The 3mbt protein adopts a mixed α/β fold with a unique triangular architecture and dimensions of 60 Å × 70 Å × 65 Å, with a thickness of 30 Å. The three-dimensional structure consists of three similar globular modules (~35 Å in radius) that assemble in a propeller-like manner, with a pseudo-3-fold rotation symmetry of ~120° (Figure 2A). The N and C termini of the 3mbt construct are proximal to each other and are positioned on the same side of the propeller. This in turn generates an elongated central cavity, which is highlighted in electrostatic surface views of the opposing faces of the three-leaved propeller-like arrangement (Figures 2B and 2C).
Figure 2. 3mbt Adopts a Triangular Propeller-like Architecture.
(A) Ribbon representation (orthorhombic crystal) of the structure (β subunit core face) of 3mbt. The first (residues 204–309), second (residues 310–416), and third (residues 417–527) mbt repeats are colored cyan, red, and yellow, respectively. Secondary structure elements are indicated for the first mbt repeat, which comprises a long N-terminal arm (αB, αC, and αD) and a β subunit core (β1, β2, β3, β1, β4, β5, α2, and β6). αA exists only in the second and third repeats and is indicated explicitly for clarity. α1 of the second repeat is shown to clarify the unique kissing α1–β 2 interaction (see text and Figure 4C).
(B) Electrostatic surface representation (orthorhombic crystal) of the β subunit core face of the triangular propeller-like 3mbt structure. Blue, electropositive; red, electronegative. This face, with its extensive acidic (red) patches, contains three ligand binding pockets (numbers 1, 2, and 3 refer to the pockets belonging to the first, second, and third repeats, respectively).
(C) Electrostatic surface representation (orthorhombic crystal) of the opposite bowl-like face of 3mbt. This face, which contains basic (blue) patches on the periphery, is comprised of N-terminal arms and predominantly exhibits nonpolar character. Some electropositive charge is clustered toward one side of the surface.
A central cavity with approximate dimensions of 10 Å × 15 Å can be clearly seen in both (B) and (C).
C-Terminal β Subunit Core and Attached N-Terminal Long Arm
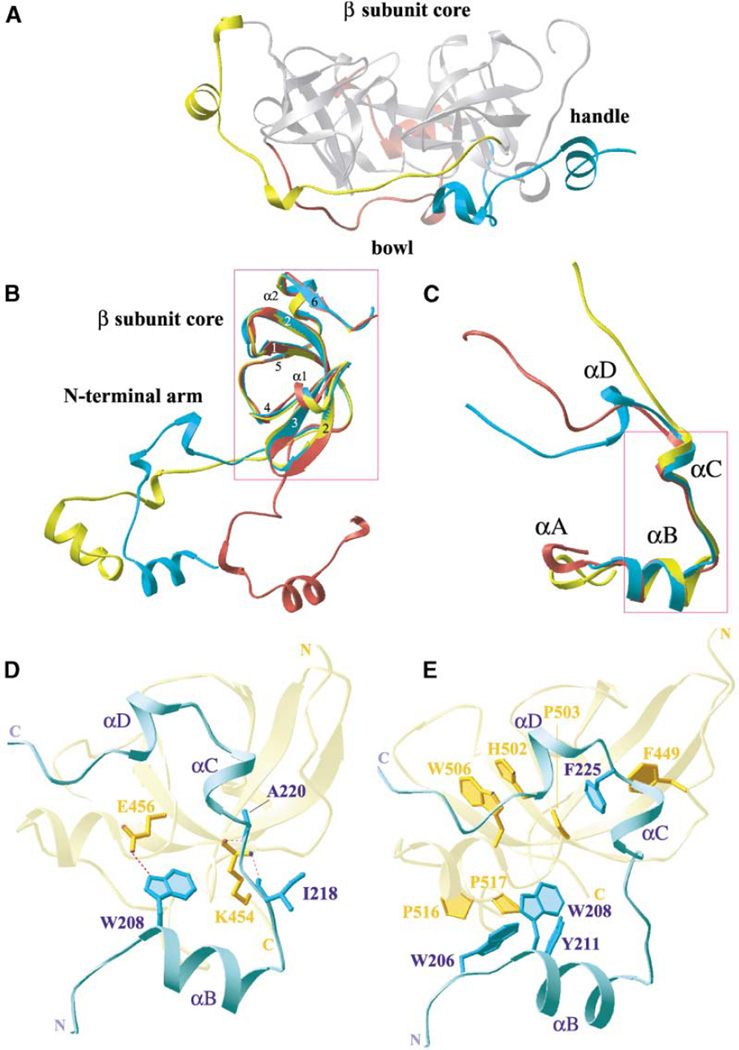
Individual mbt repeats consist of two distinctive structural motifs, a long N-terminal arm and a C-terminal β subunit core (Figure 2A). The N-terminal arm of each individual mbt repeat is extended ~35–40 Å away from the C-terminal attached core and interdigitates extensively within the core of its preceding neighbor, resulting in a stable three-leaved propeller-shaped architecture (Figure 2A). A side view of the 3mbt structure reveals that the N-terminal arms of each repeat are located along one face, forming the base of a bowl-like scaffold, while the β subunit cores are located along the opposite face, on the top of the bowl (Figure 3A).
Figure 3. Long N-Terminal Arm and β Subunit Core Motifs of 3mbt.
(A) Side view of the 3mbt structure (orthorhombic crystal) reveals that the N-terminal arms and β subunit cores are located along different faces of the molecule. The N-terminal arms of each repeat (colored as in Figure 2A) are located along one face (bottom), exhibiting a bowl-like feature, while the β subunit cores (silver) are located on the opposite face (top), being positioned on top of the bowl. The structurally conserved element αA-linker-βB forms a handle-like feature along three sides of the molecule.
(B) Superimpositions of the first (cyan), second (red), and third (yellow) repeats of 3mbt (orthorhombic crystal). The β subunit cores superposition well on each other (boxed region), with resulting rmsd’s of 1.0 Å for the Cα atoms. By contrast the attached N-terminal arms are oriented in different directions.
(C) Superimpositions of the N-terminal arms from the first (cyan), second (red), and third (yellow) repeats of 3mbt (orthorhombic crystal). The αB-linker-αC segments superposition well on each other (boxed region; rmsd’s of 0.9 for Cα atoms), while the C-terminal segments are oriented in different directions. An element of 310-helical turn, αC, is only observed for the arm of the first repeat.
(D) Ribbon drawing showing interdomain hydrogen bonding contacts between residues from the αA-linker-αB segment of the first N-terminal arm (cyan) and the β subunit core and N-terminal arm from the third mbt repeat (yellow). Hydrogen bond networks are indicated by dashed magenta lines.
(E) Ribbon drawing showing interdomain stacking interactions between aromatic and proline residues from the first N-terminal arm (cyan) and the β subunit core and the N-terminal arm from the third repeat (yellow).
Each β subunit core consists of five β strands (β1–β5), a short 310-helical turn (α1), and an α helix (α2) followed by a short β strand (β6) (Figure 2A). The β1–β5 strands form a barrel-like fold, with three of the β strands (β1–β3) forming the antiparallel component of the barrel topology. The β2 strands are of variable length; the first repeat is longer than the second and third repeats (Figures 1B and 2A). The 310-helical turn (α1) connects β3 and β4, and an α helix (α2) connects β5 and β6; the other β strands are connected by short nonhelical turns or loops. The β subunit core is closed by a parallel interaction between the β2 and β6 strands.
The arms are basically long loops with a variable number of amino acids (~30–45). All arms contain one αhelix (αB) and one 310-helical turn (αC). The arm of the first repeat is the longest, and it contains an additional 310-helical turn, αD. The arm of the third repeat is the shortest. The arms of the second and third repeats are attached to the preceding repeat and contain a 310-helical turn, αA. A similar turn is not found in the first repeat. The individual β subunit cores superposition well on each other (Figure 3B). In addition, the αB-linker-αC segments of the individual arms also superposition well on each other, while their adjacent C-terminal segments are oriented in different directions (Figure 3C). This variability in the C-terminal segments of the three arms is matched by the observed minimal amino acid sequence similarity in the C-terminal region.
Interdigitation between Arms and β Subunit Cores
A hairpin loop (containing the β6–αA segment) connects the C terminus of an individual β subunit core with the N terminus of the extended arm of the succeeding repeat (Figure 2A). This linkage is adjacent to a 310-helical turn, αA, and an α helix, αB, of the arm segment, which serves the role of a handle within the context of the bowl-like topology (Figure 3A). The β subunit cores are glued on top of the bowl-shaped scaffold formed by the N-terminal arms of the adjacent repeats. Such interfaces between repeats are mediated by stacking and hydrogen bonding interactions involving both arm and β core residues.
The hydrogen bonds between the N-terminal arm of the first repeat (cyan) and the β subunit core of the third repeat (yellow) are highlighted in Figure 3D. Residues (I218 and A220 [arm]) on the αB-linker-αC segment align through main chain hydrogen bond interactions with a conserved residue (K454 [core]) on the β1 strand (Figure 3D). In addition, a side chain-side chain hydrogen bond is observed between W208 (arm) and E456 (core), as shown in Figure 3D.
The stacking interface between the αB element of the arm of the first repeat (cyan) and both the β subunit core and the arm segment of the third repeat (yellow) is shown in Figure 3E. Phenylalanine F225 (arm segment) of the first repeat is buttressed by F449 (arm segment) of the third repeat and positions its aromatic ring over proline P503 (core segment) of the third repeat (Figure 3E). This interface is further buttressed by stacking interactions involving the third repeat core segment residues histidine H502 and tryptophan W506. In addition, prolines P516 and P517 (core segment) of the third repeat are stacked over tryptophans W206 and W208 (arm segment) of the first repeat (Figure 3E). Similar and structurally conserved residues occupy these positions in all three repeats (Figure 1). Note the orthogonal alignments of the proximally positioned F225 and F449 residues and of the proximally positioned W206 and W208 residues in Figure 3E.
Alignment and Relative Flexibility of Three Repeats
The three mbt repeats cannot be superpositioned on each other by a simple rotation around a single axis. The first (cyan) and third (yellow) repeats, but not the second (red), can be approximately related by an ~120° 3-fold symmetry rotation axis (Figure 2A). The second repeat is bent inwards and somewhat out of the plane of the other two repeats. This results in close proximity of the α1 of the second repeat and β2 of the first repeat, to generate a unique kissing α1–β2 contact.
A graphical representation of the relative flexibility within the 3mbt architecture (following superposition of seleno-met, native I, and native II crystal structures) is summarized in Figure 4A. The most flexible regions within 3mbt (indicated by increasing thickness in the worm representation in Figure 4A) are observed at the kissing α1–β2 interface and within the handle-like segments of the first and second repeats.
Figure 4. Details of Interdomain Interactions in Orthorhombic 3mbt Structure.
(A) Worm representation of the superimpositions of all determined structures (seleno-met, native I, and native II) of 3mbt repeats (Table 1). The resulting average rmsd’s for all main chain atoms are 0.75 Å Regions that superposition well are indicated with thin worm representation, while regions that superposition less well are indicated with thicker worm representation. The most flexible regions are the handle-like segments of the second and third repeats.
The 2Fo − Fc electron density map of the central cavity and interdomain interactions of 3mbt repeats at 1.55 Å resolution, contoured at 1.5 σ, with the refined atomic model in a ball and stick representation. Residues from first, second, and third mbt repeat are colored cyan, red, and yellow, as in Figure 2A. Oxygen, nitrogen, and sulfur atoms are colored red, cyan, and green, respectively.
(B) View showing inter-β subunit core contacts from the first (cyan) and second (red) repeats. An additional interaction is mediated by a kissing α1–β2 region that involves D384 (β1 from second repeat) and S253 (β2 from first repeat).
(C) View showing inter-β subunit core contacts from the first (cyan) and third (yellow) repeats. The interactions involve unique residues compared with (B).
(D) View showing interdomain contacts mediated by a unique residue, R444, from the interdigitated region of the third N-terminal arm (yellow) that pairs with residue D391 from the second β subunit core (red) and residue D497 from the third β subunit core (yellow) via hydrogen bond networks (dotted magenta lines).
(E) View focuses on the bound SO42− molecule in the central cavity of the 3mbt triangular propeller structure. The SO42− molecule is anchored in the central cavity via hydrogen bond networks (dotted green lines) with residues R460 and H498.
Interdomain Side to Side Contacts between β Subunit Cores
There are contacts between the third (yellow) and first (cyan) β subunit cores and between the first (cyan) and second (red) β subunit cores (Figure 2A). These interactions are mediated by hydrogen bonding between residues positioned on β2, β3, and β5 of the β subunit cores. Such hydrogen bonding alignments are observed between residues Y255, P252, and D289 in first β subunit core and residues D369, W389, and R374 in the second β subunit core (Figure 4B), as well as between residues R267 and N284 in the first β subunit core and residues P463, P499, and D500 in the third β subunit core (Figure 4C). In addition, the kissing α1–β2 interface mediates an additional hydrogen bond between S253 of β2 and D384 of α1 (Figure 4B), the only helical elements that are involved in inter-β subunit core interactions.
There is a variation in separation between β subunit cores, with the largest separation observed between the second and third β subunit cores (Figure 2A). This gap is bridged by R444, positioned on the C-terminal arm of the third repeat, through hydrogen bond formation with D391 of the second repeat and D497 of the third repeat (Figure 4D).
Bound Sulfate within Elongated Central Cavity
The elongated oval shape of the central cavity of 3mbt has dimensions of 15 Å × 10 Å (Figures 2B and 2C) and is predominantly lined by charged and polar residues (Figure 4E). The charged residues include four aspartic acids (D289, D495, D497, and D500) and three arginines (R356, R374, and R460), while the polar residues include a glutamine (Q250), an asparagine (N284), and a histidine (H498). Interestingly, a SO42− the molecule is bound within central cavity and anchored through hydrogen bonds to residues R460 and H498. In addition, hydration sites have also been identified in this central cavity (Figure 4E).
Charge Distribution on Opposing Propeller-like Faces
The electrostatic charge distribution patterns spanning the two faces of the propeller-like 3mbt repeat architecture are shown in Figures 2B and 2C. The face that contains the β subunit cores exhibits an uneven charge distribution, with acidic patches (red) predominantly located within the second β subunit core, spanning the kissing α1–β2 and the handle-like regions (Figure 2B). The bowl face of the triangular propeller is predominantly nonpolar but exhibits basic patches (blue) along the edge between first and second repeats (Figure 2C). One well-defined pocket per repeat is clearly visible on the β subunit core face of the 3mbt repeat architecture (labeled 1, 2, and 3 in Figure 2B).
MES Binding Sites
All three pockets in the seleno-met structure are occupied by 2-(N-morpholino) ethane sulfonic acid (MES) ligands. A superposition of the three binding pockets associated with the first (cyan), second (red), and third (yellow) repeats with bound MES ligands is shown in Figure 5A. The morpholino rings insert into the pockets, while the sulfonic acid groups are directed outward. The three MES ligands occupy somewhat different alignments within the pockets, each of which is oriented by distinctive amino acid residues that line individual pockets (Figure 5A). A more detailed analysis of MES bound in the pocket of the second mbt repeat is shown in Figure 5B; the walls of this pocket are lined by residues D355, C363, F379, W382, and Y386. Thus, the MES ligand is contained within a cage lined primarily by aromatic amino acids. A similar distribution of amino acids also lines the walls of the pockets of the first (D248, F256, F272, Y275, and H279) and third (D459, R467, F483, W486, and Y490) repeats.
Figure 5. MES Ligand Binding Pockets in Trigonal 3mbt Structures.
(A) Superimposition of three mbt repeats with bound MES molecules in the seleno-met structure. A MES molecule (labeled 1, 2, and 3) is bound to each β subunit core (colored as in Figure 2A).
(B) The mutation sites are located in and around the MES binding pocket (second mbt repeat) in the 3mbt structure (seleno-met). The morphilino ring of the bound MES molecule (red) is encapsulated by polar and aromatic residues (silver). The electron density for bound MES is shown in a chicken wire representation. Residues that are point mutated (magenta) and deleted (blue) are highlighted with the letters M and D, respectively. Residue M357 in the second repeat is included, since it occupies a position analogous to R461 in the third repeat (see [C] and [D]). Hydrogen bond networks are indicated by dotted lines (magenta). Oxygen and nitrogen atoms are colored red and blue, respectively.
(C) The surface representation of the open form of the MES (yellow) binding pocket from the third mbt repeat in the seleno-met structure. Residues lining this pocket are indicated in black. Arginine R461 is indicated in dotted surface (pink) representation with its side chain in a stick mode (white). Conserved residues among mbt repeats that are mutated in the SCM protein (Bornemann et al., 1998) are explicitly indicated in blue (deletion mutations, labeled D) and red (point mutations, labeled M).
(D) The surface representation of the closed form of the MES binding pocket from the third mbt repeat in the GTP-containing native 1 structure. The R461 arginine finger flips its long and bulky side chain into the pocket, closing this site to MES ligands. This alignment is stabilized by a single hydrogen bond with residue D459 (point mutated in mbt of SCM protein; Bornemann et al., 1998).
Open and Closed Pocket of 3mbt
The native I trigonal crystal grown with GTP-preincubated protein has MES bound only in pockets 1 and 2, while pocket 3 was closed by R461, whose bulky guanidinium side chain flaps over the entrance of the pocket. The guanidinium group is positioned perpendicular to residue W486 and, in the process, buries residue F483 and makes the binding site unavailable to morpholino ligands.
The open and closed forms of pocket 3 on 3mbt are shown in Figures 5C and 5D, respectively. The side chain of R461 is exposed to solvent in the open form but is anchored in the closed form by a hydrogen bond between its guanidinium amino group with δ1-oxygen of D459 (Figure 5D).
Crystal Packing in Trigonal and Orthorhombic Crystals
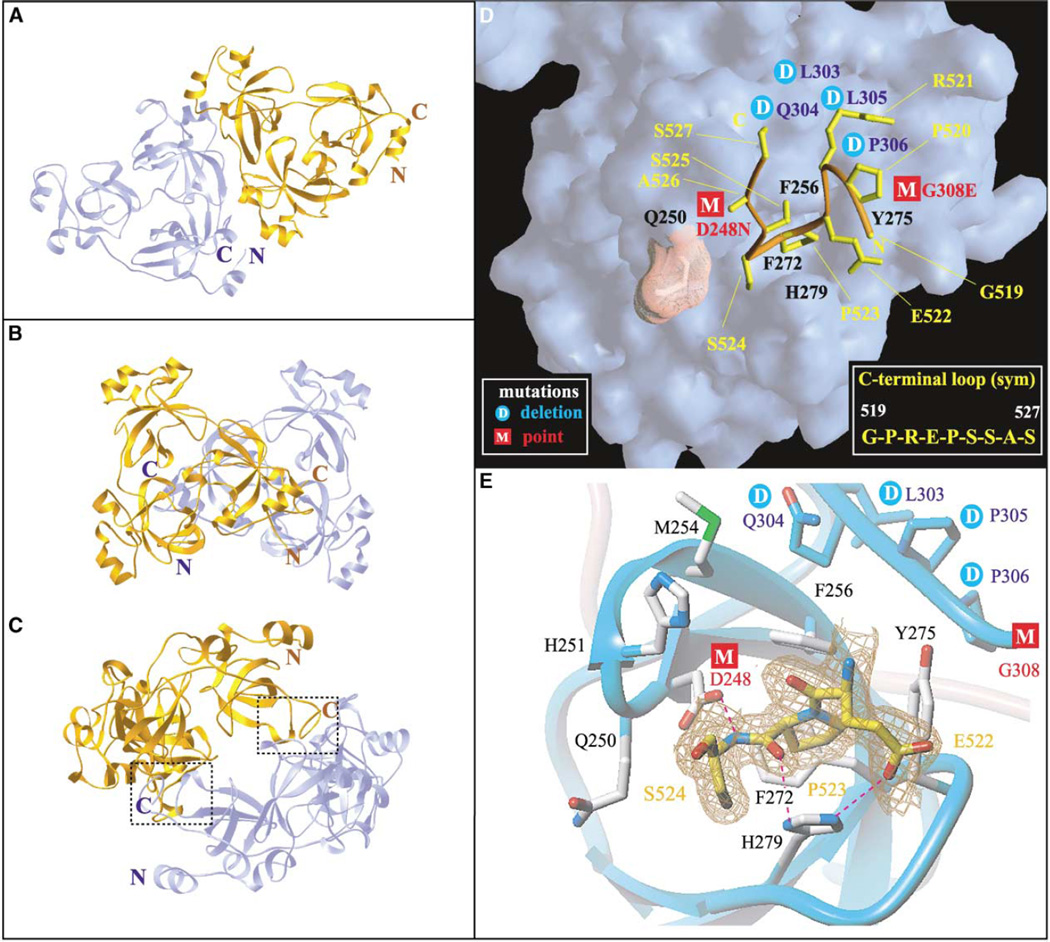
Two symmetry-related 3mbt molecules align through side by side packing in the trigonal crystal (Figure 6A). This results in a total buried surface area of 314 Å2 per monomer. This value is less than the optimum cutoff value of 856 Å2 for discriminating between monomers and dimers on the basis of a recent study with a nonredundant structure database (Ponstingl et al., 2000).
Figure 6. C-Terminal Peptide Binding Pocket in Orthorhombic 3mbt Structure.
(A) Packing of adjacent 3mbt molecules (cyan and gold) in the trigonal crystal, resulting in a total buried surface area of 314 Å2 per monomer (B and C) Two views of packing of adjacent 3mbt molecules (cyan and gold) in the GTP-preincubated native II orthorhombic structure, resulting in a total buried surface area of 1663 Å2 surface area per monomer. Note mutual insertion of C-terminal loops into ligand binding pockets of the first repeat (seen clearly in [C]).
(D) The surface representation of the binding pocket from the first repeat of 3mbt, which accommodates the C-terminal loop from a symmetry related 3mbt molecule. The peptide backbone of the C-terminal loop is colored orange, and its side chains are indicated in yellow. The side chain of residue Q250, represented as a dotted pink surface, is directed away from the pocket, analogous to the R461 arginine finger in Figure 5C. Mutations sites and cage residues are colored as in Figure 5C.
(E) The details of the first repeat pocket with the bound C-terminal loop. The residues involved in the binding pocket are shown in ball and stick representation (silver). Only residues 522–524 of the C-terminal segment are shown in the ball and stick representation in the interest of clarity, and the electron density map of this segment is shown in a chicken wire representation. Mutations sites and oxygen and nitrogen atoms are colored as in Figure 5B. The intermolecular hydrogen bond networks between C-terminal loop and cage residues are shown as dashed magenta lines. The main chain atoms of certain residues are shown explicitly in ball and stick representation for clarity.
The nine amino acids residues (GPREPSSAS) at the C terminus of 3mbt are disordered in the trigonal (seleno-met and GTP-preincubated native I) crystal forms. By contrast, this nine-amino acid segment (residues 519–527) can be observed in the 1.55 Å resolution crystal structure of the orthorhombic crystal (GTP-preincubated native II) form of 3mbt. A total of 1663 Å2 buried surface area per monomer is observed for the native II structure (Figures 6B and 6C). Significantly, the C-terminal residues 519–527 from one 3mbt molecule insert into the pocket of the first repeat of a second symmetry-related 3mbt molecule (Figure 6C).
C-Terminal Peptide Binding Pocket
The nine-residue C-terminal tail adopts a loop fold (Figure 6D), which is stabilized by four intramolecular hydrogen bond networks involving the main chain and side chain of residues P520 and S525 with E522 and P523. The C-terminal loop inserts the ring of proline P523 into the ligand binding cavity, where it is sandwiched between aromatic residues F256 and H279 (Figure 6E). Complex formation is stabilized by three intermolecular hydrogen bonds with residues that line the binding pocket. These include hydrogen bonds between the side chains of residues D248 and H279 with the backbone and side chains of residue S524 (Figure 6E). In addition, a hydrogen bond was detected between S525 of the C-terminal segment and residue H251 in the binding pocket.
Discussion
Circular Topology
The most striking feature of the 3mbt structure is its three-leaved propeller-like architecture (Figure 2A), with a SO42− ion occupying the central cavity (Figure 4E). The interdigitation of the three mbt repeats results in a compact fold, where the central hole is surrounded by the β subunit cores of three mbt repeats, while the αB helices of the N-terminal arms project furthest outward from the central cavity (Figure 3A). The extensive acidic patch along the β subunit core face (Figure 2B) and smaller basic patches on the opposing bowl-like face (Figure 2C) of the 3mbt architecture could serve as scaffolds for complex formation with as yet to be identified interacting partners. Only four-, five-, six-, seven-, and eight-bladed propeller-like structures have been reported (Fulop and Jones, 1999; Paoli, 2001); thus, to the best of our knowledge, the three-leaved propeller-like structure for 3mbt is novel.
Comparison of 3mbt Architecture with Other Cyclic Repeat Topologies
The adaptation of propeller-like topologies by repeat-containing protein modules has been observed previously for WD40 repeats (Neer et al., 1994), RCC1 (Renault et al., 1998), β subunits of heterotrimeric G proteins (Lambright et al., 1996), techylectin (Beisel et al., 1999), nitrite reductase (Fulop et al., 1995), prolyl oligopeptidase (Fulop et al., 1998), and clathrin (ter Haar et al., 1998). These proteins adopt β propeller topologies characterized by a donut-like architecture, coupled with a central deep shaft. By contrast, the 3mbt topology looks more like a flat tripedal clover (Figure 2A).
The classical β propeller donut-shaped architecture is generated when individual domains, containing four-stranded antiparallel β sheets, pack in a circular manner, thereby creating a central tunnel. This simplicity of the classical β propeller donut architecture contrasts with the circular architecture observed for 3mbt, which involves interdigitation of long N-terminal arms with adjacent β subunit cores (Figure 2A). Ring closure of the classical β propeller architecture is achieved by molecular clasp arrangements (Neer and Smith, 1996), which involves interactions between the first β strand of the N terminus and the last β strand of the C terminus. By contrast, the circular topology of the repeat units in 3mbt is mediated by stacking interactions of conserved aromatic residues that, for instance, anchor the β subunit core of the third repeat on top of the N-terminal arm of the first repeat (Figure 3E).
Correlating Conserved Sequence Elements with Structural Motifs
Most members of the β propeller fold have structural features that can be matched to internal sequence repeats. Several conserved sequence elements within the mbt repeats contribute to structural motifs that bridge the alignment of the N-terminal arms with their adjacent β subunit cores. Thus, a hydrophobic pentad motif involving primarily aromatic residues is associated with histidine, proline, and tryptophan residues in the sequence element HP–W spanning β5-linker-α2 (n repeat), a phenylalanine located adjacent to αC (n repeat), and a phenylalanine located at the C terminus (n+1 repeat) (Figure 1, residues indicated by open circles). This hydrophobic pentad motif involves H502, P503, and W506, together with F449 and F225, as shown in Figure 3E.
Another conserved structural motif contains two charged residues (K and E) from the signature sequence MKLE at β1 and three residues in the handle segment of the N-terminal arm (Figure 1, residues indicated by closed circles). This includes the side chain-side chain W208·E456 hydrogen bond and main chain hydrogen bonds involving residues A220, I218, and K454, as shown in Figure 3D.
N-Terminal Arms Bridge Adjacent mbt Repeats
The extended long N-terminal arms bridge adjacent mbt repeats by making contacts with the neighboring β subunit cores, resulting in a head to tail triangular propeller-like arrangement (Figure 2A). The αB and αC helices are common to all three arms, while αD is only present in the first arm, which is also the longest in length (Figure 3C). Interestingly, the extra αD helix present in the first arm forces the loop to make an ~90° turn prior to extending the C-terminal segment away from the αB-linker-αC segment (Figure 3C). The αD of the first arm is located at the inner side of the third β subunit core (Figure 2A), forming part of the inner shaft. The second and third arms lack αD, and the corresponding amino acid segment is also truncated for the third repeat (Figure 1A). Therefore, the trajectory adopted by the N-terminal arms of the first, second, and third repeats are quite distinct (Figure 3C), with the ends being closest for the first repeat (cyan) and furthest for the third repeat (yellow). The variation in the length of the C-terminal portion (linker-αD-linker region) of the arm segments is not only observed within the three repeats of 3mbt, but also between mbt-containing proteins across species (Figure 1A). Taken together, the above observations reinforce the conclusion that the relative orientations of the N-terminal arms make critical contributions to the formation of the three-leaved propeller-shaped architecture.
Accessibility of the Mobile Handle-like Segments
The handle segments of the bowl (αA-linker-αB; see Figures 2A and 3A) constitute the outer segments of the 3mbt architecture, contain conserved charged residues, mainly glutamic acids and lysines, positioned between the conserved hydrophobic and aromatic residues at both ends (Figure 1A), and are exposed to solvent. It has been proposed that these accessible charged residues, especially the conserved glutamic acids, are important for function (refer to the Protein families [Pfam] database). Note that three successive glutamic acids (E311, E312, and E313) project from the αA helix of the second repeat. The handle segments are among the most mobile regions of the architecture, since their positions exhibit the largest variability amongst the seleno-met, native I, and native II 3mbt structures (Figure 4A).
Structural and Functional Comparison between mbt Cores, Tudor Domains, and Chromodomains
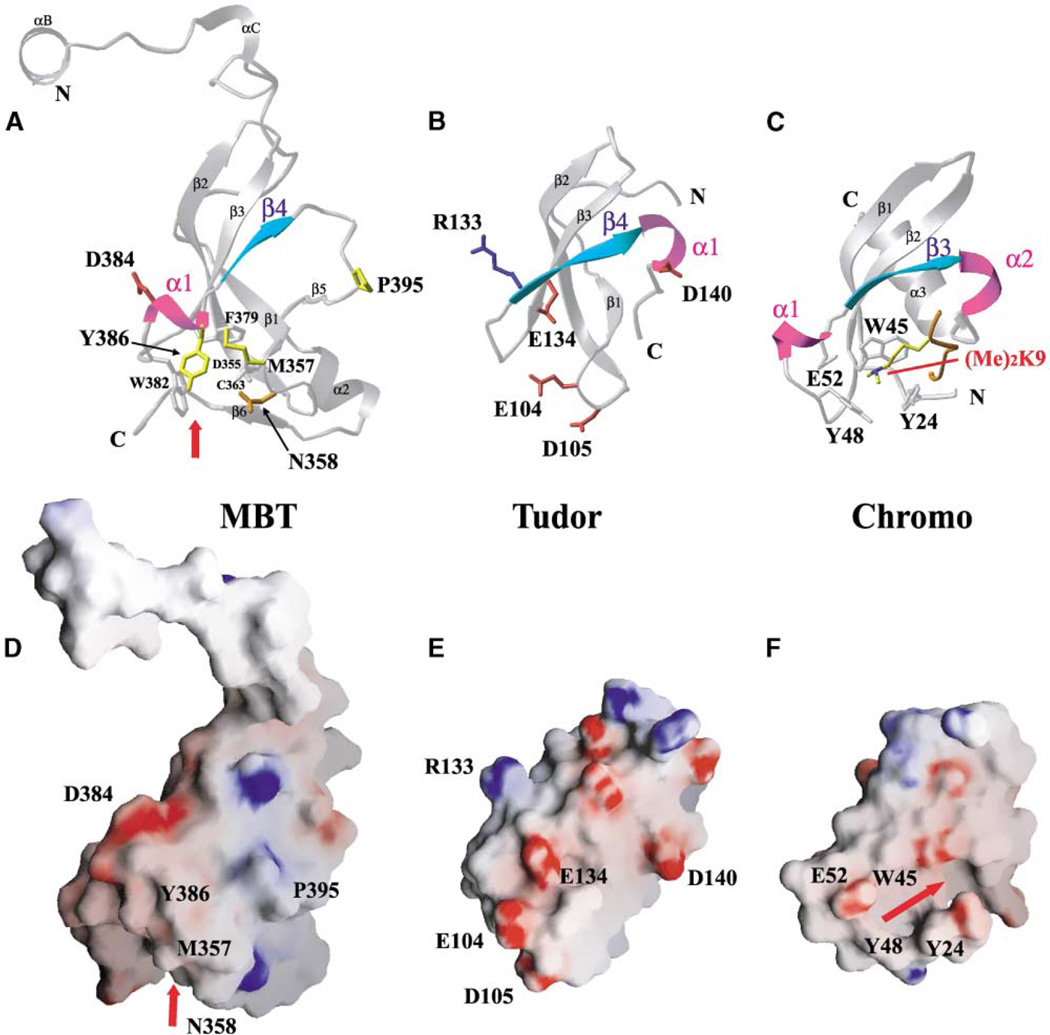
We conducted a structural homology search in order to identify proteins that exhibit folds similar to the mbt repeats. The Tudor domain (Figure 7B; Selenko et al., 2001) gave the closest match to the mbt β subunit core architecture (Figure 7A) (z score, 5.8). By contrast, the chromodomain (Figure 7C) is a more distant structural relative of the mbt β subunit core (z score, 3.0). We note that only 11%–13% of amino acids are identical between the mbt β subunit core and the Tudor domain (Figure 1B). The signature sequence M-K/R-L/F/V-E at β1, which is shared among all mbt repeats, is not observed in the Tudor domain (Figure 1B). In addition, the α1 helical turn (cyan) is located between β3 and β4 in 3mbt (Figure 7A, second repeat), whereas it is located after β4 in the Tudor domain (Figure 7B). The α1 helical turn plays a critical role in defining the shape of the ligand binding pockets in the individual repeats within 3mbt (Figures 5B and 6E). In addition, the linker segment between α1 and β4 in the mbt repeats exhibits species-dependent length variations, with the first mbt repeat from two C. elegans proteins having ~15 extra amino acids inserted between α1 and β4 (Figure 1B). The influence that these additional sequences have on secondary structure elements and their impact on the topology of the binding pocket will be the subject of future study.
Figure 7. Structural Homology between the 3mbt β Subunit Core, Tudor Domain, and Chromodomain.
(A) Ribbon representation showing the β subunit core of the second repeat of the seleno-met 3mbt structure.
(B) Ribbon representation of the Tudor domain (Selenko et al., 2001). Note the difference in the position of the α1 helical turn between Tudor domain (B) and mbt repeat (A).
(C) Ribbon representation of the chromodomain of HP1 bound to a dimethylated lysine 9 histone H3 tail (Jacobs and Khorasanizadeh, 2002).
(D) Electrostatic surface representation of the second mbt repeat of the seleno-met 3mbt structure. Refer to Figure 2B for the electrostatic surface of the ligand binding cage.
(E) Electrostatic surface representation of the Tudor domain. Note the differences in surface charge distributions between the Tudor domain (E) and mbt repeat (D).
(F) Electrostatic surface representation of the chromodomain of HP1. Note the cavity and the aromatic cage that accommodates the histone H3 peptide tail and dimethylated lysine 9.
The secondary-structural elements of α1 and β4 (A and B) are colored magenta and cyan, respectively. Hydrophobic (yellow), positively charged (red), negatively charged (blue), and polar (orange) side chains are shown in the ball and stick representation for 3mbt in (A) and the Tudor domain in (B). Other related residues involved in the cage are indicated in silver for clarity. The electrostatic surface in 3mbt (D) and the Tudor domain (E) is colored as in Figures 2B and 2C. The red arrow indicates the location of the ligand binding cage in the mbt structure and the binding cavity for the histone H3 peptide tail in the chromodomain, respectively.
The Tudor domain has been shown to interact with small nuclear RNA-associated proteins (snRNPs), and the failure to form these interactions presumably contributes to spinal muscular atrophy (SMA) disease. The interactions between Tudor and Sm proteins (Selenko et al., 2001) and between Tudor proteins and the Gar domain of fibrillarin (Jones et al., 2001) are facilitated by the acidic patches on Tudor’s surface (Figure 7E). By contrast, no negatively charged patches are observed on the structurally equivalent surfaces of individual subunits of the 3mbt protein (Figure 7D, second repeat). This implies that the β subunit core of the mbt repeats and the corresponding Tudor domains will not bind the same partners.
The chromodomain has been found in proteins from a wide variety of organisms and has a common mechanism of chromosomal organization for gene silencing (Eissenberg, 2001). The chromodomain of HP1 has been shown to recognize dimethylated lysine K9 in histone H3 (Figure 7C), which, in turn, directs the binding of other proteins to control chromatin structure and gene expression (Bannister et al., 2001; Lachner et al., 2001; Nakayama et al., 2001). Unlike both mbt cores and Tudor, the chromodomain has only three β strands, which pack against a C-terminal helix (Jacobs and Khorasanizadeh, 2002; Nielsen et al., 2002) (Figure 7C). The absence of one β strand (β1 in mbt cores and Tudor) in the chromodomain results in a channel and a distinct aromatic amino acid-lined cavity perpendicular to the three β strands (Figure 7F), which is targeted by dimetylated lysine-containing peptides (Figure 7C).
Open and Closed States of the Binding Pocket
We have observed both open (either bound MES in the trigonal seleno-met crystals or bound C-terminal peptide segment in the GTP-preincubated native II orthorhombic dimer crystals) and closed (in GTP-preincubated native I trigonal crystal) ligand binding pockets within individual repeats of 3mbt. The most striking consequence of these observations is the concept of a gate, controlled by a single arginine residue (R461 in the case of the third repeat), which is fully accessible to solvent in the open configuration (Figure 5C) but blocks entrance into the ligand binding cavity in the closed configuration, by hydrogen bonding to D459 (Figure 5D).
Bound Morpholino Ligands
The ligand binding pockets within individual mbt repeats are rich in aromatic and polar amino acids (Figure 5B) and readily accommodate MES molecules within a narrow range of orientations, as shown in Figure 5A. MES is a popular buffer in protein crystallization, and a number of crystallized proteins have been found to contain bound MES ligands. The Protein Data Bank contains 19 MES-protein complexes (Berman et al., 2000). Although binding of MES molecules does not always reflect the presence of a ligand binding site, MES has been found in spaces normally occupied by the protein’s natural ligand (Brown et al., 1999; Focia et al., 1998; Hohenester et al., 1994; Knochel et al., 1996; Teplyakov et al., 1998), where it can even function as an inhibitor (Fitzgerald et al., 1998).
Morpholino-like ring structures are found in several clinically active drugs, including an acetylcholinesterase inhibitor [8-(cis-2,6-dimethylmorpholino)-octylcarbamoyleseroline] developed as a treatment for Alzheimer’s disease (Bartolucci et al., 1999) and a leukocyte elastase inhibitor (pyridone-based trifluoromethyl ketone) (Bernstein et al., 1995).
Bound C-Terminal Peptide Segment
One pocket (third repeat) that binds MES in the seleno-met and GTP-preincubated native I crystals also accommodates the C-terminal peptide (G519–S527) in the orthorhombic GTP-preincubated native II crystal of 3mbt (Figure 6D). This C-terminal peptide adopts a turn motif, with P523 located at the apex of the turn, inserting into the pocket, and being anchored in place through hydrogen bonds involving two serine residues (S524 and S525). Thus, the ligand binding pocket is able to accommodate both heterocyclic six-membered (MES) and cyclic five-membered proline (P523) rings, indicative of its adaptability in accommodating ligand shape and hydrogen bonding potential.
Mutations in Binding Pocket Region
The biological implications of the MES/peptide binding pocket can be evaluated from existing data on mbt repeat mutations. SCM proteins contain a pair of mbt repeats and a SAM domain. Several mutations in SCM impair its function as a PcG protein (Bornemann et al., 1998). Two hypomorphic mutations (D215N and G275E) within the first mbt repeat of Drosophila SCM (Figure 1B, boxed amino acids) map in the vicinity of the identified ligand/peptide binding pocket. These residues exhibit a high degree of conservation among mbt repeats, and it is likely that the mutations change the charge distribution and distort the pocket. Another SCM mutant, which has a four-residue deletion (L270-Q271-P272-P273) in the first repeat of SCM (Figure 1B, boxed segment), should perturb the binding pocket because of its location along the rim of the pocket, spanning short β6 and the following loop segment.
The counterparts of the mutated D215 SCM residue are D248, D355, and D459 in the first, second, and third repeats of 3mbt. These aspartic acid residues are involved in key hydrogen bond networks that determine the open versus closed state of the binding pocket. In the open state, D459 is hydrogen bonded with R467, an adjacent cage residue (Figure 5C). However, additional hydrogen bond networks involving D459 and R461 are established to stabilize the R461 “arginine finger” alignment in the closed state of the binding pocket (Figure 5D). The counterparts of the mutated G519 SCM residue are G308, D415, and G519 in the first, second, and third repeats of 3mbt. Replacement of the glycine by a bulkier charged residue could result in both structural hindrance with adjacent residues and a charge redistribution, thereby distorting the nature of the binding pocket. The observed clustering of mutation sites in the neighborhood of the binding pocket (projected onto the first 3mbt repeat in Figure 6D, the second 3mbt repeat in Figure 5B, and the third 3mbt repeat in Figure 5C) suggests that these ligand/peptide binding pockets have a potential for playing a functional role in recognition events.
Putative Phosphate Binding Site within Central Cavity
The central cavity of other circular propeller-like proteins has been previously shown to harbor ions important for binding or enzymatic activity. Molecular recognition in several protein-nucleotide structures, including p21Ras-GTP (Scheffzek et al., 1997) and T7 helicase-ATP (Sawaya et al., 1999), have revealed that the nucleotide is recognized by a P loop segment and that the γ-phosphate of the bound nucleotide is hydrogen bonded to arginine and histinine residues within the protein. Residues H498 and R460, which are coordinated to sulfate in 3mbt (Figure 4E), could serve as the γ-phosphate sensors and potentially propagate conformational changes upon nucleotide binding. The central cavity in 3mbt has a diameter (~15 Å × ~10 Å) large enough to accommodate nucleotide triphosphate ligands or single-stranded oligonucleotides. Therefore, the location of the bound sulfate could reflect a putative phosphate binding site.
Biological Implications
The regulation of DNA replication, transcription, recombination, and repair in eukaryotes is mediated by subunits of chromatin-remodeling complexes. These subunits range from chromodomains, which recognize methylated lysine-containing histone H3 peptides, to bromodomains, which recognize their acetylated lysine-containing counterparts. Recently, bioinformatics searches have identified a family of homologous motifs containing a three-β-stranded core element, which includes mbt repeats, a chromodomain, a Tudor domain, and SAND and PWWP motifs (Maurer-Stroh et al., 2003).
Currently, little is known about the contribution of mbt repeats, as a component of the PcG group, in the dynamic remodeling of chromatin. It is conceivable that interactions between h-l(3)mbt and chromatin could be mediated by the mbt repeats. Further progress will require a systematic identification of proteins that interact with h-l(3)mbt and impact on its function. X-ray and NMR structures have been reported for the complex between the chromodomain of heterochromatin protein 1 (HP1) and a histone H3 peptide dimethylated at lysine 9 (Jacobs and Khorasanizadeh, 2002; Nielsen et al., 2002) (Figure 7C). The N-methyl groups of the modified lysine insert into an aromatic amino acid-lined pocket (Figure 7F), providing functional insights into the recognition of methylated proteins by the β sheet-containing chromodomain. A related study has provided functional insights into the recognition of an acetylated lysine-containing protein by an α helix-containing bromodomain (Mujtaba et al., 2002). Therefore, our future efforts will focus on the characterization of putative complexes between 3mbt and modified histone peptides. Such an approach holds promise for expanding on our current understanding of the role of mbt repeats in PcG-based transcriptional repression (Francis and Kingston, 2001; Simon and Tamkun, 2002) and, ultimately, for repairing transcriptional regulatory defects in the cell.
Experimental Procedures
Cloning, Expression, and Purification
The 3mbt region was amplified from a full-length human l(3)mbt cDNA clone (MacGrogan et al., 2001) with the Expand High Fidelity PCR System (Roche, Germany) and primers prim-for (5′-AGGAATTCGGCCTGCAACAGGTGAGAAG-3′) and prim-rev (5′-GCTGTCGACGGCAGAGCTGGGCTCTCT-3′), where the underlined bases are the restriction enzyme sites. The PCR product was digested with EcoRI/Sal1 and cloned into GST-fusion vector pGEX-4T-3 (Amersham). After confirming the insert sequence, the plasmid was designated 3mbt-prim-1 and used for expression. 3mbt-prim-1 was transformed into E.coli BL21(RP). Cells were grown in LB plus 100 µg ml−1 ampicillin at 37°C. At an OD600 of 0.6, expression was induced with 0.5 mM IPTG at 30°C. Cells were harvested 16 hr after induction, resuspended in 10 ml buffer A (50 mM Tris [pH 8.0], 5 mM DTT, and 150 mM NaCl) per liter of culture and stored at −80°C. For purification, cells were thawed at room temperature and lysed by sonication. After centrifugation at 15,000 rpm, the soluble fraction was purified by glutathione-Sepharose 4B column chromatography. The column was washed with 5× column volume of buffer B (50 mM Tris [pH 8.0], 5 mM DTT, and 250 mM NaCl), and the eluate was collected with buffer C (50 mM Tris [pH 8.0], 5 mM DTT, and 10 mM glutathione). The GST tag was cleaved by the addition of thrombin (10 units/ml) to the fusion protein at 4°C. Then the protein was dialyzed against buffer D (50 mM Tris [pH 8.0] and 5 mM DTT). The cleaved protein was then purified from the GST tag by reloading into glutathione-Sepharose 4B and then by loading into an anion exchange monoQ column with buffer D. The protein was eluted with a 30× column volume gradient that reached 100% of buffer E (25 mM Tris [pH 8.0], 5 mM DTT, and 1 M NaCl). The protein was then further purified by gel filtration on a Superdex-75 26/60 column equilibrated with 25 mM Tris [pH 8.0] and 5 mM DTT. Pure 3mbt was concentrated to 10 mg ml−1 and stored at −80°C.
The selenium-methionine (seleno-met)-substituted protein was expressed in the same host cell. Five milliliters of overnight cell culture grown in LB was added into 100 ml of minimal (M9 plus 20% glucose plus 1 mM MgSO4 plus 0.3 mM CaCl2) media and further grown to an OD600 of ~0.6–1.0. Cells were spun down, resuspended, and added into 1 liter of minimal media. Cells were allowed to grow at 37°C to an OD600 of ~0.5, and then lysine, phenylalanine, threonine at 50 mg ml−1 and isoleucine, leucine, valine, and L-selenomethionine at 100 mg ml−1 were added into the culture. Expression was induced 30 min after the addition of these amino acids, and cells were harvested and purified as described above.
Crystallization, Data Collection, and Processing
Crystals of native and seleno-met 3mbt were grown by vapor diffusion in sitting drops at 20°C. Drops were generated by mixing 1 µl of protein solution with 1 µl of reservoir solution and 0.2 µl of 100 mM DTT. The trigonal crystal form was obtained from a reservoir solution containing 100 mM MES (pH 6.5), 0.15 mM (NH4)2SO4, 20% PEG MME 5000, and 10 mM DTT. The orthorhombic crystal form was obtained from a reservoir solution containing 100 mM MES (pH 6.5), 0.1 mM (NH4)2SO4, 15% PEG MME 5000, and 10 mM DTT. For the GTP-containing 3mbt crystallization trials, protein was preincubated with 10 mM GTP at room temperature for about 1 hr before crystallization. Trigonal crystals formed within 7 days and typically grew to dimensions of 0.7 × 0.3 × 0.3 mm3 in 30 days. Orthorhombic crystals formed within 4 days and grew to dimensions of 0.5 × 0.5 × 0.05 mm in 10 days. For low-temperature data collection, crystals were transferred to the cryoprotectant (reservoir solution plus 30% [v/v] PEG 400) for a few seconds and flash-frozen in a liquid nitrogen stream. MAD data sets of seleno-met crystal were collected on a MAR CCD at BioCARS (beamline 14 ID) at the Advanced Photon Source (APS) in Argonne, Illinois. Native I and native II data sets from GTP-preincubated 3mbt were collected on MAR CCD at SBC (beamline 19BM) at the APS. Images were processed and scaled with HKL2000 (Otwinowski and Minor, 1997).
Structure Determination and Refinement
The trigonal seleno-met structure was determined by the MAD technique. Heavy-atom search, Patterson map, as well as electron density map, calculations and MAD phasing were performed with the program CNS1.1 (Adams et al., 1997), and maps were displayed with the program TURBO-FRODO (Cambillau and Roussel, 1997). The heavy-atom search was applied to anomalous difference Patterson maps calculated at 3.5 Å, and ten well-defined selenium sites were located. MAD phasing was then performed with data to 2.5 Å resolution. In order to find more Se sites, we used the above SAD phases in combination with the anomalous difference structure factors to compute anomalous difference Fourier and double difference (or log likelihood gradient) maps. Five additional selenium sites were found. Density modification by the solvent flipping method and phase extension to 1.9 Å were performed. Values for phasing power and figure of merit are listed in Table 1. Model building was manually completed for one 3mbt molecule with TURBO-FRODO. Two other 3mbt molecules present in the asymmetric unit of seleno-met crystals were built with a noncrystallographic symmetry option implemented in CNS. The trigonal native I crystal is isomorphous to seleno-met crystal and, starting from the seleno-met structure, was directly subject to refinement.
The orthorhombic native II structure was determined by the molecular replacement method with the refined trigonal structure as a starting model and the program AmoRe (Navaza, 2001). Rigid-body, positional refinement and stimulated annealing were performed in CNS1.1 (Adams et al., 1997). The TLS and bulk solvent parameters, restrained temperature factor, and final positional refinement were completed with the program REFMAC (Murshudov et al., 1997). Rfree was monitored by setting aside 5% of the reflection as a test set. The selected refinement parameters for seleno-met, native I, and native II structures are listed in Table 1. A Ramachandran plot calculated with PROCHECK (CCP4, 1994) indicates that 100% of the non-Gly and non-Pro residues in the final models lie in the most favored and additional allowed regions. For native I and native II crystals, which were grown with GTP-preincubated protein, we did not detect any difference electron density belonging to GTP molecule.
Figures showing three-dimensional structures and molecular surfaces were prepared with RIBBONS (Carson, 1997), MOLMOL (Koradi et al., 1996), TURBO-FRODO (Cambillau and Roussel, 1997), and GRASP (Nicholls et al., 1991).
Acknowledgments
This work was supported by funding from the NIH to D.J.P and by a grant from the Leukemia and Lymphoma Society Specialized Center of Research and funding from the Gabrielle Rich Leukemia Foundation to S.D.N. We acknowledge helpful discussions with Dr. Nagesh Kalakonda. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract number W-31-109-Eng-38. Portions of this work were performed at the Consortium for Advanced Radiation Sources (BioCARS) and at the Structural Biology Center (SBC-CAT) located at Sector 14 and Sector 19, respectively, of the Advanced Photon Source. Use of BioCARS was supported by the National Institutes of Health, National Center for Research Resources, under grant number RR07707. SBC-CAT is supported by the U.S. Department of Energy, Office of Biological and Environmental Research, under contract number W-31-109-ENG-38.
Footnotes
Accession Numbers
Coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 1OYX, 1OZ3, and 1OZ2 for seleno-met, native I, and native II.
References
- Adams PD, Pannu NS, Read RJ, Brünger AT. Cross-validated maximum likelihood enhances crystallographic simulated annealing refinement. Proc. Natl. Acad. Sci. USA. 1997;94:5018–5023. doi: 10.1073/pnas.94.10.5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asimakopoulos FA, Green AR. Deletions of chromosome 20q and the pathogenesis of myeloproliferative disorders. Br. J. Haematol. 1996;95:219–226. doi: 10.1046/j.1365-2141.1996.d01-1896.x. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Bartolucci C, Perola E, Cellai L, Brufani M, Lamba D. “Back door” opening implied by the crystal structure of a carbamoylated acetylcholinesterase. Biochemistry. 1999;38:5714–5719. doi: 10.1021/bi982723p. [DOI] [PubMed] [Google Scholar]
- Beisel HG, Kawabata S, Iwanaga S, Huber R, Bode W. Tachylectin-2: crystal structure of a specific GlcNAc/GalNAc-binding lectin involved in the innate immunity host defense of the Japanese horseshoe crab Tachypleus tridentatus. EMBO J. 1999;18:2313–2322. doi: 10.1093/emboj/18.9.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bench AJ, Nacheva EP, Hood TL, Holden JL, French L, Swanton S, Champion KM, Li J, Whittaker P, Stavrides G, et al. Chromosome 20 deletions in myeloid malignancies: reduction of the common deleted region, generation of a PAC/BAC contig and identification of candidate genes. UK Cancer Cytogenetics Group (UKCCG) Oncogene. 2000;19:3902–3913. doi: 10.1038/sj.onc.1203728. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein PR, Gomes BC, Kosmider BJ, Vacek EP, Williams JC. Nonpeptidic inhibitors of human leukocyte elastase. 6. Design of a potent, intratracheally active, pyridone-based trifluoromethyl ketone. J. Med. Chem. 1995;38:212–215. doi: 10.1021/jm00001a028. [DOI] [PubMed] [Google Scholar]
- Boccuni P, MacGrogan D, Scandura JM, Nimer SD. The human L(3)MBT polycomb group protein is a transcriptional repressor and interacts physically and functionally with TEL (ETV6) J. Biol. Chem. 2003;22:15412–15420. doi: 10.1074/jbc.M300592200. [DOI] [PubMed] [Google Scholar]
- Bornemann D, Miller E, Simon J. The Drosophila Polycomb group gene Sex comb on midleg (Scm) encodes a zinc finger protein with similarity to polyhomeotic protein. Development. 1996;122:1621–1630. doi: 10.1242/dev.122.5.1621. [DOI] [PubMed] [Google Scholar]
- Bornemann D, Miller E, Simon J. Expression and properties of wild-type and mutant forms of the Drosophila sex comb on midleg (SCM) repressor protein. Genetics. 1998;150:675–686. doi: 10.1093/genetics/150.2.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen TR, Duncan IM. Maternal expression of genes that regulate the bithorax complex of Drosophila melanogaster. Dev. Biol. 1986;118:442–456. doi: 10.1016/0012-1606(86)90015-1. [DOI] [PubMed] [Google Scholar]
- Brown K, Pompeo F, Dixon S, Mengin-Lecreulx D, Cambillau C, Bourne Y. Crystal structure of the bifunctional N-acetylglucosamine 1-phosphate uridyltransferase from Escherichia coli: a paradigm for the related pyrophosphorylase superfamily. EMBO J. 1999;18:4096–4107. doi: 10.1093/emboj/18.15.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunker CA, Kingston RE. Transcriptional repression by Drosophila and mammalian Polycomb group proteins in transfected mammalian cells. Mol. Cell. Biol. 1994;14:1721–1732. doi: 10.1128/mcb.14.3.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambillau C, Roussel A. Turbo Frodo, version OpenGL.1. Marseille, France: University Aix-Marseille II; 1997. [Google Scholar]
- Carson M. Ribbons. Methods Enzymol. 1997;277:493–505. [PubMed] [Google Scholar]
- (CCP4) Collaborative Computational Project 4. The CCP4 suite: programs for protein crystallography, Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Eissenberg JC. Molecular biology of the chromo domain: an ancient chromatin module comes of age. Gene. 2001;275:19–29. doi: 10.1016/s0378-1119(01)00628-x. [DOI] [PubMed] [Google Scholar]
- Fitzgerald PM, Wu JK, Toney JH. Unanticipated inhibition of the metallo-beta-lactamase from Bacteroides fragilis by 4-morpholineethanesulfonic acid (MES): a crystallographic study at 1.85-Å resolution. Biochemistry. 1998;37:6791–6800. doi: 10.1021/bi9730339. [DOI] [PubMed] [Google Scholar]
- Focia PJ, Craig SP, 3rd, Nieves-Alicea R, Fletterick RJ, Eakin AE. A 1.4 Å crystal structure for the hypoxanthine phosphoribosyltransferase of Trypanosoma cruzi. Biochemistry. 1998;37:15066–15075. doi: 10.1021/bi981052s. [DOI] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE. Mechanisms of transcriptional memory. Nat. Rev. Mol. Cell Biol. 2001;2:409–421. doi: 10.1038/35073039. [DOI] [PubMed] [Google Scholar]
- Fulop V, Jones DT. Beta propellers: structural rigidity and functional diversity. Curr. Opin. Struct. Biol. 1999;9:715–721. doi: 10.1016/s0959-440x(99)00035-4. [DOI] [PubMed] [Google Scholar]
- Fulop V, Moir JW, Ferguson SJ, Hajdu J. The anatomy of a bifunctional enzyme: structural basis for reduction of oxygen to water and synthesis of nitric oxide by cytochrome cd1. Cell. 1995;81:369–377. doi: 10.1016/0092-8674(95)90390-9. [DOI] [PubMed] [Google Scholar]
- Fulop V, Bocskei Z, Polgar L. Prolyl oligopeptidase: an unusual beta-propeller domain regulates proteolysis. Cell. 1998;94:161–170. doi: 10.1016/s0092-8674(00)81416-6. [DOI] [PubMed] [Google Scholar]
- Gateff E, Loffler T, Wismar J. A temperature-sensitive brain tumor suppressor mutation of Drosophila melanogaster: developmental studies and molecular localization of the gene. Mech. Dev. 1993;41:15–31. doi: 10.1016/0925-4773(93)90052-y. [DOI] [PubMed] [Google Scholar]
- Hohenester E, Keller JW, Jansonius JN. An alkali metal ion size-dependent switch in the active site structure of dialkylglycine decarboxylase. Biochemistry. 1994;33:13561–13570. doi: 10.1021/bi00250a008. [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- Jones KW, Gorzynski K, Hales CM, Fischer U, Badbanchi F, Terns RM, Terns MP. Direct interaction of the spinal muscular atrophy disease protein SMN with the small nucleolar RNA-associated protein fibrillarin. J. Biol. Chem. 2001;276:38645–38651. doi: 10.1074/jbc.M106161200. [DOI] [PubMed] [Google Scholar]
- Knochel TR, Hennig M, Merz A, Darimont B, Kirschner K, Jansonius JN. The crystal structure of indole-3-glycerol phosphate synthase from the hyperthermophilic archaeon Sulfolobus solfataricus in three different crystal forms: effects of ionic strength. J. Mol. Biol. 1996;262:502–515. doi: 10.1006/jmbi.1996.0531. [DOI] [PubMed] [Google Scholar]
- Koga H, Matsui S, Hirota T, Takebayashi S, Okumura K, Saya H. A human homolog of Drosophila lethal(3)malignant brain tumor (l(3)mbt) protein associates with condensed mitotic chromosomes. Oncogene. 1999;18:3799–3809. doi: 10.1038/sj.onc.1202732. [DOI] [PubMed] [Google Scholar]
- Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- Kurtin PJ, Dewald GW, Shields DJ, Hanson CA. Hematologic disorders associated with deletions of chromosome 20q: a clinicopathologic study of 107 patients. Am. J. Clin. Pathol. 1996;106:680–688. doi: 10.1093/ajcp/106.5.680. [DOI] [PubMed] [Google Scholar]
- Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Lambright DG, Sondek J, Bohm A, Skiba NP, Hamm HE, Sigler PB. The 2.0 Å crystal structure of a heterotrimeric G protein. Nature. 1996;379:311–319. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- MacGrogan D, Alvarez S, DeBlasio T, Jhanwar SC, Nimer SD. Identification of candidate genes on chromosome band 20q12 by physical mapping of translocation breakpoints found in myeloid leukemia cell lines. Oncogene. 2001;20:4150–4160. doi: 10.1038/sj.onc.1204540. [DOI] [PubMed] [Google Scholar]
- Maurer-Stroh S, Dickens NJ, Hughes-Davies L, Kouzarides T, Eisenhaber F, Ponting CP. The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem. Sci. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- Mori N, Morosetti R, Lee S, Spira S, Ben-Yehuda D, Schiller G, Landolfi R, Mizoguchi H, Koeffler HP. Allelotype analysis in the evolution of chronic myelocytic leukemia. Blood. 1997;90:2010–2014. [PubMed] [Google Scholar]
- Mujtaba S, He Y, Zeng L, Farooq A, Carlson JE, Ott M, Verdin E, Zhou MM. Structural basis of lysine-acetylated HIV-1 Tat recognition by PCAF bromodomain. Mol. Cell. 2002;9:575–586. doi: 10.1016/s1097-2765(02)00483-5. [DOI] [PubMed] [Google Scholar]
- Muller J. Transcriptional silencing by the Polycomb protein in Drosophila embryos. EMBO J. 1995;14:1209–1220. doi: 10.1002/j.1460-2075.1995.tb07104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Navaza J. Implementation of molecular replacement in AMoRe. Acta Crystallogr. D Biol. Crystallogr. 2001;57:1367–1372. doi: 10.1107/s0907444901012422. [DOI] [PubMed] [Google Scholar]
- Neer EJ, Smith TF. G protein heterodimers: new structures propel new questions. Cell. 1996;84:175–178. doi: 10.1016/s0092-8674(00)80969-1. [DOI] [PubMed] [Google Scholar]
- Neer EJ, Schmidt CJ, Nambudripad R, Smith TF. The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Nielsen PR, Nietlispach D, Mott HR, Callaghan J, Bannister A, Kouzarides T, Murzin AG, Murzina NV, Laue ED. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature. 2002;416:103–107. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Paoli M. Protein folds propelled by diversity. Prog. Biophys. Mol. Biol. 2001;76:103–130. doi: 10.1016/s0079-6107(01)00007-4. [DOI] [PubMed] [Google Scholar]
- Peterson AJ, Kyba M, Bornemann D, Morgan K, Brock HW, Simon J. A domain shared by the Polycomb group proteins Scm and ph mediates heterotypic and homotypic interactions. Mol. Cell. Biol. 1997;17:6683–6692. doi: 10.1128/mcb.17.11.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V. Chromatin-silencing mechanisms in Drosophila maintain patterns of gene expression. Trends Genet. 1997a;13:314–318. doi: 10.1016/s0168-9525(97)01178-5. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. PcG complexes and chromatin silencing. Curr. Opin. Genet. Dev. 1997b;7:249–258. doi: 10.1016/s0959-437x(97)80135-9. [DOI] [PubMed] [Google Scholar]
- Ponstingl H, Henrick K, Thornton JM. Discriminating between homodimeric and monomeric proteins in the crystalline state. Proteins. 2000;41:47–57. doi: 10.1002/1097-0134(20001001)41:1<47::aid-prot80>3.3.co;2-#. [DOI] [PubMed] [Google Scholar]
- Poux S, McCabe D, Pirrotta V. Recruitment of components of Polycomb Group chromatin complexes in Drosophila. Development. 2001;128:75–85. doi: 10.1242/dev.128.1.75. [DOI] [PubMed] [Google Scholar]
- Renault L, Nassar N, Vetter I, Becker J, Klebe C, Roth M, Wittinghofer A. The 1.7 Å crystal structure of the regulator of chromosome condensation (RCC1) reveals a seven-bladed propeller. Nature. 1998;392:97–101. doi: 10.1038/32204. [DOI] [PubMed] [Google Scholar]
- Sawaya MR, Guo S, Tabor S, Richardson CC, Ellenberger T. Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7. Cell. 1999;99:167–177. doi: 10.1016/s0092-8674(00)81648-7. [DOI] [PubMed] [Google Scholar]
- Scheffzek K, Ahmadian MR, Kabsch W, Wiesmuller L, Laut-wein A, Schmitz F, Wittinghofer A. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- Selenko P, Sprangers R, Stier G, Buhler D, Fischer U, Sattler M. SMN tudor domain structure and its interaction with the Sm proteins. Nat. Struct. Biol. 2001;8:27–31. doi: 10.1038/83014. [DOI] [PubMed] [Google Scholar]
- Simon J. Locking in stable states of gene expression: transcriptional control during Drosophila development. Curr. Opin. Cell Biol. 1995;7:376–385. doi: 10.1016/0955-0674(95)80093-x. [DOI] [PubMed] [Google Scholar]
- Simon JA, Tamkun JW. Programming off and on states in chromatin: mechanisms of Polycomb and trithorax group complexes. Curr. Opin. Genet. Dev. 2002;12:210–218. doi: 10.1016/s0959-437x(02)00288-5. [DOI] [PubMed] [Google Scholar]
- Simon J, Chiang A, Bender W. Ten different Polycomb group genes are required for spatial control of the abdA and AbdB homeotic products. Development. 1992;114:493–505. doi: 10.1242/dev.114.2.493. [DOI] [PubMed] [Google Scholar]
- Struhl G, Akam M. Altered distributions of Ultrabithorax transcripts in extra sex combs mutant embryos of Drosophila. EMBO J. 1985;4:3259–3264. doi: 10.1002/j.1460-2075.1985.tb04075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplyakov A, Obmolova G, Badet-Denisot MA, Badet B, Polikarpov I. Involvement of the C terminus in intramolecular nitrogen channeling in glucosamine 6-phosphate synthase: evidence from a 1.6 Å crystal structure of the isomerase domain. Structure. 1998;6:1047–1055. doi: 10.1016/s0969-2126(98)00105-1. [DOI] [PubMed] [Google Scholar]
- ter Haar E, Musacchio A, Harrison SC, Kirchhausen T. Atomic structure of clathrin: a beta propeller terminal domain joins an alpha zigzag linker. Cell. 1998;95:563–573. doi: 10.1016/s0092-8674(00)81623-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui H, Ichikawa T, Kobayashi K, Kumanishi T. Cloning of a novel murine gene Sfmbt, Scm-related gene containing four mbt domains, structurally belonging to the Polycomb group of genes. Gene. 2000;248:127–135. doi: 10.1016/s0378-1119(00)00131-1. [DOI] [PubMed] [Google Scholar]
- Wedeen C, Harding K, Levine M. Spatial regulation of Antennapedia and bithorax gene expression by the Polycomb locus in Drosophila. Cell. 1986;44:739–748. doi: 10.1016/0092-8674(86)90840-8. [DOI] [PubMed] [Google Scholar]
- White NJ, Nacheva E, Asimakopoulos FA, Bloxham D, Paul B, Green AR. Deletion of chromosome 20q in myelodysplasia can occur in a multipotent precursor of both myeloid cells and B cells. Blood. 1994;83:2809–2816. [PubMed] [Google Scholar]
- Wismar J. Molecular characterization of h-l(3)mbt-like: a new member of the human mbt family. FEBS Lett. 2001;507:119–121. doi: 10.1016/s0014-5793(01)02959-3. [DOI] [PubMed] [Google Scholar]
- Wismar J, Loffler T, Habtemichael N, Vef O, Geissen M, Zirwes R, Altmeyer W, Sass H, Gateff E. The Drosophila melanogaster tumor suppressor gene lethal(3)malignant brain tumor encodes a proline-rich protein with a novel zinc finger. Mech. Dev. 1995;53:141–154. doi: 10.1016/0925-4773(95)00431-9. [DOI] [PubMed] [Google Scholar]