Abstract
The nucleotide excision repair (NER) machinery excises a variety of bulky DNA lesions, but with varying efficiencies. The structural features of the DNA lesions that govern these differences are not well understood. An intriguing model system for studying structure–function relationships in NER is the major adduct derived from the reaction of the highly tumorigenic metabolite of benzo[a]pyrene, (+)-anti-benzo[a]pyrene diol epoxide, with the exocyclic amino group of guanine ((+)-trans-anti-[BP]-N2-dG, or G*). The rates of incision of the stereochemically identical lesions catalyzed by the prokaryotic UvrABC system was shown to be greater by a factor of 2.3±0.3 in the TG*T than in the CG*C sequence context [Biochemistry 46 (2007) 7006–7015]. Here we employ molecular dynamics simulations to elucidate the origin of the greater excision efficiency in the TG*T case and, more broadly, to delineate structural parameters that enhance NER. Our results show that the BP aromatic ring system is 5′-directed along the modified strand in the B-DNA minor groove in both sequence contexts. However, the TG*T modified duplex is much more dynamically flexible, featuring more perturbed and mobile Watson–Crick hydrogen bonding adjacent to the lesion, a greater impairment in stacking interactions, more dynamic local roll/bending, and more minor groove flexibility. These characteristics explain a number of experimental observations concerning the (+)-trans-anti-[BP]-N2-dG adduct in double-stranded DNA with the TG*T sequence context: its conformational heterogeneity in NMR solution studies, its highly flexible bend, and its lower thermal stability. By contrast, the CG*C modified duplex is characterized by a single BP conformation and a rigid bend. While current recognition models of bulky lesions by NER factors have stressed the importance of impaired Watson–Crick pairing/stacking and bending, our results highlight the likelihood of an important role for the local dynamics in the vicinity of the lesion.
Keywords: DNA repair, carcinogenesis, benzo[a]pyrenyl–guanine lesion, sequence-dependent mobility, molecular dynamics
Introduction
Nucleotide excision repair (NER) is one of the most important mammalian defense mechanisms against bulky promutagenic DNA lesions and is critically important for maintaining the integrity of DNA and preventing carcinogenesis. If this form of DNA repair is successful, there are no further consequences. However, if the lesions are resistant to NER, they may survive until DNA replication occurs; error-prone translesion bypass, causing mutations and potentially cancer, may result (e.g., Refs. [1,2]). The basic hypothesis in the field of NER is that the mammalian DNA repair apparatus recognizes structural distortions and thermodynamic destabilization in the DNA duplex caused by the lesions, followed by a verification step that detects the presence of a chemically modified nucleobase and the excision of a fragment 24–32 nucleotides long that contains the damaged base.3–11 It is now established that the first and rate-determining step in NER is the recognition of the bulky lesions by the XPC/HR23B protein heterodimer complex.12 Since the NER machinery processes a diverse array of bulky lesions with different efficiencies, the conformational features that invoke NER are of great interest. Disturbed Watson–Crick pairing, impaired base stacking, and DNA bending are among the recognition hallmarks that have been proposed.13–16
NMR solution studies of bulky carcinogen–DNA adducts have provided considerable insights into adduct conformations.17,18 An intriguing finding is the importance of base sequence context in governing the conformations of such DNA adducts. Indeed, equilibria between different adduct conformations can be governed by the sequences of adjacent base pairs, and even next nearest neighbors can influence the balance of populations in different conformers.17,19–21 Thus, the biological processing of such lesions by DNA polymerases and the cellular DNA repair machinery may be determined by the sequence-dependent conformational state of the DNA lesion.20,22–24 The relative rate of NER of a DNA adduct and therefore its mutagenic potential can depend on the nature of the neighboring bases, producing mutational hotspots and coldspots.17,19,20,22–25
Remarkable sequence-dependent conformational equilibria have been noted in solution NMR studies17 of mutagenic DNA adducts derived from the polycyclic aromatic hydrocarbon benzo[a]pyrene (BP), an environmental precarcinogen.1 Particularly intriguing are the properties of the major 10S (+)-trans-anti-[BP]-N2-dG adduct (G*) (Fig. 1) derived from the reaction of the most tumorigenic metabolite of BP,26 (+)-(7R,8S,9S,10R)-7,8-dihydroxy-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene, with the exocyclic amino group of guanine in DNA.27 For this adduct in a CG*C sequence context in a DNA 11-mer duplex, the BP rings reside in the DNA minor groove (MG), 5′-directed along the modified strand with no evidence for any conformational heterogeneity.28 However, in the TG*T sequence context, a similar MG motif is dominant, but a full structural characterization by NMR was not feasible due to severe conformational heterogeneity.29 In the TG*C sequence context, the dominant conformation is of the same MG type, but a minor conformation may involve insertion of the BP moiety into the duplex with disruption of Watson–Crick pairing at the lesion site.30 More subtle sequence effects on the MG conformations of the 10S (+)-trans-anti-[BP]-N2-dG adduct in 5′-CG*G and 5′-GG*C sequence contexts have been recently reported31 and have been attributed to the effects of steric hindrance between the exocyclic amino groups of guanines and the BP residues that compete for the same space in the MG. Interestingly, for the same 10S adduct in a CG*C sequence context in an 11/10-mer duplex in which the G* had no partner, a base-displaced/intercalated (BD) conformation was adopted in which the BP rings were inserted into the helix with the modified guanine residue, as well as the puckered benzylic ring positioned on the major groove side of the helix.32
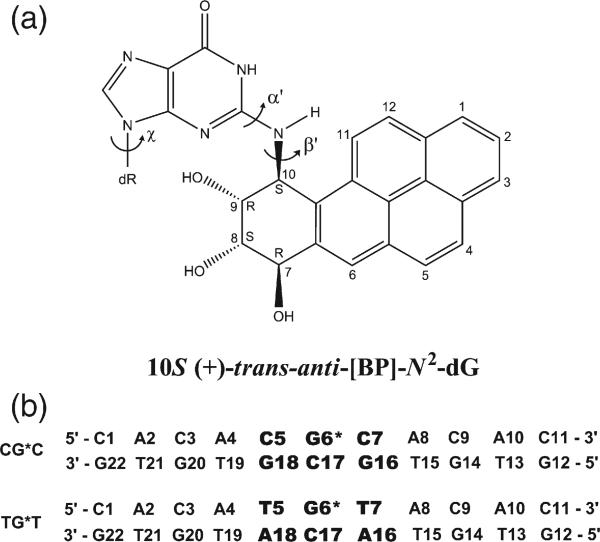
Fig. 1.
(a) Structure of the 10S (+)-trans-anti-[BP]-N2-dG adduct. The torsion angles α′ and β′ are defined as follows: α′, N1-C2-N2-C10(BP); β′, C2-N2-C10(BP)-C9 (BP). χ is the glycosidic torsion angle, defined as O4′-C1′-N9-C4. (b) Sequence context, where G6* represents the lesion-modified guanine.
The CG*C and TG*T sequences are of unusual interest because the nature of the flanking bases affects not only the adduct conformations in double-stranded DNA, but also a number of other properties. The thermodynamic characteristics in the two sequence contexts differ: as expected from the thermodynamic properties of T:A and C:G base pairs,33,34 the TG*T 11-mer duplex has a lower thermal melting point than the 11-mer CG*C duplex (by about 14–15 °C).25 Furthermore, gel electrophoresis studies have shown that 11-mer CG*C duplexes are characterized by bends that are comparatively rigid, whereas the bends in the TG*T duplexes are more flexible.35 In the TG*T case, small minicircles 77–144 bp in size are formed in self-ligation experiments of 11-mer duplexes catalyzed by T4 ligase.29 While similar-size minicircle distributions are observed in analogous 11-mer CG*C self-ligation experiments, the yield of minicircles is approximately three times lower (R. Xu and N. E. Geacintov, unpublished observations), consistent with the higher rigidity of the bends in CG*C.29,35
NER studies in vitro have revealed that the lesions are excised with greater efficiency (by factors of 2.3±0.3) by prokaryotic UvrABC proteins in the TG*T than in the CG*C sequence contexts in fully complementary 43-mer duplexes.25 The UvrABC proteins in this study came from the thermophilic Bacillus caldotenax, which allowed a detailed investigation of the effect of the temperature on incision efficiency. The enhanced efficiency factor of 2.3±0.3 was observed at 37 °C.25 This finding represents a case where the same lesion is excised with different efficiencies in two different, natural DNA sequence contexts. A similar sequence context dependence was observed in the excision of C8-guanine adducts of 2-aminofluorene and N-acetyl-2-aminofluorene catalyzed by UvrABC from Escherichia coli,22 and other base sequence effects on the excision of these lesions have also been reported.23 Ruan et al. attributed their observed sequence effects to the overall weaker base stacking and weaker hydrogen bonding interactions associated with A:T rather than G:C base pairs flanking the lesion.25
The sequence-dependent differences in NER excision efficiencies provide an excellent opportunity for analyzing in detail the perturbations in structural and dynamic properties of the DNA caused by these lesions. Our goal is to identify those factors, or combination of factors, that account for the higher susceptibilities of TG*T relative to CG*C duplex sequences to prokaryotic NER. Conformational heterogeneity has precluded full NMR characterization of the TG*T duplex.29 More broadly, using this interesting pair of damaged sequences as a model, we wished to attain new insights on the structural distinctions that serve as signals of differential recognition and/or subsequent processing of lesions by the prokaryotic NER machinery.
A combination of molecular modeling, molecular dynamics (MD) simulations, and free energy calculations was employed to address these questions. The 10S (+)-trans-anti-[BP]-N2-dG adducts are known to be more susceptible to NER by prokaryotic36 and eukaryotic16 DNA repair factors when they are in BD rather than in MG conformations in CG*C sequence contexts. Furthermore, in the case of a 2-aminofluorene-C8-guanine adduct, BD conformations have recently been shown to manifest greater susceptibility to prokaryotic NER, as compared to orientations of the adduct in a groove of B-DNA.24 We therefore explored the hypothesis that a contribution to the population balance of a BD conformation in the TG*T context accounts for the higher susceptibility to NER of the TG*T duplex sequence. Surprisingly, this appears not to be the case, since we find that both sequences strongly favor MG conformations with, however, significantly greater dynamic flexibility of the duplex in the region of the lesion in the TG*T case. We propose that this higher dynamic flexibility in the TG*T sequence context plays a significant role in its greater susceptibility to prokaryotic NER incision efficiency. More generally, greater dynamic flexibility of lesions may enhance their NER susceptibility.
Results
Using the AMBER 8.0 simulation package,37 we have carried out 10.0 ns MD simulations for the 10S (+)-trans-anti-[BP]-N2-dG adduct in both the CG*C and TG*T 11-mer duplex sequences, respectively (Fig. 1). Both MG and BD conformers were considered in each case. In addition, we studied the unmodified control CGC and TGT 11-mer duplexes containing a normal G:C base pair instead of the modified G*:C pair. Detailed analyses of the MD-generated ensembles were performed to delineate conformational and dynamic characteristics of each modified and unmodified duplex. Specifically, our objectives were to determine the conformational origins of the differences in NER susceptibilities between the two modified sequence contexts.25
Minor groove conformers are favored in both duplex sequence contexts
Since BD conformers are highly susceptible to excision in prokaryotic24,36 and human16 NER assays, we investigated the population balance between MG and BD conformations in the two sequence contexts. For the thermodynamic analyses, we employed the ensembles of structures derived from the last 7.0 ns of the MD simulations, using the molecular mechanics Poisson–Boltzmann surface area (MM-PBSA) methodology38 in the AMBER 8.0 simulation package.37 To estimate the relative stability of the MG and BD conformational families, we calculated the relative free energies ΔG of the two conformational states in the CG*C and TG*T sequence contexts. Table 1 gives the ΔG values, and Table S1, Supplementary Data, provides full thermodynamic analyses. Our results indicate that the MG conformers are favored energetically in both the CG*C and TG*T 11-mer duplex sequences by 5.9±1.1 kcal mol−1 in the case of CG*C, and by 4.7±1.0 kcal mol−1 in the case of TG*T. This is consistent with the NMR data for CG*C,28 which showed a single MG conformation, as well as for TG*T29 where a structurally uncharacterized MG conformer predominated. Our calculations indicate no substantial contribution to the population balance by a BD conformation. Therefore, since the MG conformations are favored in both sequence contexts, only these conformations will be considered in the following sections.
Table 1.
Relative free energies for minor groove (MG) and base-displaced/intercalated (BD) conformations
| ΔG (kcal·mol–1) | |||
|---|---|---|---|
| CG*C |
TG*T |
||
| MG | BD | MG | BD |
| 0 | 5.9 (1.1) | 0 | 4.7 (1.0) |
For each sequence context, the conformation with the lower free energy is assigned a ΔG of 0. See Table S1, Supplementary Data, for full MM-PBSA analyses over the 3.0–10.0 ns windows. Errors of means (defined in Table S1), given in parentheses, are based on Table S1.
The BP ring systems positioned in the MG in TG*T are more flexible than in the CG*C duplexes
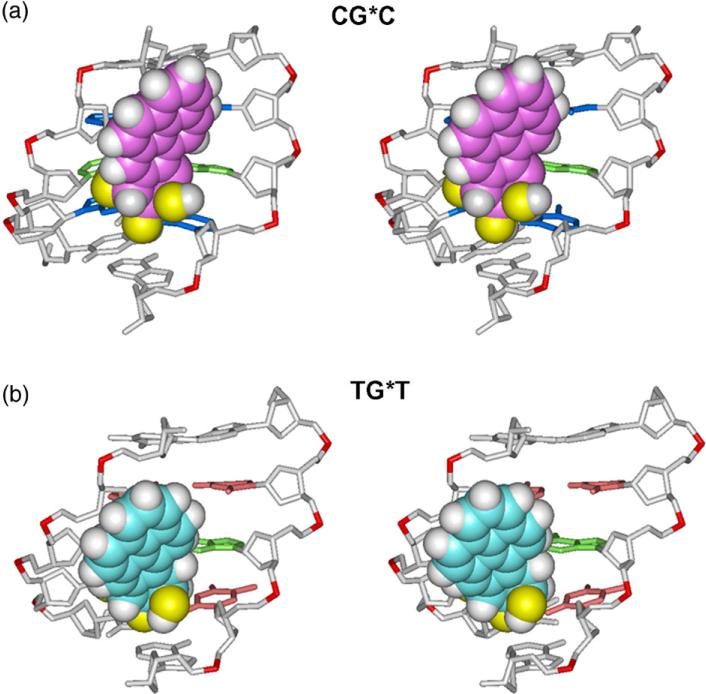
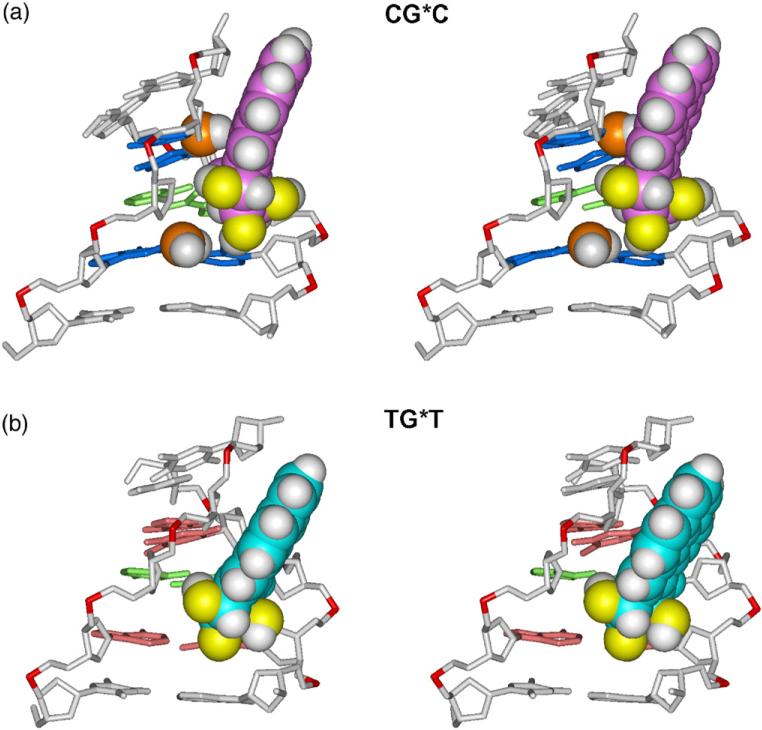
The best representative structures of the 10S (+)-trans-anti-[BP]-N2-dG adduct are of the MG type in both sequence contexts and are shown in Fig. 2. The best representative structure is a real frame from the ensemble, as determined by a cluster analysis, and represents the most populated conformation or the conformation that is the closest to all other snapshots in the ensemble.39 The views in Fig. 2 are into the MG of the central 5-mer segments of CG*C and TG*T. In both cases, the BP moiety in the B-DNA MG is 5′-directed along the modified strand. However, there are subtle differences in the orientations of the BP ring systems: in the case of CG*C, the BP rings are rigidly positioned to avoid collisions with the exocyclic amino groups of the guanines from the complementary strand (Fig. 3). In contrast, in the case of TG*T there are no nearby exocyclic amino groups in the MG and the BP rings have more conformational freedom (Fig. 3).
Fig. 2.
Stereo views of the 10S (+)-trans-anti-[BP]-N2-dG adduct looking into the minor groove in the (a) CG*C and (b) TG*T sequence contexts. The structures shown are the best representative conformations39 for the last 7.0 ns of the MD simulations. Only the central 5-mers are shown. In the CG*C sequence context, for the BP moiety, the carbon atoms are magenta, oxygen atoms yellow, and hydrogen atoms white. The modified guanine and its partner cytosine are green. The flanking base pairs are marine. In the TG*T sequence context, for the BP moiety, the carbon atoms are cyan, oxygen atoms yellow, and hydrogen atoms white. The modified guanine and its partner cytosine are green. The flanking base pairs are pink. The DNA duplexes are white, except for the phosphorus atoms, which are red. Hydrogen atoms and pendant phosphate oxygen atoms in the DNA duplexes are not displayed. The BP moieties are in CPK representation. The backbones of the duplexes were aligned to be optimally superimposed in order to highlight the different orientations of the BP rings.
Fig. 3.
Effect of guanine amino groups in the minor groove on positioning of the 10S (+)-trans-anti-[BP]-N2-dG adduct in (a) CG*C and (b) TG*T sequence contexts. Only the central 5-mers are shown. The structures shown are the best representative conformations39 from the last 7.0 ns of the MD simulation. The BP moiety and relevant guanine amino groups are in CPK representation. Hydrogen atoms in the rest of the DNA duplexes are not displayed. The color scheme is the same as in Fig. 2, except that guanine amino groups are in orange (N) and white (H). Supplementary data movie_a and movie_b show these structures as they statically rotate, and movie_c and movie_d show their dynamics.
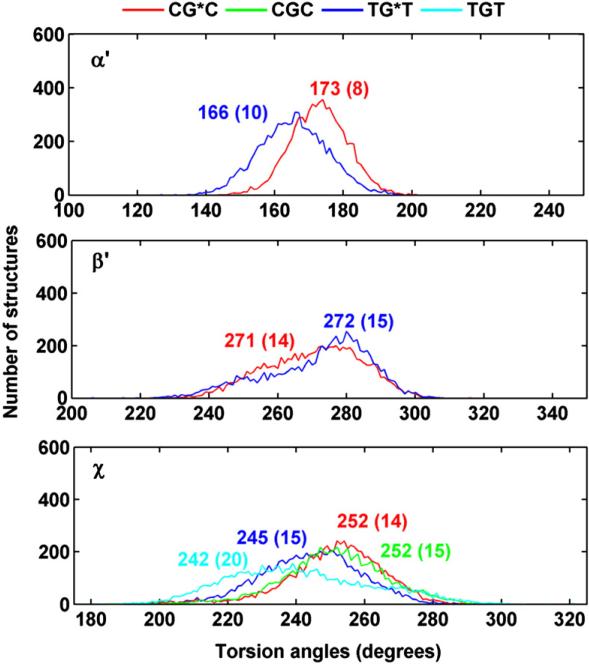
The differential positioning of the BP moieties in the CG*C and TG*T sequence contexts is manifested in differences in the carcinogen–DNA linkage site torsion angle α′ and the glycosidic torsion angle χ (Fig. 1a). The distributions in populations of all three torsion angles α′, β′, and χ (Fig. 1) are shown in Fig. 4, and the time dependence of each is given in Fig. S1, Supplementary Data. The torsion angle α′ (Fig. 1a) in TG*T is somewhat more flexible, as shown by the broader population distribution and greater standard deviation from the mean value (Fig. 4 and Fig. S1, Supplementary Data); this difference reflects a less rigid placement of the BP moiety in the TG*T than in the CG*C case.
Fig. 4.
Population distribution of the carcinogen–DNA linkage site and glycosidic torsion angles α′, β′ and χ. CG*C is red and TG*T is blue; CGC is green and TGT is cyan. Ensemble averages and standard deviations (in parentheses) are given. Time dependence of these torsion angles are given in Fig. S1, Supplementary Data.
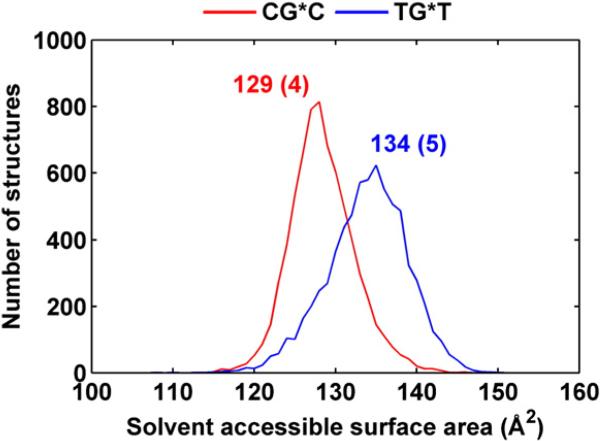
The glycosidic torsion angle χ in TG*T is also more disturbed relative to the unmodified sequence than in CG*C, primarily because of the greater flexibility. Reflecting in part the orientational difference, the solvent-accessible surface area (SASA) of the BP aromatic rings is ~5 Å2 greater in TG*T than in CG*C. Shielding of the BP rings by the amino groups in CG*C (Fig. 3) also contributes to the lower SASA value. Figure 5 shows the population distributions and Fig. S2, Supplementary Data, shows the time dependence of the SASA.
Fig. 5.
Population distribution of the SASA of the BP moiety for CG*C (red) and TG*T (blue) sequence contexts. Ensemble averages and standard deviations (in parentheses) are given.
Watson–Crick hydrogen bonding is more disturbed and dynamically flexible in TG*T than in CG*C
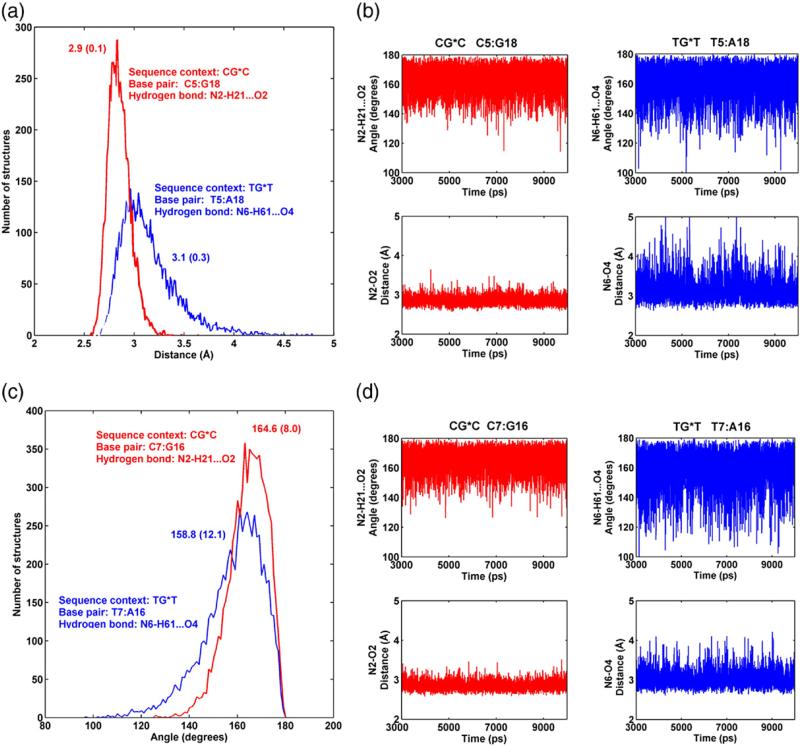
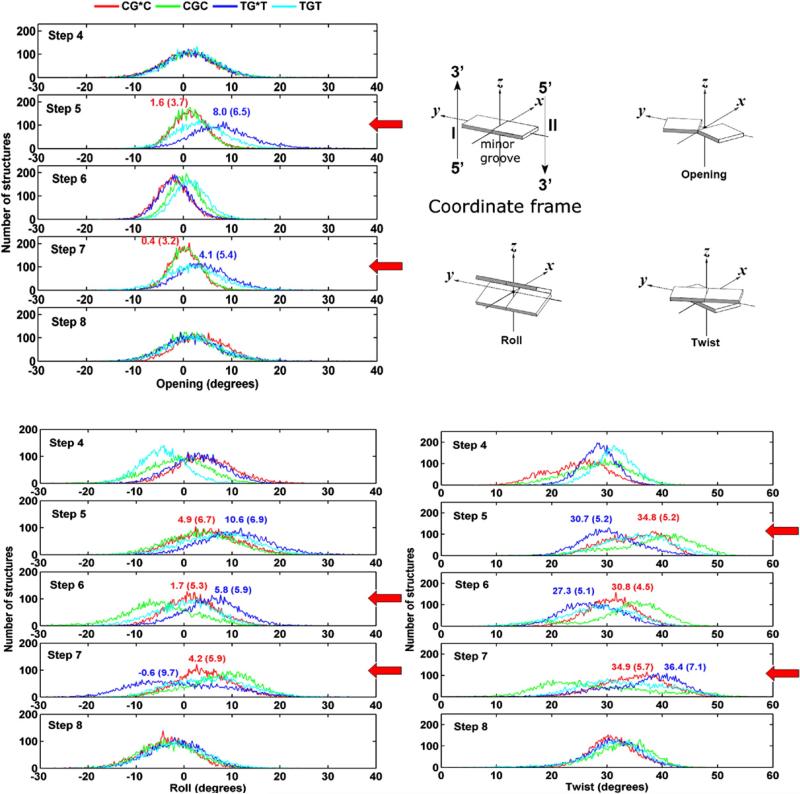
Our results show that the T5:A18 pair in TG*T is perturbed, while the analogous C5:G18 pair is much less altered, as compared to their respective unmodified controls (Table 2). Specifically, it is the (T5) N6-H6···O4 (A18) hydrogen bond, on the major groove side that is disrupted in 23% of the population, while all the hydrogen bonds at C5:G18 are intact in over 95% of the population (Table 2). In addition, the (T7) N6-H61···O4 (A16) hydrogen bond on the 3′ side of the lesion in TG*T was disrupted 10% of the population (Table 2). For the unmodified cases, all the base pairs are intact in over 96% of the population for CGC and 93% for TGT (Table 2). Furthermore, Fig. 6 shows that Watson–Crick hydrogen bonding is more dynamic at steps 5 and 7 in the case of TG*T. The time dependence of all hydrogen bonding distances and angles at/near the lesion site is given in Fig. S3, Supplementary Data. Correlated with the more dynamic Watson–Crick pairing, the parameter Opening (see Fig. 7 for definition) is greater and more dynamic for the T: A pairs flanking the lesion in TG*T than for the lesion-flanking C:G pairs in CG*C, as shown in Fig. 7 and Fig. S4 and Table S2, Supplementary Data. Moreover, Opening is greatest for the most disrupted T5:A18 pair and the positive value reflects the disturbance at the (T5) N6-H6···O4 (A18) hydrogen bond on the major groove side. Opening towards the major groove is also greater for the unmodified TGT sequence at the T:A base pairs, but to a lesser extent than in the modified case (Fig. 7).
Table 2.
Disruption of Watson–Crick base pairs near the lesion site for the CG*C, CGC, TG*T, and TGT sequence contexts
| Sequence context | Watson–Crick base pair | Hydrogen bond | % of population disrupted |
|---|---|---|---|
| TG*T | T5:A18 | N6-H61···O4 | ~23 |
| N3-H3···N1 | ~1 | ||
| T7:A16 | N6-H61···O4 | ~10 | |
| N3-H3···N1 | ~1 | ||
| TGT | T5:A18 | All | <7 |
| T7:A16 | All | <4 | |
| CG*C | C5:G18 | All | <5 |
| C7:G16 | All | <2 | |
| CGC | C5:G18 | All | <4 |
| C7:G16 | All | <3 |
Criteria for disruption of Watson–Crick base pairs: heavy atom–heavy atom distance >3.3 Å or donor-hydrogen-acceptor angle <140°. The percent of disrupted populations was computed from data in Fig. 6 and Fig. S3, Supplementary Data.
Fig. 6.
(a) Population distribution and (b) time dependence of hydrogen bond distances and angles for base pair 5:18; (c) population distribution and (d) time dependence of hydrogen bond distances and angles for base pair 7:16. CG*C is red and TG*T is blue. Ensemble averages and standard deviations (in parentheses) are given for (a) and (c). Only the hydrogen bond on the major groove side for either step is shown. Full analyses of hydrogen bonds at the lesion site for CG*C and TG*T and their unmodified sequence contexts are shown in Fig. S3, Supplementary Data.
Fig. 7.
Population distribution of Opening, Roll, and Twist near/at the lesion site for CG*C, CGC, TG*T, and TGT. Illustrations are adapted from 69. For Opening, C1:G22 is step 1, A2:T21 is step 2, and so on; for Roll and Twist, C1:G22 to A2:T21 is step 1, A2:T21 to C3:G20 is step 2, and so on. The most significant differences are indicated with a red arrow. Ensemble averages and standard deviations (in parentheses) are given.
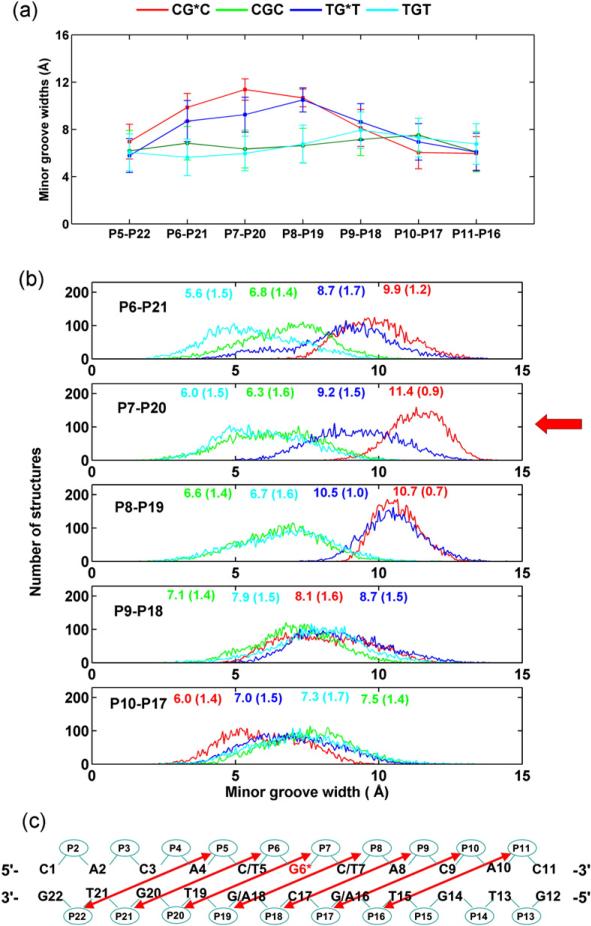
The MG width near the lesion site is more dynamic in TG*T, although greater in CG*C
The BP rings cause MG enlargement in both sequence contexts compared to their unmodified controls (Fig. 8). However, the local enlargement near the lesion is greater in CG*C (11.4±0.9 Å) than in TG*T (9.2±1.5 Å) (Fig. 8). On the other hand, the time dependence (Fig. S5, Supplementary Data) and the population distribution data (Fig. 8) for the MG widths reveal that the MG width at the lesion site (P7–P20) is more dynamic in the TG*T than in the CG*C sequence context. The smaller MG widths and larger flexibility at the lesion site in TG*T correlate with its larger and more dynamic Opening into the major groove and its disturbed (T5) N6-H6···O4 (A18) hydrogen bond on the major groove side; as Opening on the major groove side increases, the MG, on the opposite side, becomes narrower.
Fig. 8.
(a) Trajectory average MG widths of the modified duplexes and unmodified controls. (b) Population distribution of the MG width near/at the lesion site for CG*C, CGC, TG*T, and TGT. Ensemble averages and standard deviations (in parentheses) are given. (c) MG widths are the distance between P5 and P22, P6 and P21, P7 and P20, and so on, less 5.8 Å to account for the van der Waals radius of the phosphorus atoms.67 The most significant difference is indicated with a red arrow.
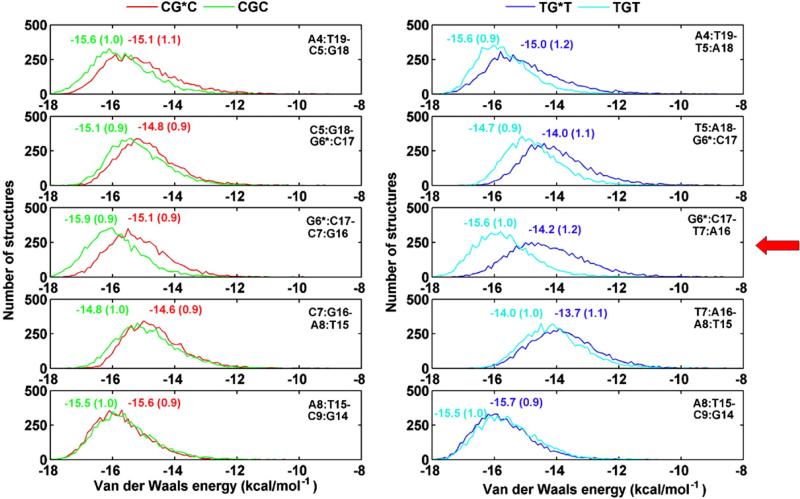
Base stacking is more perturbed in TG*T than in CG*C, allowing for greater flexibility
The van der Waals interaction energies between adjacent base pairs indicate that pairwise base stacking interactions near the lesion are more disturbed as compared to the unmodified sequence in the TG*T than in the CG*C duplex; this is shown in Fig. 9, which gives van der Waals interaction energy population distributions of the modified and unmodified duplexes. The greatest relative destabilization is at the (G6*:C17–T7:A16)/(G6*:C17–C7:G16) steps, followed by the (T5:A18–G6*:C17)/(C5:G18–G6*: C17) steps. The sum of these interactions within the central 5-bp sequence (from A4 to A8) is reduced by ~2.8 kcal mol−1 in the TG*T duplex and by ~1.7 kcal mol−1 in the CG*C duplex. The more weakened stacking in TG*T contributes to its greater flexibility.
Fig. 9.
Population distribution of the van der Waals interaction energies between adjacent base pairs near/at the lesion site for CG*C, CGC, TG*T, and TGT. Ensemble averages and standard deviations are given. The step with the greatest relative destabilization of TG*T is indicated with a red arrow.
Roll is greater and more dynamic in TG*T than in CG*C
It is well established that DNA bending occurs primarily via Roll.40–42 Our results in Fig. 7 show that in the TG*T duplex Roll is greater at steps 5 and 6 than in the CG*C case. The greater Roll is accompanied by untwisting (Fig. 7), since Twist and Roll are anticorrelated.40–42 In addition, the distribution plots of Roll show greater flexibility in Roll at step 7 (Fig. 7). The high standard deviation in the mean value of Roll at this step also reflects its greater dynamic flexibility (Fig. 7) (Table S2, Supplementary Data).
Hydrogen bonding between BP hydroxyl groups and adjacent bases tends to influence the BP orientational dynamics
Hydroxyl groups of the BP benzylic ring can form hydrogen bonds with adjacent base or backbone atoms (Table 3). Figure S6, Supplementary Data, shows the time dependence of these hydrogen bond distances and angles. Of particular interest is a hydrogen bond between the BP 8-OH and O2 of T7 in the TG*Tsequence, and the same BP 8-OH with O2 of C7 in CG*C. Occupancies are 64% in the former case, and 54% in the latter. We note that when the hydrogen bond is broken (heavy atom–heavy atom distance >3.3 Å, or donor-hydrogen-acceptor angle <140°), in either sequence, the torsion angle β′ tends to be lower (Fig. S7, Supplementary Data). Thus, the dynamics of these hydrogen bonds contribute to the orientational dynamics of the BP rings. Possibly, the more rigid placement of the BP rings in CG*C allows less opportunity for formation of this hydrogen bond, accounting for its somewhat lower occupancy.
Table 3.
Hydrogen bonds between BP benzylic ring and adjacent base or backbone atoms
| Sequence context | Hydrogen bond | Occupancy (%) |
|---|---|---|
| CG*C | (BP)O7-HO7···O2(C17) | 85 |
| (BP)O8-HO8···O2(C7) | 54 | |
| (BP)O7-HO7···O4′(G18) | 12 | |
| TG*T | (BP)O7-HO7···O2(C17) | 91 |
| (BP)O8-HO8···O2(T7) | 64 | |
| (BP)O9-HO9···O4′(A8) | 18 |
Criteria for formation of hydrogen bonding: heavy atom–heavy atom distance <3.3 Å and donor-hydrogen-acceptor angle >140°.
Discussion
More dynamically flexible TG*T duplex: Connecting computation with experiment
We found notable structural differences in the TG*T and CG*C duplexes, although both sequences were modified with the same lesion and the lesion adopted the same MG conformational family. Our results reveal a more dynamic, flexible TG*T duplex than in the CG*C case. The greater flexibility is manifested in more disrupted and more mobile hydrogen bonding at the flanking T:A base pairs than the flanking C:G pairs, and is accompanied by greater and more flexible local Roll and larger and more flexible local Opening into the major groove in the TG*T case. In concert, the MG width is more flexible in the TG*T sequence, although it is smaller.
The underlying origin of these dynamic differences between the TG*T and the CG*C duplexes lies in the combined structural differences between T:A and C:G base pairs. An obvious difference is the weaker hydrogen bonding in the T:A pairs than the C:G pairs, with only two hydrogen bonds in the former, but three in the latter. These differences are reflected in the lower thermal melting points of TG*T versus CG*C duplexes.25 The weaker local base–base stacking interactions at TG* and G*T steps than at CG* and G*C dinucleotide steps is a further contributing factor.33,34 The greater destabilization caused by the lesion in the TG*T case is also consistent with the lower thermodynamic stability of the GT and TG dinucleotide steps as compared to the analogous GC and CG steps in unmodified duplexes.33,34 These intrinsically less stable and more flexible GT and TG steps are thus more affected by the presence of the bulky BP residue. In fact, the TG step in unmodified DNA is known to be among the most flexible dinucleotide steps.33,34
Another important factor is the absence of guanine exocyclic amino groups in the MG in the TG*T case, producing a narrower local MG width. In the CG*C sequence, these amino groups compete sterically with the BP rings for the same space in the MG. This results in a more constricted environment for the accommodation of the BP residue. Consequently, the TG*T duplex is more dynamically flexible. A lower exposure of the BP aromatic ring system to solvent in the CG*C than the TG*T sequence is partly due to its greater confinement, and the guanine exocyclic amino groups in CG*C also contribute to the smaller SASA by shielding the BP aromatic ring system (Fig. 3). Experimental observations involving fluorescence quenching (Y. Tang and N. E. Geacintov, to be published) and UV absorption spectra25 substantiate that the aromatic BP residue is more solvent-accessible in the TG*T than in the CG*C sequence context in double-stranded DNA molecules.
The enhanced dynamic flexibility accounts for the much more flexible bend observed in circularization experiments for the TG*T sequence.29 DNA bending is known to be largely due to Roll.40–42 Also, bending and base opening are coupled according to free-energy calculations,43 with the concomitant unstacking facilitating the bending of the DNA duplex. Therefore the greater local Roll and its greater local flexibility, together with the flexible Opening and weaker stacking, permit circularization with greater efficiency in TG*T than in CG*C.29
These considerations can account for the conformational heterogeneity manifested in the NMR solution studies of the TG*T duplex sequence.29 The heterogeneity in this case does not appear to originate from a significant population of BD conformations. By contrast, only a single well-characterized MG conformer is found by NMR in the CG*C context.28 Interestingly, the NMR studies for TG*T indicated a disturbance of Watson–Crick base pairing on the 5′ side of the lesion, both at the immediate neighboring base pair, as seen here, and also at the next nearest 5′ neighboring base pair.29 On the other hand, we found a modest disturbance on the 3′ side of the lesion, which was not observed in the NMR data. A possible explanation lies in the different time scales (nanoseconds versus milliseconds) under observation in MD versus NMR, respectively.
Dynamic flexibility and NER damage recognition
The present study highlights the likely importance of increased dynamic flexibility in enhancing lesion repair susceptibility by the prokaryotic NER machinery. It is of interest to place this greater mobility in the context of our current understanding of NER lesion recognition. The DNA damage recognition step is believed to be a rate-limiting step in NER.44 Since a variety of lesions are recognized, it has been suggested that the NER factors do not recognize the lesion itself, but the local distortions in the DNA that are associated with the lesions.15,44–46 In this connection, we have shown recently that the human NER factor XPC/HR23B is the first mammalian NER factor that recognizes anti-[BP]-N2-dG lesions; this factor distinguishes between the BD (+)-cis-, and MG (+)-trans- and (−)-trans-[BP]-N2-dG lesions by opening the duplex to different extents and at different sites in the vicinity of the lesions.47
Gunz et al. have proposed a thermodynamic probing mechanism based on their observations that differences of more than 3 orders of magnitude are observed in the efficiency by which helix-destabilizing and helix-stabilizing adducts are excised by the NER mechanism.15 According to their bipartite model of NER, the NER machinery first recognizes disruptions of Watson–Crick hydrogen bonding, followed by an unspecified second verification step that ensures that a chemically modified nucleobase is indeed present.7,8,16,48,49 This has been called an “indirect readout mechanism” by Dip et al.48 Our detailed structural analyses based on MD simulations have previously provided further insights into the nature of DNA distortions that provoke NER susceptibility.50–53 We found that a number of other structural parameters, besides Watson–Crick hydrogen bonding, are compromised by bulky lesions, a phenomenon we termed the multipartite mechanism of lesion recognition.52
Recent observations suggest that NER activity is triggered by a flipped out deoxyribonucleotide in the unmodified complementary strand.49,54,55 The crystal structure containing UvrB bound to a DNA duplex55 also showed that UvrB inserts a β-hairpin through the DNA helix. Strand opening and bending at the site of the lesion for the TG*Tsequence is consistent with this structure and the fact that UvrABC acts upon the TG*T sequence ~2.3-fold more efficiently. Nucleotide flipping would be facilitated by distorted Watson–Crick hydrogen bonding and/or weakened base–base stacking interactions.14 Molecular simulations have recently shown large and long-lived strand-separated “bubbles” opposite a cis–syn thymine dimer in duplex DNA.56 Various models of lesion recognition by the NER apparatus have been considered in a review by Isaacs and Spielmann57 who stressed the importance of local conformational flexibility associated with lesions. Our MD results highlight the important role that base sequence context surrounding the lesion can play in producing enhanced dynamic motions, which would facilitate nucleotide flipping and consequently enhance NER efficiency. The recent crystal structure of the yeast XPC orthologue Rad4 bound to DNA containing a cyclobutane pyrimidine dimer reveals flipping of the damaged bases as well as the undamaged opposite bases.58 This structure emphasizes the importance of duplex destabilization by the lesion in facilitating recognition.59
Conclusion
We have demonstrated the concept that the identical bulky lesion has a different impact on the local dynamics of the DNA duplex when positioned in two different base sequence contexts. Our study provides strong evidence on the molecular level, based on MD simulations, for a significantly greater dynamic mobility in the TG*T duplex than in the CG*C duplex. This flexibility around the site of the lesion is correlated with a greater susceptibility to excision of the stereochemically identical (+)-trans-anti-[BP]-N2-dG lesion in TG*T duplexes by prokaryotic UvrABC proteins. The enhanced dynamics that we find in the TG*T sequence context suggests that sequence-dependent dynamic perturbations facilitate the recognition and excision of the lesion. The enhanced dynamic motions around the lesion site in the TG*T sequence produce coupled structural distortions including bending, diminished base stacking, and impaired Watson–Crick pairing that facilitate nucleotide flipping.49,54,55,58 More globally, enhanced lesion dynamics may play an important role in facilitating NER. Future experimental and computational studies are needed to provide further evidence for this suggestion.
Methods
Starting structures
The MG conformation of the NMR solution structure for the 10S (+)-trans-anti-[BP]-N2-dG adduct in the CG*C sequence context28 was used as the starting structure. Base sequences were modified to reflect those shown in Fig. 1b. Since there was no solution structure available for the 10S (+)-trans-anti-[BP]-N2-dG BD conformer in a full duplex, we modeled it from the base-displaced 10S (−)-cis-anti-[BP]-N2-dG adduct structure60 by interchanging the hydrogen atom and the hydroxyl group at both C9 and C8 in the benzylic ring. Also, since high-resolution MG or BD NMR solution structures for the TG*T sequence (Fig. 1b) are not available, we modeled the starting structures for the TG*T sequence from the corresponding CG*C models by replacing the base pairs flanking the lesion. The starting structures for the unmodified DNA duplexes in both sequence contexts were energy-minimized B-DNA structures computed with DUPLEX.61 The initial models are shown in Fig. S8, Supplementary Data.
Force field
MD simulations were carried out using SANDER in the AMBER 8.0 simulation package,37 the Cornell et al. force field,62 with parm99.dat parameter set.63 Partial charges and all parameters for the 10S (+)-trans-anti-[BP]-N2-dG adduct employed in this work were the same as detailed previously.51
Molecular dynamics computation protocols and best representative structures
Details of the MD protocols and the computation of the best representative structures, using the cluster analyses option in MOIL-View39 are given in the Supplementary Data. The stability of the MD simulation was evaluated for each model. For each sequence context, the RMSD of each snapshot in the trajectory relative to its respective initial structure was plotted as a function of time and is shown in Fig. S9, Supplementary Data. The average RMSD of all atoms, excluding 1 bp at each end, in the current structure against the initial model for the 10.0 ns simulation was 1.7±0.3 Å for CG*C (MG), 2.1±0.5 Å for CG*C (BD), 1.7±0.3 Å for unmodified CGC, 1.6±0.2 Å for TG*T (MG), 2.4±0.5 Å for TG*T (BD), and 1.8±0.3 Å for unmodified TGT. For all cases, the MD achieved stability, fluctuating around the mean after 3.0 ns of simulation, and we employed the structural ensembles from the 3.0 to 10.0 ns time frame for further analyses.
Free-energy analyses
The MM-PBSA method in AMBER was employed to perform the thermodynamics analyses.38 Details of this method and the protocols we employed are available in Supplementary Data.
Structural analyses
The PTRAJ module of the AMBER 8.0 package37 and the CARNAL module of the AMBER 7.0 package64 were employed for structural analyses. Frames were selected at 1-ps intervals from the last 7.0 ns of simulation. DNA duplex groove dimensions were analyzed using MD Toolchest65,66 and 5.8 Å was subtracted from the pairwise phosphorus–phosphorus distances to account for the van der Waals radius of the phosphorus atoms.67 The first and last base pairs were excluded in this analysis due to possible end effects. Stacking interactions for the MG conformation were estimated by computing the van der Waals interaction energies between all adjacent base pairs. The ANAL module of the AMBER 8.0 package was employed. SASA was calculated with the Connolly algorithm68 in the INSIGHTII 2000 (Accelrys Software, Inc.), adopting default parameters: probe radius of 1.4 Å and atom radius scale of 1.0 Å.
INSIGHTII 2000 was employed for visualization and model building. Computations were carried out on our own cluster of Silicon Graphic Origin and Altix high-performance computers.
Supplementary Material
Acknowledgments
This research was supported by NIH Grants CA-28038 (S.B.), CA-099194 (N.E.G.) and CA-046533 (D.J.P.).
Abbreviations used
- BP
benzo[a]pyrene
- MD
molecular dynamics
- NER
nucleotide excision repair
- dG
deoxyguanosine
- MG
minor groove
- BD
base-displaced/intercalated
- MM-PBSA
molecular mechanics Poisson–Boltzmann surface area
- SASA
solvent-accessible surface area
Footnotes
Supplementary Data
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.jmb.2007.09.034
References
- 1.Luch A. Nature and nurture—lessons from chemical carcinogenesis. Nat. Rev. Cancer. 2005;5:113–125. doi: 10.1038/nrc1546. [DOI] [PubMed] [Google Scholar]
- 2.Greenblatt MS, Bennett WP, Hollstein M, Harris CC. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- 3.Riedl T, Hanaoka F, Egly JM. The comings and goings of nucleotide excision repair factors on damaged DNA. EMBO J. 2003;22:5293–5303. doi: 10.1093/emboj/cdg489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tapias A, Auriol J, Forget D, Enzlin JH, Scharer OD, Coin F, et al. Ordered conformational changes in damaged DNA induced by nucleotide excision repair factors. J. Biol. Chem. 2004;279:19074–19083. doi: 10.1074/jbc.M312611200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sugasawa K, Ng JM, Masutani C, Iwai S, van der Spek PJ, Eker AP, et al. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell. 1998;2:223–232. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]
- 6.Sugasawa K, Ng JM, Masutani C, Maekawa T, Uchida A, van der Spek PJ, et al. Two human homologs of Rad23 are functionally interchangeable in complex formation and stimulation of XPC repair activity. Mol. Cell Biol. 1997;17:6924–6931. doi: 10.1128/mcb.17.12.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sugasawa K, Shimizu Y, Iwai S, Hanaoka F. A molecular mechanism for DNA damage recognition by the xeroderma pigmentosum group C protein complex. DNA Repair (Amst.) 2002;1:95–107. doi: 10.1016/s1568-7864(01)00008-8. [DOI] [PubMed] [Google Scholar]
- 8.Sugasawa K, Okamoto T, Shimizu Y, Masutani C, Iwai S, Hanaoka F. A multistep damage recognition mechanism for global genomic nucleotide excision repair. Genes Dev. 2001;15:507–521. doi: 10.1101/gad.866301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kusumoto R, Masutani C, Sugasawa K, Iwai S, Araki M, Uchida A, et al. Diversity of the damage recognition step in the global genomic nucleotide excision repair in vitro. Mutat. Res. 2001;485:219–227. doi: 10.1016/s0921-8777(00)00082-3. [DOI] [PubMed] [Google Scholar]
- 10.Chen Z, Xu XS, Yang J, Wang G. Defining the function of XPC protein in psoralen and cisplatin-mediated DNA repair and mutagenesis. Carcinogenesis. 2003;24:1111–1121. doi: 10.1093/carcin/bgg051. [DOI] [PubMed] [Google Scholar]
- 11.Ng JM, Vermeulen W, van der Horst GT, Bergink S, Sugasawa K, Vrieling H, Hoeijmakers JH. A novel regulation mechanism of DNA repair by damage-induced and RAD23-dependent stabilization of xeroderma pigmentosum group C protein. Genes Dev. 2003;17:1630–1645. doi: 10.1101/gad.260003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thoma BS, Vasquez KM. Critical DNA damage recognition functions of XPC-hHR23B and XPA-RPA in nucleotide excision repair. Mol. Carcinog. 2003;38:1–13. doi: 10.1002/mc.10143. [DOI] [PubMed] [Google Scholar]
- 13.Gillet LC, Scharer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem. Rev. 2006;106:253–276. doi: 10.1021/cr040483f. [DOI] [PubMed] [Google Scholar]
- 14.Yang W. Poor base stacking at DNA lesions may initiate recognition by many repair proteins. DNA Repair (Amst.) 2006;5:654–666. doi: 10.1016/j.dnarep.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 15.Gunz D, Hess MT, Naegeli H. Recognition of DNA adducts by human nucleotide excision repair. Evidence for a thermodynamic probing mechanism. J. Biol. Chem. 1996;271:25089–25098. doi: 10.1074/jbc.271.41.25089. [DOI] [PubMed] [Google Scholar]
- 16.Hess MT, Gunz D, Luneva N, Geacintov NE, Naegeli H. Base pair conformation-dependent excision of benzo[a]pyrene diol epoxide–guanine adducts by human nucleotide excision repair enzymes. Mol. Cell. Biol. 1997;17:7069–7076. doi: 10.1128/mcb.17.12.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geacintov NE, Cosman M, Hingerty BE, Amin S, Broyde S, Patel DJ. NMR solution structures of stereoisometric covalent polycyclic aromatic carcinogen–DNA adduct: principles, patterns, and diversity. Chem. Res. Toxicol. 1997;10:111–146. doi: 10.1021/tx9601418. [DOI] [PubMed] [Google Scholar]
- 18.Lukin M, de Los Santos C. NMR structures of damaged DNA. Chem. Rev. 2006;106:607–686. doi: 10.1021/cr0404646. [DOI] [PubMed] [Google Scholar]
- 19.Patel DJ, Mao B, Gu Z, Hingerty BE, Gorin A, Basu AK, Broyde S. Nuclear magnetic resonance solution structures of covalent aromatic amine–DNA adducts and their mutagenic relevance. Chem. Res. Toxicol. 1998;11:391–407. doi: 10.1021/tx9702143. [DOI] [PubMed] [Google Scholar]
- 20.Cho BP. Dynamic conformational heterogeneities of carcinogen–DNA adducts and their mutagenic relevance. J. Environ. Sci. Health, Part C; Environ. Carcinog. Ecotoxicol. Rev. 2004;22:57–90. doi: 10.1081/LESC-200038217. [DOI] [PubMed] [Google Scholar]
- 21.Elmquist CE, Wang F, Stover JS, Stone MP, Rizzo CJ. Conformational differences of the C8-deoxyguanosine adduct of 2-amino-3-methylimidazo[4,5-f]quinoline (IQ) within the NarI recognition sequence. Chem. Res. Toxicol. 2007;20:445–454. doi: 10.1021/tx060229d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zou Y, Shell SM, Utzat CD, Luo C, Yang Z, Geacintov NE, Basu AK. Effects of DNA adduct structure and sequence context on strand opening of repair intermediates and incision by UvrABC nuclease. Biochemistry. 2003;42:12654–12661. doi: 10.1021/bi034446e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mekhovich O, Tang M, Romano LJ. Rate of incision of N-acetyl-2-aminofluorene and N-2-aminofluorene adducts by UvrABC nuclease is adduct- and sequence-specific: comparison of the rates of UvrABC nuclease incision and protein–DNA complex formation. Biochemistry. 1998;37:571–579. doi: 10.1021/bi971544p. [DOI] [PubMed] [Google Scholar]
- 24.Meneni S, Shell SM, Zou Y, Cho BP. Conformation-specific recognition of carcinogen–DNA adduct in Escherichia coli nucleotide excision repair. Chem. Res. Toxicol. 2007;20:6–10. doi: 10.1021/tx600273h. [DOI] [PubMed] [Google Scholar]
- 25.Ruan Q, Liu T, Kolbanovskiy A, Liu Y, Ren J, Skorvaga M, et al. Sequence context- and temperature-dependent nucleotide excision repair of a benzo[a]pyrene diol epoxide–guanine DNA adduct catalyzed by thermophilic UvrABC proteins. Biochemistry. 2007;46:7006–7015. doi: 10.1021/bi700294k. [DOI] [PubMed] [Google Scholar]
- 26.Conney AH. Induction of microsomal enzymes by foreign chemicals and carcinogenesis by polycyclic aromatic hydrocarbons: G. H. A. Clowes Memorial Lecture. Cancer Res. 1982;42:4875–4917. [PubMed] [Google Scholar]
- 27.Szeliga J, Dipple A. DNA adduct formation by polycyclic aromatic hydrocarbon dihydrodiol epoxides. Chem. Res. Toxicol. 1998;11:1–11. doi: 10.1021/tx970142f. [DOI] [PubMed] [Google Scholar]
- 28.Cosman M, de los Santos C, Fiala R, Hingerty BE, Singh SB, Ibanez V, et al. Solution conformation of the major adduct between the carcinogen (+)-anti-benzo[a]pyrene diol epoxide and DNA. Proc. Natl. Acad. Sci. USA. 1992;89:1914–1918. doi: 10.1073/pnas.89.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu R, Mao B, Amin S, Geacintov NE. Bending and circularization of site-specific and stereoisomeric carcinogen–DNA adducts. Biochemistry. 1998;37:769–778. doi: 10.1021/bi971785x. [DOI] [PubMed] [Google Scholar]
- 30.Fountain MA, Krugh TR. Structural characterization of a (+)-trans-anti-benzo[a]pyrene–DNA adduct using NMR, restrained energy minimization, and molecular dynamics. Biochemistry. 1995;34:3152–3161. doi: 10.1021/bi00010a004. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez FA, Cai Y, Lin C, Tang Y, Kolbanovskiy A, Amin S, et al. Exocyclic amino groups of flanking guanines govern sequence-dependent adduct conformations and local structural distortions for minor groove-aligned benzo[a]pyrenyl–guanine lesions in a GG mutation hotspot context. Nucleic Acids Res. 2007;35:1555–1568. doi: 10.1093/nar/gkm022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cosman M, Fiala R, Hingerty BE, Amin S, Geacintov NE, Broyde S, Patel DJ. Solution conformation of the (+)-trans-anti-[BP]dG adduct opposite a deletion site in a DNA duplex: intercalation of the covalently attached benzo[a]pyrene into the helix with base displacement of the modified deoxyguanosine into the major groove. Biochemistry. 1994;33:11507–11517. doi: 10.1021/bi00204a013. [DOI] [PubMed] [Google Scholar]
- 33.Breslauer KJ, Frank R, Blocker H, Marky LA. Predicting DNA duplex stability from the base sequence. Proc. Natl. Acad. Sci. USA. 1986;83:3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.SantaLucia J, Jr, Allawi HT, Seneviratne PA. Improved nearest-neighbor parameters for predicting DNA duplex stability. Biochemistry. 1996;35:3555–3562. doi: 10.1021/bi951907q. [DOI] [PubMed] [Google Scholar]
- 35.Tsao H, Mao B, Zhuang P, Xu R, Amin S, Geacintov NE. Sequence dependence and characteristics of bends induced by site-specific poly-nuclear aromatic carcinogen–deoxyguanosine lesions in oligonucleotides. Biochemistry. 1998;37:4993–5000. doi: 10.1021/bi980291c. [DOI] [PubMed] [Google Scholar]
- 36.Jiang GH, Skorvaga M, Croteau DL, Van Houten B, States JC. Robust incision of benzo[a]pyrene-7,8-dihyrodiol-9,10-epoxide–DNA adducts by a recombinant thermoresistant interspecies combination UvrABC endonuclease system. Biochemistry. 2006;45:7834–7843. doi: 10.1021/bi052515e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, et al. AMBER 8. University of California; San Francisco, CA: 2004. [Google Scholar]
- 38.Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, et al. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc. Chem. Res. 2000;33:889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 39.Simmerling C, Elber R, Zhang J. MOIL-View—a program for visualization of structure and dynamics of biomolecules and STO—a program for computing stochastic paths. In: Pullman, editor. Modeling of Biomolecular Structure and Mechanisms. Kluwer; Netherlands: 1995. [Google Scholar]
- 40.Dickerson RE. DNA bending: the prevalence of kinkiness and the virtues of normality. Nucleic Acids Res. 1998;26:1906–1926. doi: 10.1093/nar/26.8.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gorin AA, Zhurkin VB, Olson WK. B-DNA twisting correlates with base-pair morphology. J. Mol. Biol. 1995;247:34–48. doi: 10.1006/jmbi.1994.0120. [DOI] [PubMed] [Google Scholar]
- 42.Olson WK, Gorin AA, Lu XJ, Hock LM, Zhurkin VB. DNA sequence-dependent deformability deduced from protein–DNA crystal complexes. Proc. Natl. Acad. Sci. USA. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giudice E, Varnai P, Lavery R. Base pair opening within B-DNA: free energy pathways for GC and AT pairs from umbrella sampling simulations. Nucleic Acids Res. 2003;31:1434–1443. doi: 10.1093/nar/gkg239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wood RD. DNA damage recognition during nucleotide excision repair in mammalian cells. Biochimie. 1999;81:39–44. doi: 10.1016/s0300-9084(99)80036-4. [DOI] [PubMed] [Google Scholar]
- 45.Evans E, Moggs JG, Hwang JR, Egly JM, Wood RD. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J. 1997;16:6559–6573. doi: 10.1093/emboj/16.21.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujiwara Y, Masutani C, Mizukoshi T, Kondo J, Hanaoka F, Iwai S. Characterization of DNA recognition by the human UV-damaged DNA-binding protein. J. Biol. Chem. 1999;274:20027–20033. doi: 10.1074/jbc.274.28.20027. [DOI] [PubMed] [Google Scholar]
- 47.Mocquet V, Kropachev K, Kolbanovskiy M, Kolbanovskiy A, Tapias A, Cai Y, et al. The human DNA repair factor XPC-HR23B distinguishes stereoisomeric benzo[a]pyrenyl–DNA lesions. EMBO J. 2007;26:2923–2932. doi: 10.1038/sj.emboj.7601730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dip R, Camenisch U, Naegeli H. Mechanisms of DNA damage recognition and strand discrimination in human nucleotide excision repair. DNA Repair. 2004;3:1409–1423. doi: 10.1016/j.dnarep.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 49.Buterin T, Meyer C, Giese B, Naegeli H. DNA quality control by conformational readout on the undamaged strand of the double helix. Chem. Biol. 2005;12:913–922. doi: 10.1016/j.chembiol.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 50.Yan S, Shapiro R, Geacintov NE, Broyde S. Stereochemical, structural, and thermodynamic origins of stability differences between stereoisomeric benzo[a]pyrene diol epoxide deoxyadenosine adducts in a DNA mutational hot spot sequence. J. Am. Chem. Soc. 2001;123:7054–7066. doi: 10.1021/ja0043035. [DOI] [PubMed] [Google Scholar]
- 51.Yan S, Wu M, Patel DJ, Geacintov NE, Broyde S. Simulating structural and thermodynamic properties of carcinogen-damaged DNA. Biophys. J. 2003;84:2137–2148. doi: 10.1016/S0006-3495(03)75020-6. (Erratum in: Biophys. J., 92, 697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Geacintov N, Naegeli H, Patel DJ, Broyde S. Structural aspects of polycyclic aromatic carcinogen-damaged DNA and its recognition by NER proteins. In: Siede W, Kow YW, Doetsch PW, editors. DNA Damage and Recognition. Taylor and Francis; London: 2006. [Google Scholar]
- 53.Wu M, Yan S, Patel DJ, Geacintov NE, Broyde S. Relating repair susceptibility of carcinogen-damaged DNA with structural distortion and thermodynamic stability. Nucleic Acids Res. 2002;30:3422–3432. doi: 10.1093/nar/gkf427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Malta E, Moolenaar GF, Goosen N. Base flipping in nucleotide excision repair. J. Biol. Chem. 2006;281:2184–2194. doi: 10.1074/jbc.M508901200. [DOI] [PubMed] [Google Scholar]
- 55.Truglio JJ, Karakas E, Rhau B, Wang H, DellaVecchia MJ, Van Houten B, Kisker C. Structural basis for DNA recognition and processing by UvrB. Nat. Struct. Mol. Biol. 2006;13:360–364. doi: 10.1038/nsmb1072. [DOI] [PubMed] [Google Scholar]
- 56.Blagoev KB, Alexandrov BS, Goodwin EH, Bishop AR. Ultra-violet light induced changes in DNA dynamics may enhance TT-dimer recognition. DNA Repair (Amst.) 2006;5:863–867. doi: 10.1016/j.dnarep.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 57.Isaacs RJ, Spielmann HP. A model for initial DNA lesion recognition by NER and MMR based on local conformational flexibility. DNA Repair (Amst.) 2004;3:455–464. doi: 10.1016/j.dnarep.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 58.Min JH, Pavletich NP. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature. 2007;449:570–575. doi: 10.1038/nature06155. [DOI] [PubMed] [Google Scholar]
- 59.Geacintov NE, Broyde S, Buterin T, Naegeli H, Wu M, Yan S, Patel DJ. Thermodynamic and structural factors in the removal of bulky DNA adducts by the nucleotide excision repair machinery. Biopolymers. 2002;65:202–210. doi: 10.1002/bip.10239. [DOI] [PubMed] [Google Scholar]
- 60.Cosman M, Hingerty BE, Luneva N, Amin S, Geacintov NE, Broyde S, Patel DJ. Solution conformation of the (–)-cis-anti-benzo[a]pyrenyl-dG adduct opposite dC in a DNA duplex: intercalation of the covalently attached BP ring into the helix with base displacement of the modified deoxyguanosine into the major groove. Biochemistry. 1996;35:9850–9863. doi: 10.1021/bi9605346. [DOI] [PubMed] [Google Scholar]
- 61.Hingerty BE, Figueroa S, Hayden TL, Broyde S. Prediction of DNA structure from sequence: a build-up technique. Biopolymers. 1989;28:1195–1222. doi: 10.1002/bip.360280703. [DOI] [PubMed] [Google Scholar]
- 62.Cieplak P, Cornell WD, Bayly C, Kollman PA. Application of the multimolecule and multi-conformational RESP methodology to biopolymers—charge derivation for DNA, RNA, and proteins. J. Comput. Chem. 1995;16:1357–1377. [Google Scholar]
- 63.Cheatham TE, Cieplak P, Kollman PA. A modified version of the Cornell et al. force field with improved sugar pucker phases and helical repeat. J. Biomol. Struct. Dyn. 1999;16:845–862. doi: 10.1080/07391102.1999.10508297. [DOI] [PubMed] [Google Scholar]
- 64.Case DA, Pearlman DA, Caldwell JW, Cheatham TE, III, Wang J, Ross WS, et al. AMBER 7. University of California; San Francisco, CA: 2002. [Google Scholar]
- 65.Ravishanker G, Swaminathan S, Beveridge DL, Lavery R, Sklenar H. Conformational and helicoidal analysis of 30 PS of molecular dynamics on the d(CGCGAATTCGCG) double helix: “curves,” dials and windows. J. Biomol. Struct. Dyn. 1989;6:669–699. doi: 10.1080/07391102.1989.10507729. [DOI] [PubMed] [Google Scholar]
- 66.Ravishanker G, Beveridge DL. Toolchest 2.0. Wesleyan University; Middletown, CT 06459: 1993. [Google Scholar]
- 67.Fratini AV, Kopka ML, Drew HR, Dickerson RE. Reversible bending and helix geometry in a B-DNA dodecamer: CGCGAATTBrCGCG. J. Biol. Chem. 1982;257:14686–14707. [PubMed] [Google Scholar]
- 68.Connolly ML. Solvent-accessible surfaces of proteins and nucleic acids. Science. 1983;221:709–713. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- 69.Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.