Abstract
We demonstrate by NMR that the two-repeat human telomeric sequence d(TAGGGTTAGGGT) can form both parallel and antiparallel G-quadruplex structures in K+-containing solution. Both structures are dimeric G-quadruplexes involving three stacked G-tetrads. The sequence d(TAGGGUTAGGGT), containing a single thymine-to-uracil substitution at position 6, formed a predominantly parallel dimeric G-quadruplex with double-chain-reversal loops; the structure was symmetric, and all guanines were anti. Another modified sequence, d(UAGGGTBrUAGGGT), formed a predominantly antiparallel dimeric G-quadruplex with edgewise loops; the structure was asymmetric with six syn guanines and six anti guanines. The two structures can coexist and interconvert in solution. For the latter sequence, the antiparallel form is more favorable at low temperatures (<50 °C), while the parallel form is more favorable at higher temperatures; at temperatures lower than 40 °C, the antiparallel G-quadruplex folds faster but unfolds slower than the parallel G-quadruplex.
Introduction
Telomeres, the ends of eukaryotic chromosomes, are essential for stable chromosome maintenance and appear to play an important role in cellular aging and cancer. Human telomeric DNA contains tandem repeats of the sequence TTAGGG. The guanine-rich strand can fold into four-stranded G-quadruplex structures involving G-tetrads, which have become currently an attractive target for development of anticancer drugs.1
The solution structure of the four-repeat human telomere TTAGGG sequence has been characterized in our laboratory by NMR in Na+-containing solution.2 This sequence forms an intramolecular quadruplex, where four GGG segments are involved in formation of three stacked G-tetrads with anti•syn•anti•syn alignments around each tetrad, while three TTA connecting loops adopt successive edgewise, diagonal, and edgewise alignments, such that adjacent strands are aligned antiparallel to each other. By contrast, the NMR-based solution structure of the four-repeat Tetrahymena TTGGGG sequence, which differs from the human sequence by a single G-for-A substitution, adopts a distinctly different fold in Na+-containing solution.3 This quadruplex also contains three stacked G-tetrads, but individual tetrads adopt either syn•syn•syn•anti or anti•anti•anti•syn alignments, and the successive connecting loops adopt edgewise, edgewise, and double-chain-reversal, such that three strands are oriented in one direction and the fourth in the opposite direction. The observation of a double-chain-reversal loop connecting two adjacent parallel-aligned strands3 was unanticipated but has since been observed within additional quadruplexes investigated in our laboratory in solution and appears to be a robust folding topology.4
Recently, Parkinson et al.5 reported crystal structures of parallel G-quadruplexes from human telomeric sequences consisting of two or four TTAGGG repeats in the presence of K+. These structures were very different from all G-quadruplexes reported previously and could readily facilitate higher order telomere folding and unfolding.5 All four strands are parallel (G-stretches are oriented in the same direction), the connecting TTA loops are double-chain-reversal (they are located in the grooves and connect adjacent parallel strands), and all guanines adopt anti glycosidic conformations.
We questioned whether Parkinson et al.’s crystal structures could also exist in solution. Using NMR spectroscopy we showed that the two-repeat telomeric sequence d(TAGGGT-TAGGGT) forms a similar parallel G-quadruplex structure in K+-containing solution. In addition, we found that the same sequence can also form an antiparallel G-quadruplex structure (see below). These two structures can coexist and interconvert in solution. They have different thermodynamic properties and different kinetics of folding and unfolding.
Materials and Methods
Sample Preparation
The unlabeled and the site-specific low-enrichment (2% 15N,13C-labeled) oligonucleotides were synthesized using solid-phase β-cyanoethyl phosphoramidite chemistry and purified by HPLC as previously described.2,6 They were dialyzed successively against 50 mM KCl solution and against water. Unless otherwise stated, the strand concentration of the NMR samples was typically 0.5–3 mM; the solutions contained 0.1 mM EDTA, 0.01 mM DSS, and 90 mM of KCl. The sample pH was adjusted to 7 with HCl and KOH solutions. The samples were heated at 90 °C for 3 min and annealed to room temperature over several days.
Nuclear Magnetic Resonance
NMR experiments were performed on a 600 MHz Varian Unity INOVA spectrometer. Experiments in H2O used the jump-and-return (JR) water suppression7,8 for detection. For experiments in D2O, low-power irradiation at water frequency was used during the recovery delay to saturate the residual HDO signal and was switched off 0.1–0.3 s before excitation to avoid saturation of H3′ resonances.8b
Resonances were assigned using site-specific low-enrichment labeling,6 through-bond correlations at natural abundance8 (imino-H8 by JRHMBC;9a H8–H2 by HMBC;9b H8/6-H1′ by HSQC and sHMBC;9c H1′-H2′/2”/3′/4′/5′/5” by COSY9d and TOCSY;9e H5′/5”/4′/3′-P by hetero-TOCSY9f), and NOESY.
Determination of Stoichiometry
The molecular size was determined by gel filtration chromatography using a Synchropack GPC100 column (Synchrom, Lafayette, IN) calibrated with oligonucleotides of known molecular weight as previously described.8a
NMR determination of stoichiometry was based on the titration of the concentration-dependent equilibrium between structured and unfolded forms.8a The unfolded monomer form was identified as the species, which is predominant at low concentration and/or high temperature, showing sharp nonexchangeable protons but no imino proton signals. We note that the measurement at a high temperature (50 °C) allowed to have both structured and unfolded forms at high equilibrium concentrations, which could be reached quickly after 1 h in the NMR spectrometer (see below the results for kinetic measurements). The titration was performed on samples in H2O on the basis of signals of different imino and aromatic protons, using the JR pulse sequence with a repetition delay of 5 s. The determination of the absolute concentrations of different forms was helped by the comparison of peak intensities with the internal DSS signal.
Kinetic Measurements
Kinetics of folding and unfolding of different dimeric G-quadruplexes was studied in real-time experiments by following characteristic imino protons (see Results). For clarity, we assumed a simple model, where transitions between the parallel G-quadruplex (P), the unfolded single strand (S), and the antiparallel G-quadruplex (A) resulted in equilibrium among the three states (although other states such as a prefolded single strand may be also involved). The direct interconversion between the two G-quadruplexes is improbable.
| (1) |
We will describe below the measurements of folding and unfolding kinetics only for the parallel G-quadruplex. These measurements were also similarly applied to the antiparallel G-quadruplex.
The rate equation for the change in the parallel G-quadruplex concentration can be written as
| (2) |
In the right-hand side, the first term represents the parallel G-quadruplex-to-single strand transition (the unfolding) and the second term represents the single strand-to-parallel G-quadruplex transition (the folding).
(a) Kinetics of Folding
The folding process at a given temperature (e.g. 25 °C) was followed immediately after all structures had been unfolded by heating at 90 °C for 3 min. However, at millimolar concentration there was a fast formation of G-quadruplex structures, which likely occurred at higher temperatures (>25 °C) upon cooling from 90 °C, so that some amount of DNA was already in the form of G-quadruplexes at time t = 0. The initial concentration of the single strand [S(0)] could be therefore normalized to the total DNA concentration [S0]:
| (3) |
Here α was obtained by comparing the amount of the two dimeric G-quadruplex structures at equilibrium and at time t = 0 (other structures, e.g. higher molecular aggregation, were not considered). Typically, in our experimental conditions α ≈ 0.5–0.7.
In most cases we could still neglect the unfolding reaction, or
| (4) |
From (2)–(4), the folding time (τfold) can be obtained from the slope of the initial buildup curve using
| (5) |
where 1/τfold = k−1[S0].
(b) Kinetics of Unfolding
We used three different methods to characterize the unfolding time (τunfold = 1/k1) of the parallel G-quadruplex.
(1) Complementary-Strand-Trap Method
At a given temperature, the complementary C-strand d(ACCCTAACCCTA), which was likely unstructured at pH 7 as indicated by the absence of imino protons in NMR spectra, was added to the G-strand in order to trap the latter into a Watson–Crick duplex. Only the unfolded G-strand would be trapped;10,11 the concentrations of G-quadruplex structures would decay through G-quadruplex-to-single strand transitions, and the reverse reaction was inhibited because the unfolded G-strand was trapped. The unfolding of each G-quadruplex and the formation of the duplex were followed in real time by their imino protons, respectively. In our experimental conditions (strand concentrations of the G-strand and of the C-strand were 0.04–0.4 and 0.2–2.0 mM, respectively), the hybridization between the two strands occurred quickly and was not a limiting step for the unfolding. The term k−1[S]2 in eq 2 was suppressed; the concentration of the parallel G-quadruplex could be described as a single-exponential decaying toward zero:
| (6) |
Here [P(0)] and [P(t)] were the concentration of the parallel form at time zero and at time t, respectively.
(2) Concentration-Jump Method
In this method, we diluted largely (about 100-fold) a concentrated sample and observed how the equilibrium between each G-quadruplex form and the single strand was reached at a given temperature. The initial concentration of the single strand [S(0)] could be normalized to the equilibrium concentration [S∞]:
| (7) |
The relation between the equilibrium concentrations of the parallel G-quadruplex [P∞] and of the single strand [S∞] can be written as
| (8) |
The concentrations of each form at different times were derived from the intensities of their respective characteristic protons. From eqs 2, 7, and 8, the unfolding time can be determined from the slope of the initial buildup curves with
or
| (9) |
where βP = [P(0)]/[P∞] − γ2.
(3) Temperature-Jump Method
The antiparallel G-quadruplex or the parallel G-quadruplex could become overpopulated when the sample temperature was jumped up or down, respectively (see Results). The system would then evolve to reach the new equilibrium between the two G-quadruplex forms and the single strand. In principle, the same approach as the concentration-jump method could be applied. Unfortunately, in many cases, peaks corresponding to the single strand were too small to be observed for the initial slope analysis as just described. However, an analysis of the decay of each G-quadruplex concentration, e.g. a determination of its time constant, was still useful. Although interpretation of this time constant was difficult due to the involvement of both forward and reverse reactions, it provided a boundary for comparison of different G-quadruplex unfolding kinetics.
Results
Two-Repeat Human Telomeric Sequence d(TAGGGT-TAGGGT) Forms Different Structures in Solution
For the natural two-repeat human telomeric sequence d(TAGGGT-TAGGGT), the number of proton resonances (Figure 1a) indicated the presence of multiple G-quadruplex forms in K+-containing solution. In Parkinson et al.’s sequence, where two thymines (T1 and T6) were substituted by bromouracils (BrU), we also observed more than one form. Furthermore, we found that thymine-to-uracil (T → U) and thymine-to-bromouracil (T → BrU) substitutions at different positions of the sequence could shift the equilibrium toward a predominant form. We tested and combined many such substitutions at different positions to favor one or the other conformations in order to get “clean” NMR spectra for more detailed structural analysis. Finally, we have chosen two sequences, denoted U6 and U1B7, where a single (T6 → U6) and double (T1 → U1; T7 → BrU7) substitutions have been made, respectively.
Figure 1.
Imino proton spectra of the human telomeric sequences at 600 MHz: (a) natural sequence d(TAGGGTTAGGGT); (b) U6 sequence d(TAGGGUTAGGGT); (c) U1B7 sequence d(UAGGGTBrUAGGGT). Experimental conditions: strand concentration, 3 mM; temperature, 20 °C; 90 mM KCl; pH 7.
U6 and U1B7 Form Parallel and Antiparallel Dimeric G-Quadruplexes
Imino proton spectra of U6 (Figure 1b) and of U1B7 (Figure 1c) show sharp peaks at 10.5–12.5 ppm corresponding to guanine imino protons of G-quadruplex structures. In each case, there are a set of minor peaks and a set of major peaks. The stoichiometry of 2 for the major forms (Figure 2; see legend) was determined by gel filtration chromatography (Figure 2a) and equilibrium NMR titration (Figure 2b). Their imino proton spectra are very different (Figures 1 and 3): U6 shows 6 major peaks corresponding to 6 guanine imino protons of a symmetric dimer;12 U1B7 shows 12 peaks corresponding to 12 guanine imino protons from two different strands (red and green) of an asymmetric dimer.12
Figure 2.
(a) Determination of stoichiometry by gel filtration chromatography on an analytical Synchropak GPC 100 column with the elution buffer being 70 mM KCl and 20 mM potassium phosphate, pH 7, at room temperature. The known number of residues of several nucleic acid reference samples (in black) (including single-stranded oligonucleotides, duplexes, and quadruplexes) is plotted in log scale as a function of the elution time. The straight line is interpolation through these data. The elution time of U6 and U1B7 (~1 mM) is consistent with a molecular size of 24 nucleotides, corresponding to a dimer (open data point with open arrow). The elution time of heated diluted samples of U6 and U1B7 (~1 µM) is consistent with a molecular size of 12 nucleotides, corresponding to a single strand (open data point with black arrow). (b) Determination of stoichiometry by NMR titration of the equilibrium strand concentrations of the multimer structure and of the unstructured monomer. Squares and triangles represent U6 and U1B7, respectively. Lines of slope 2 are drawn through the data points. Insert: Proton spectra of U6, showing the peaks of the structured form (grey) and the peaks of the unstructured single strand (black), whose relative intensities are dependent on DNA concentration. Experimental conditions: temperature, 50 °C; 70 mM KCl; potassium phosphate, 20 mM; pH 7.
Figure 3.
Guanine imino proton spectra of (a) U6 and (a′) U1B7. Assignments are indicated over the reference spectra (ref): blue for U6; red and green for two different strands in U1B7. Imino protons were assigned in 15N-filtered spectra of samples, 2% 15N,13C-labeled at the indicated positions. (b, b′) Imino proton spectra after 1 day in D2O showing G4 and G10 peaks from the central G-tetrads. Experimental conditions: strand concentration, 0.5–3 mM; temperature, 20 °C; 90 mM KCl; pH 7.
We performed unambiguous resonance assignments using site-specific low-enrichment labeling approach.6 For example, the guanine imino proton of each site-specific labeled residue was assigned on the basis of its intensity in the 15N-filtered spectrum (Figure 3a,a′). The assignments were also independently obtained in natural-abundance through-bond correlation and NOESY experiments (see Materials and Methods).
Figure 3b,b′ shows two and four slowly exchanging imino protons in the spectra of U6 and U1B7, respectively, which remained after 1 day in D2O. They were assigned to the imino protons of G4 and G10, consistent to their positions in the central G-tetrad (see below).
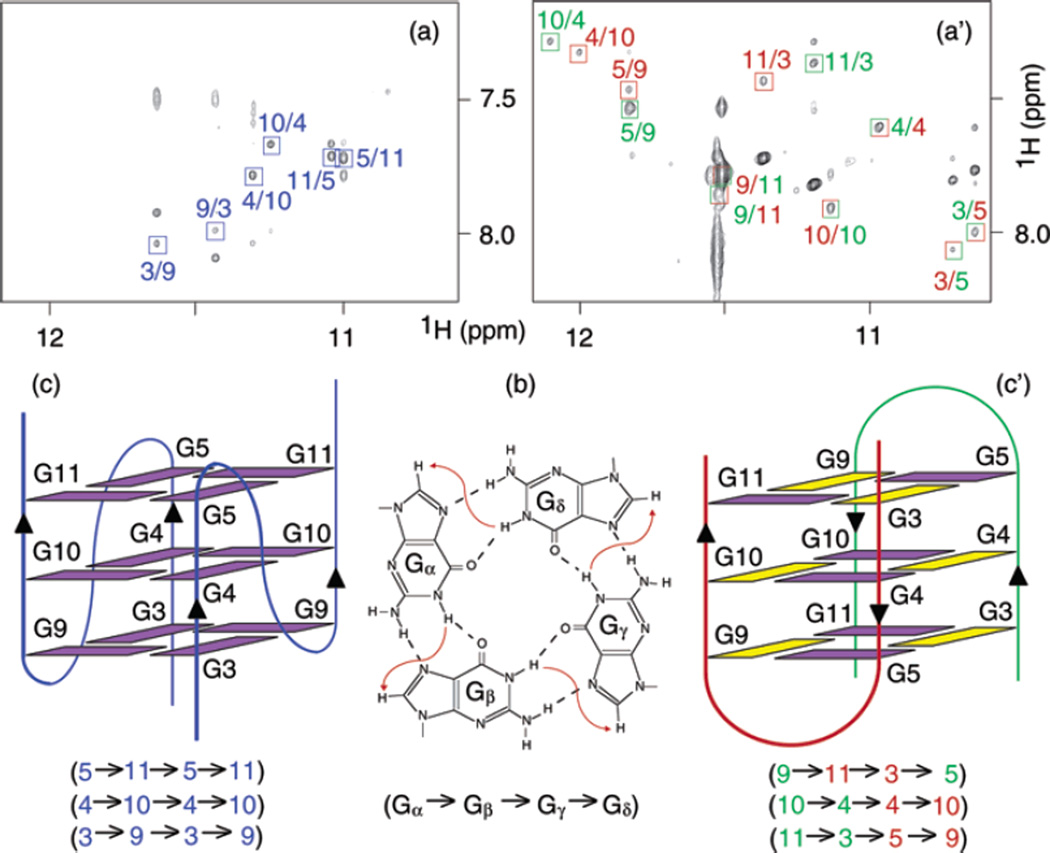
The G-tetrad arrangements could be then defined from NOESY spectra (Figure 4a,a′) on the basis of the specific imino-H8 connectivity pattern in a G-tetrad (Figure 4b). These arrangements were further confirmed with guanine-to-inosine substitutions (Figure S1).
Figure 4.
NOESY spectra (mixing time, 200 ms) of (a) U6 and (a′) U1B7 (same conditions as in Figure 1). The imino-H8 cross-peaks are framed and labeled with the number of imino protons in the first position and that of H8 in the second position. (b) Specific imino-H8 connectivity pattern around a G-tetrad (Gα•Gβ•Gγ•Gδ) indicated with arrows (connectivity between Gδ and Gα implied). (c, c′) Schematic structures of U6 and U1B7 that satisfy NOE connectivities shown in parentheses. Violet and yellow represent anti and syn guanines, respectively.
The folding topologies of U6 and U1B7 (Figure 4c,c′) were drawn from G-tetrad arrangements. U6 formed a parallel dimeric G-quadruplex with double-chain-reversal loops (Figure 4c), similar to the crystal structure.5 All guanines were anti: no strong H8–H1′ NOE cross-peaks were observed (Figure S2). U1B7 formed a completely different antiparallel dimeric G-quadruplex with edgewise loops (Figure 4c′). Six guanines were anti, and six guanines were syn: 6 strong H8–H1′ NOE cross-peaks was observed (Figure S2). The topologies of U6 and U1B7 shown in Figure 4c,c′ were also supported by NOE cross-peaks observed in other spectral regions such as imino–imino, aromatic–aromatic, and aromatic–sugar proton regions (data not shown).
Parallel and Antiparallel G-Quadruplexes Can Coexist in Solution
We note that the minor form of U6 (or U1B7) has NMR features similar to those of the major form of U1B7 (or U6): number of peaks; chemical shift; NOE patterns. In the case of the natural sequence, both structures could be formed; however, the U6-like parallel G-quadruplex was predominant (Figure 1).
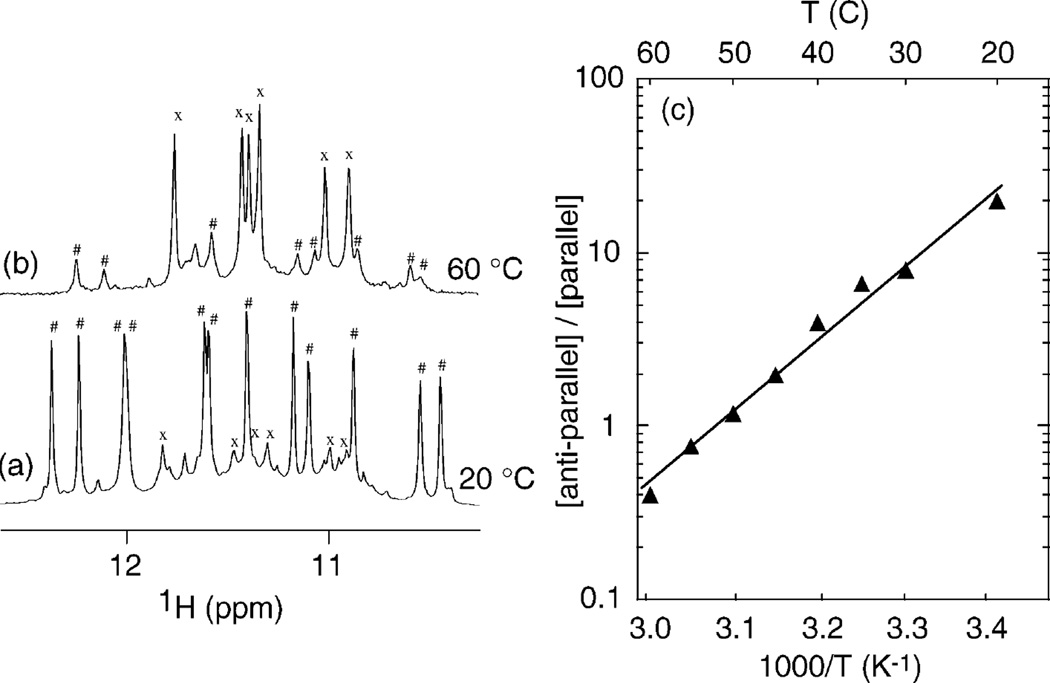
To study the interconversion between the two forms, we focus on the sequence U1B7. The proportions of the two forms in U1B7 depend on the temperature (Figure 5). The parallel and antiparallel G-quadruplexes are characterized by 6 imino protons (labeled with “x”) and 12 imino protons (labeled with “#”), respectively. The antiparallel form is more favorable at lower temperatures, whereas the parallel form is more favorable at higher temperatures. The parallel G-quadruplex, which is a minor species at 20 °C (Figure 5a), becomes predominant at 60 °C (Figure 5b). The ratio of antiparallel to parallel equilibrium concentrations is plotted as a function of temperature in Figure 5c. The corresponding enthalpy is 77 kJ/mol.
Figure 5.
Proportion of parallel and antiparallel G-quadruplexes in U1B7 depending on the temperature. (a, b) Imino proton spectra of U1B7 at equilibrium at (a) 20 °C and (b) 60 °C. The parallel form is characterized by 6 peaks (labeled with “x”); the antiparallel form is characterized by 12 peaks (labeled with “#”). The parallel G-quadruplex is a minor form at 20 °C; the antiparallel G-quadruplex is a minor form at 60 °C. (c) Equilibrium ratio of antiparallel G-quadruplex to parallel G-quadruplex concentrations versus temperature. Experimental conditions: strand concentration, 0.7 mM; 90 mM KCl; pH 7.
Kinetics of Folding and Unfolding of Different G-Quadruplexes
It has been suggested that the simpler fold of the parallel structural models, compared to the antiparallel ones, would facilitate the folding and unfolding processes of G-quadruplex structures in the telomere.5 We present in this section the measurements of folding and unfolding rates for the two G-quadruplex forms of U1B7.
(a) Antiparallel G-Quadruplex Folds Faster Than Parallel G-Quadruplex
To study the folding of different G-quadruplex forms in U1B7, the structures were first unfolded by heating at 90 °C for 3 min and the sample was then quickly cooled to a given temperature (e.g. 25 °C). The formation of the two G-quadruplex forms at 25 °C was followed by their imino protons. The initial slope of the buildup curves (Figure 6) allowed determination of the folding time (see Materials and Methods).
Figure 6.
Formation of parallel and antiparallel G-quadruplexes of U1B7 in real time. Squares and triangles represent concentrations (in arbitrary units, au) of parallel and antiparallel G-quadruplexes, respectively, which were derived from the intensities of imino protons of each form. Plot b is an expansion of a region of plot a. The initial slopes for buildup curves are drawn with dashed lines. Experimental conditions: strand concentration, 0.7 mM; temperature, 25 °C; 90 mM KCl; pH 7.
The folding times obtained at different temperatures were plotted in Figure 7. The antiparallel G-quadruplex folded slightly faster than the parallel G-quadruplex. The activation enthalpy was 138 and 161 kJ/mol for the antiparallel and the parallel form, respectively.
Figure 7.
Folding time of parallel and antiparallel G-quadruplexes of U1B7 as a function of temperature. Squares and triangles represent parallel and antiparallel G-quadruplexes, respectively. Experimental conditions: strand concentration, 0.7 mM; 90 mM KCl; pH 7.
We note that in our experimental conditions, despite being not favorable at equilibrium, the parallel G-quadruplex folded only at a slightly lower rate than the antiparallel one. The former could exceed its equilibrium concentration at the beginning of the folding experiment (Figure 6). For example, at 20 °C the amount of the parallel form was comparable to that of the antiparallel form after 1 h but became minor after 86 h and at equilibrium (Figure S3).
(b) Parallel G-Quadruplex Unfolds Faster Than Antiparallel G-Quadruplex
Previously, unfolding times of an intramolecular G-quadruplex from a human telomeric fragment could be determined by measurements of exchange cross-peaks in NOESY-type experiments at 50–60 °C, conditions where the structured and unfolded forms were of comparable concentrations and exchanged with a time constant of a few seconds.10 Here, in the case of U6 and U1B7, no exchange peaks were observed in NOESY experiments at 60 °C, even when the structured and unfolded forms were of comparable concentrations, suggesting slower unfolding.
We then measured unfolding times of U1B7 G-quadruplexes in real time. In the first method, we added the excess of the complementary C-strand into the solution of the G-strand, which formed G-quadruplex structures. The unfolded fraction of the G-strand would be trapped by the C-strand into a Watson–Crick duplex.10,11 The amount of the two G-quadruplex forms decayed to zero due to their transitions to the single strand. We could measure the unfolding time of each G-quadruplex structure by following guanine imino protons at 10.5–12.5 ppm; the newly formed Watson–Crick duplex was also followed by imino protons at 13–14 ppm (Figure 8a). The unfolding time of each G-quadruplex was obtained from a single-exponential fit of the decay (Figure 8b). In contrast to previous studies,11 where the decay was fitted to double exponential, a single exponential fitted our data perfectly well, since we could monitor individually each form at high resolution. Under the conditions of our experiment (at millimolar concentration), the hybridization between the two strands was not the rate-limiting step and the measured unfolding time was independent of the concentration of the C- or G-strands (data not shown).
Figure 8.
Measurement of unfolding time for parallel and antiparallel G-quadruplexes of U1B7 by complementary strand trapping. (a) Imino proton spectra of the mixture at time t = 0.1 h and t = 6 h. The concentrations of the Watson–Crick duplex and two G-quadruplexes were monitored by imino protons (labeled with circle, triangle, and square, respectively). (b) Evolution of concentrations (in arbitrary units, au) of two G-quadruplexes and Watson–Crick duplex as a function of time. The decay of each G-quadruplex was fitted with a single-exponential curve; the rise of the Watson–Crick duplex was fitted with a double exponential. Experimental conditions: strand concentration, 0.4 mM for the G-strand and 2 mM for the C-strand; temperature, 35 °C; 70 mM KCl; 20 mM potassium phosphate; pH 7.
Unfolding times were plotted as a function of the temperature in Figure 9. At temperatures lower than 40 °C, the parallel G-quadruplex unfolds faster than the antiparallel G-quadruplex. The activation enthalpy was 180 kJ/mol for the antiparallel form, slightly higher than for the parallel form (143 kJ/mol).
Figure 9.
Unfolding time of parallel and antiparallel G-quadruplexes of U1B7 as a function of temperature. Squares and triangles represent parallel and antiparallel G-quadruplex, respectively. Filled and open data represent measurements by complementary-strand-trap and by concentration-jump methods, respectively. Experimental conditions: strand concentration, 0.04–0.4 mM; 70 mM KCl; 20 mM potassium phosphate; pH 7.
To confirm that in the measurements just described the addition of the C-strand does not affect the unfolding time of G-quadruplexes, we also performed other independent measurements of G-quadruplex unfolding using the concentration-jump method. We diluted quickly (about 100-fold) a concentrated sample and observed how the fraction of the G-quadruplex structures decreased to reach the new equilibrium with the single strand (Figure S4). The unfolding time can be determined from the initial rate of the change in concentration of the G-quadruplexes (see Materials and Methods). Unfolding times measured from 40 to 52 °C were plotted in Figure 9. These results were consistent with the unfolding times measured above by complementary strand trapping. Data from temperature-jump experiments (see Material and Methods) also supported that the parallel G-quadruplex unfolded faster than the antiparallel G-quadruplex at temperatures below 40 °C (Figure S5).
Discussion
In this study we demonstrated that the two-repeat human telomeric sequence d(TAGGGTTAGGGT) can form both parallel and antiparallel G-quadruplex structures in K+-containing solution. The two structures can coexist and interconvert in solution. For the modified U1B7 sequence, the antiparallel form is more favorable at lower temperatures, whereas the parallel form is more favorable at higher temperatures. There are several proteins that bind G-quadruplex structures. A protein could selectively recognize and use one of the two forms for biological functions in telomeres. It would be also interesting to design drugs that specifically target one form and modulate such biological functions.
These two G-quadruplex structures have different kinetics of folding and unfolding. For the U1B7 sequence, the antiparallel G-quadruplex folds faster but unfolds slower than the parallel G-quadruplex at physiological temperatures. This folding and unfolding kinetics could be a factor that determines the relative amounts of different G-quadruplex forms in telomeres during the cell cycle. Again, intermolecular interactions of either forms with proteins and drugs could modulate the rate of telomeric G-quadruplex folding and unfolding.13
The four-repeat human telomere d[AGGG(TTAGGG)3] sequence has been found to adopt completely different G-quadruplex architectures (different looping, strand orientations, and glycosidic conformations of guanines) in Na+-containing solution2 and in K+-containing crystal.5 We have also examined the same sequence in K+-containing solution and observed multiple forms (unpublished results). The results from this study of the two-repeat sequence suggest that both Na+-solution and K+-crystal structures are likely to coexist in K+-containing solution and could potentially be in equilibrium with additional forms.
Questions about differences between crystal and solution structures have been asked for the closely related 1.5-repeat telomeric sequence d(G4T4G4) from Oxytricha, which has been examined both by X-ray and NMR in different laboratories.14 Our results suggest that different G-quadruplex topologies may differ very slightly in stability and the same sequence may form different G-quadruplex topologies under different experimental conditions. We note that for technical reasons both X-ray and NMR studies often use analogue substitutions such as (T → BrU), (T → U), (G → BrG), or (G → I), which may favor one or the other conformation.
Conclusion
The G-rich strand from the human telomere can form both parallel and antiparallel G-quadruplex structures in K+-containing solution. They have different thermodynamic properties and different kinetics of folding and unfolding. Several G-quadruplex topologies can be present at the same time, and their proportion is very sensitive to the sequence in the loops connecting G-stretches.
Supplementary Material
Acknowledgment
This research was supported by NIH Grant GM34504.
Footnotes
Supporting Information Available: Figures S1–S5 (H2–H8 and H1′–H8 regions of the NOESY spectra confirming G-quadruplex topologies and studies of unfolding kinetics by concentration- and temperature-jump methods). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Neidle S, Parkinson G. Nat. Rev. Drug. Des. 2002;1:383–393. doi: 10.1038/nrd793. [DOI] [PubMed] [Google Scholar]; (b) Hurley LH. Nat. Rev. Cancer. 2002;2:188–200. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]; (c) Mergny JL, Hélène C. Nat. Med. 1998;4:1366–1367. doi: 10.1038/3949. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y, Patel DJ. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y, Patel DJ. Structure. 1994;2:1141–1156. doi: 10.1016/s0969-2126(94)00117-0. [DOI] [PubMed] [Google Scholar]
- 4.(a) Kettani A, Gorin A, Majumdar A, Hermann T, Skripkin E, Zhao H, Jones R, Patel DJ. J. Mol. Biol. 2000;297:627–644. doi: 10.1006/jmbi.2000.3524. [DOI] [PubMed] [Google Scholar]; (b) Kuryavyi V, Majumdar A, Shallop A, Chernichenko N, Skripkin E, Jones R, Patel DJ. J. Mol. Biol. 2001;310:181–194. doi: 10.1006/jmbi.2001.4759. [DOI] [PubMed] [Google Scholar]; (c) Zhang N, Gorin A, Majumdar A, Kettani A, Chernichenko N, Skripkin E, Patel DJ. J. Mol. Biol. 2001;311:1063–1079. doi: 10.1006/jmbi.2001.4916. [DOI] [PubMed] [Google Scholar]
- 5.Parkinson GN, Lee MPH, Neidle S. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 6.(a) Phan AT, Patel DJ. J. Am. Chem. Soc. 2002;124:1160–1161. doi: 10.1021/ja011977m. [DOI] [PubMed] [Google Scholar]; (b) Phan AT, Patel DJ. J. Biomol. NMR. 2002;23:257–262. doi: 10.1023/a:1020277223482. [DOI] [PubMed] [Google Scholar]
- 7.Plateau P, Guéron M. J. Am. Chem. Soc. 1982;104:7310–7311. [Google Scholar]
- 8.(a) Phan AT, Guéron M, Leroy JL. Methods Enzymol. 2001;338:341–371. doi: 10.1016/s0076-6879(02)38228-4. [DOI] [PubMed] [Google Scholar]; (b) Phan AT, Guéron M, Leroy JL. J. Mol. Biol. 2000;299:123–144. doi: 10.1006/jmbi.2000.3613. [DOI] [PubMed] [Google Scholar]
- 9.(a) Phan AT. J. Biomol. NMR. 2000;16:175–178. doi: 10.1023/a:1008355231085. [DOI] [PubMed] [Google Scholar]; (b) van Dongen MJP, Wijmenga SS, Eritja R, Azorin F, Hilbers CW. J. Biomol. NMR. 1996;8:207–212. doi: 10.1007/BF00211166. [DOI] [PubMed] [Google Scholar]; (c) Phan AT. J. Magn. Res. 2001;153:223–226. doi: 10.1006/jmre.2001.2445. [DOI] [PubMed] [Google Scholar]; (d) Piantini U, Sorensen OW, Ernst RR. J. Am. Chem. Soc. 1982;104:6800–6801. [Google Scholar]; (e) Bax A, Davis DG. J. Magn. Res. 1985;65:355–360. [Google Scholar]; (f) Kellogg GW. J. Magn. Res. 1992;98:176–182. [Google Scholar]
- 10.Phan AT, Mergny JL. Nucleic Acids Res. 2002;30:4618–4625. doi: 10.1093/nar/gkf597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.(a) Raghuraman MK, Cech TR. Nucleic Acids Res. 1990;18:4543–4552. doi: 10.1093/nar/18.15.4543. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Green JJ, Ying L, Klenerman D, Balasubramanian S. J. Am. Chem. Soc. 2003;125:3763–3767. doi: 10.1021/ja029149w. [DOI] [PubMed] [Google Scholar]
- 12.The number of all (major) peaks in U6 corresponds to 12 residues, indicating a symmetric dimer. Major peaks of U1B7 correspond to 24 spin systems. Their equal intensity under different conditions and the observed NOE patterns indicate an asymmetric dimer.
- 13.(a) Fang G, Cech TR. Biochemistry. 1993;32:11646–11657. doi: 10.1021/bi00094a022. [DOI] [PubMed] [Google Scholar]; (b) Han H, Cliff CL, Hurley LH. Biochemistry. 1999;38:6981–6986. doi: 10.1021/bi9905922. [DOI] [PubMed] [Google Scholar]
- 14.(a) Kang C, Zhang X, Ratliff R, Moyzis R, Rich A. Nature. 1992;356:126–131. doi: 10.1038/356126a0. [DOI] [PubMed] [Google Scholar]; (b) Smith FW, Feigon J. Nature. 1992;356:164–168. doi: 10.1038/356164a0. [DOI] [PubMed] [Google Scholar]; (c) Horvath MP, Schultz SC. J. Mol. Biol. 2001;310:367–377. doi: 10.1006/jmbi.2001.4766. [DOI] [PubMed] [Google Scholar]; (d) Haider S, Parkinson GN, Neidle S. J. Mol. Biol. 2002;320:189–200. doi: 10.1016/S0022-2836(02)00428-X. [DOI] [PubMed] [Google Scholar]; (e) Haider S, Parkinson GN, Neidle S. J. Mol. Biol. 2003;326:117–125. doi: 10.1016/s0022-2836(02)01354-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.