Abstract
Phenotypes resulting from mutations in genetic model organisms can help reveal candidate genes for evolutionarily important phenotypic changes in related taxa. Although testing candidate gene hypotheses experimentally in nonmodel organisms is typically difficult, ontology-driven information systems can help generate testable hypotheses about developmental processes in experimentally tractable organisms. Here, we tested candidate gene hypotheses suggested by expert use of the Phenoscape Knowledgebase, specifically looking for genes that are candidates responsible for evolutionarily interesting phenotypes in the ostariophysan fishes that bear resemblance to mutant phenotypes in zebrafish. For this, we searched ZFIN for genetic perturbations that result in either loss of basihyal element or loss of scales phenotypes, because these are the ancestral phenotypes observed in catfishes (Siluriformes). We tested the identified candidate genes by examining their endogenous expression patterns in the channel catfish, Ictalurus punctatus. The experimental results were consistent with the hypotheses that these features evolved through disruption in developmental pathways at, or upstream of, brpf1 and eda/edar for the ancestral losses of basihyal element and scales, respectively. These results demonstrate that ontological annotations of the phenotypic effects of genetic alterations in model organisms, when aggregated within a knowledgebase, can be used effectively to generate testable, and useful, hypotheses about evolutionary changes in morphology.
Keywords: molecular evolution, gene expression, evolutionary phenotypes, catfish, nonmodel organism
Introduction
Identifying the genetic and developmental changes that underlie the morphological diversification of life is an ultimate goal of many research programs in developmental and evolutionary biology. Some of the most spectacular advances in biology in the past decade have been gained through this approach, including the discovery that genes, genetic architectures, expression patterns, networks, and developmental processes are highly conserved, well beyond expectation, even across very distantly related organisms (Degnan 2010).
Although our knowledge has deepened about the bases of phenotypic divergence along major aspects of organismal architectures (e.g., flower patterning; body axes and segment patterning), the developmental genetic basis for most of the known phenotypic transitions in the evolution of life remains unexplored nonetheless. One reason for this is that laboratories often focus on a single gene or gene network in one (or few) model taxa to pinpoint candidate genes that are known from other (model) species. Ideally, however, an investigator could take the set of novel features for a clade and query model organism databases for similar phenotypes resulting from genetic perturbations. The roles of these genes in the evolutionary phenotype could then be tested experimentally, provided an experimentally tractable system exists.
A data-driven approach for generating candidate gene hypotheses requires the capacity to process computationally the vast number of model organism phenotypes and large ontologies that allow software to recognize similarities and relationships among phenotypes regardless of what vocabulary researchers use to describe them (Mabee, Ashburner, et al. 2007). In recent years, inspired by the success of the Gene Ontology (GO) in enabling computation over descriptive information about gene function (Blake and Harris 2008), ontologies have been developed and fruitfully applied to phenotypic data (Deans et al. 2015). Though developed initially for use in disease gene discovery (Washington et al. 2009; Hoehndorf et al. 2011; Oellrich et al. 2012), ontologies such as Uberon (Haendel et al. 2014) and Phenotype and Trait Ontology (PATO) (Gkoutos, Green, Mallon, Blake, et al. 2004) empower researchers to imagine a much wider variety of computational applications for descriptive phenotype information.
Here, we demonstrate that compilations of data describing the phenotypic effects of genetic alterations in model organisms can be leveraged to generate hypotheses about evolutionary changes in morphology. To do this, we took advantage of the Phenoscape Knowledgebase (http://kb.phenoscape.org), an ontology-driven data resource containing information about both mutant zebrafish (Danio rerio) phenotypes curated by the zebrafish model organism database (ZFIN, http://zfin.org; Ruzicka et al. 2015) and evolutionary phenotype variation in ostariophysan fishes, the group of fishes that includes zebrafish, as documented in the comparative morphology and systematics literature (Mabee, Arratia, et al. 2007). We used the Phenoscape Knowledgebase to generate hypotheses about candidate genes for experimentally tractable evolutionary phenotypes (see Materials and Methods), which yielded candidate genes for two striking phenotypes that are characteristic of the diverse, ecologically and economically important clade of catfishes (Siluriformes) with over 3,600 living species: 1) Loss of the “tongue” or basihyal element (i.e., cartilage and bone; hereafter referred to as basihyal; Arratia and Schultze 1990; de Pinna 1993) and 2) loss of scales (Fink SV and Fink WL 1981). Here we present, for the first time, endogenous expression patterns of candidate genes potentially involved with the evolutionary loss of basihyal (bromodomain and PHD finger containing 1, brpf1; Laue et al. 2008) and scales (ectodysplasin A, eda; ectodysplasin A receptor, edar; Harris et al. 2008) using whole mount and cryosection in situ hybridization, respectively.
We tested the ability of the Phenoscape Knowledgebase to identify candidate genes for evolutionary phenotypes in experimentally accessible organisms (fig. 1). For this, we used the commercially important channel catfish (Ictalurus punctatus) as a tractable representative of Siluriformes. Although no catfish species are routinely used for developmental studies, I. punctatus is widespread throughout North America, commercially bred in aquaculture (USDA 2005), and has EST (expressed sequence tag) sequences (Li et al. 2007) as well as database resources (cBARBEL; Lu et al. 2011) available. Moreover, I. punctatus has been used previously for molecular (Waldbieser et al. 2001; Li and Waldbieser 2006; Xu et al. 2006) and immune system investigations (Kocabas et al. 2002; Bao et al. 2006; Baoprasertkul et al. 2006; Peatman et al. 2007). Importantly, I. punctatus can be considered a viable candidate for in situ gene expression studies (Steinke et al. 2006; Chen et al. 2010; Wang et al. 2010; Liu 2011; Liu et al. 2011; Ninwichian et al. 2012; Jiang et al. 2013; Zhang et al. 2013).
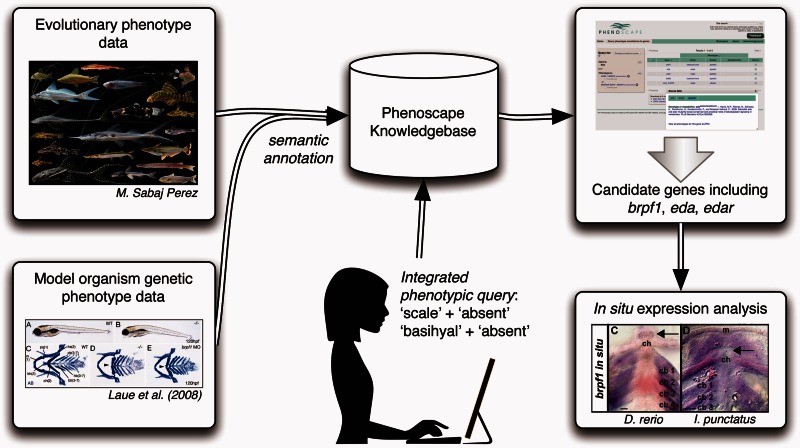
Fig. 1.
Flow chart showing computational and experimental steps used to propose and test candidate genes for evolutionary phenotypic novelties. Evolutionary phenotype data for fish species and model organism genetic phenotype data for zebrafish (from ZFIN) are semantically annotated and housed in the Phenoscape Knowledgebase. A user query to the Knowledgebase for genes associated with evolutionary phenotypes of interest (here, scales absent and basihyal absent) returns a list of candidate genes based on the model organism data. These in silico candidates can be experimentally assessed (e.g., in situ gene expression analysis using Danio rerio and Ictalurus punctatus).
Results
In Silico Analysis Identifies Candidate Genes for Evolutionary Phenotypes
As is the case for most nonmodel taxa, necessary baseline information about developmental staging and morphology (Gilbert 2009; Rowan et al. 2011), specifically the timing of pharyngeal arch chondrification, was unavailable. In light of this, samples were collected across developmental time-points for the channel catfish (I. punctatus), South American cave-dwelling suckermouth armored catfish (A. cf. triradiatus), and armored bronze corydoras catfish (Corydoras aeneus) to establish the species-specific developmental timing of pharyngeal arch chondrification. Both A. cf. triradiatus and C. aeneus were readily available and have been used previously in studies of jaw development (Geerinckx et al. 2005, 2007; Geerinckx and Adriaens 2007, 2008) and jaw morphology (Huysentruyt et al. 2007, 2008, 2009, 2011), respectively.
Two prominent phenotypic changes that distinguish catfishes from other ostariophysan fishes are the absence of a basihyal (“tongue”; fig. 2) and the absence of elasmoid scales that characterize most actinopterygian fishes (fig. 3; Fink SV and Fink WL 1981; Arratia and Schultze 1990; de Pinna 1998; Sire and Akimenko 2004). The scutes (i.e., postcranial dermal plates of armored catfishes, e.g., Callichthyidae, Loricariidae, Doradidae, etc.; Sire and Huysseune 2003) develop differently from elasmoid scales (Sire 1993) and are a derived condition within catfishes (Fink SV and Fink WL 1996). Using the Phenoscape Knowledgebase, we sought genes that could be candidates responsible for these phenotypic differences in the zebrafish model based on genetic and phenotype data from ZFIN. Gene phenotype–taxon phenotype associations were generated from the Phenoscape Knowledgebase using matching anatomical entities, and then examined by hand. The gene phenotype annotations we obtained from ZFIN associate an aplastic or absent basihyal phenotype with the disruption of 11 zebrafish genes, including bromodomain and PHD finger containing 1 (brpf1; Laue et al. 2008), disrupted in schizophrenia 1 (disc1; Wood et al. 2009), dispatched homolog 1 (disp1; Schwend and Ahlgren 2009), facelift (fac; Schilling et al. 1996), forkhead box D3 (foxd3; Neuhauss et al. 1996), heart and neural crest derivatives expressed 2 (hand2; Miller et al. 2003), K(lysine) acetyltransferase 6A (kat6a; Laue et al. 2008), SRY (sex determining region Y)-box 9a (sox9a; Yan et al. 2002), unnamed th9 (unm_th9), unnamed tn20c (unm_tn20c), and unnamed ty5 (unm_ty5). We selected brpf1 as a candidate gene because mutation or knockdown in D. rerio results in phenotypes similar to known features of catfishes (e.g., loss of basihyal; Laue et al. 2008). An aplastic or absent scale is associated with the disruption of three zebrafish genes, including eda and edar (Harris et al. 2008), and unnamed t31273 (unm_t31273), which is a gene of unknown function; therefore, we selected eda and edar as candidates for investigating scale loss in catfish because they are experimentally tractable given their known functions in zebrafish.
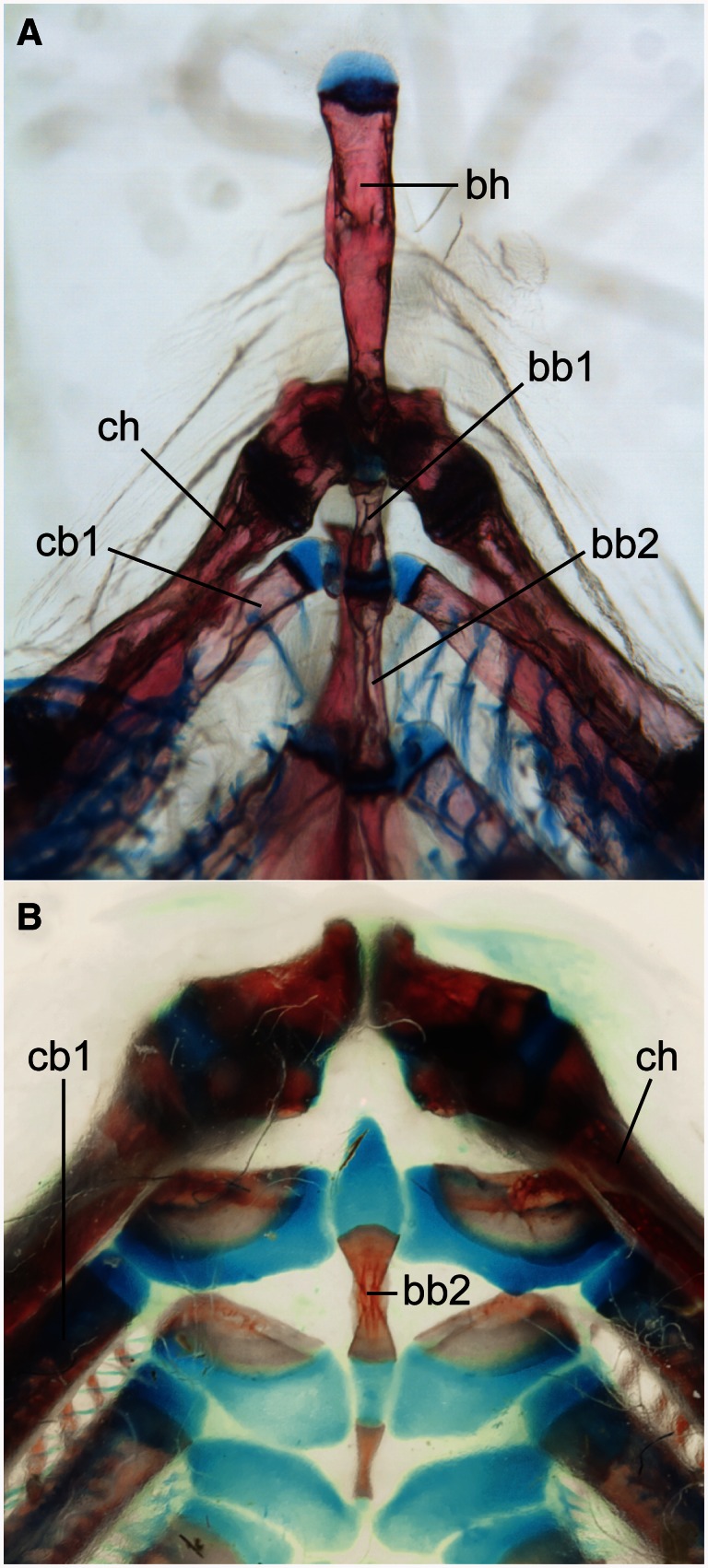
Fig. 2.
Loss of basihyal element in catfishes. Catfishes (Siluriformes) are characterized by the loss of the basihyal element in contrast to relatives, including zebrafish. (A) Zebrafish, Danio rerio (purchased from an aquarium fish store by P. Mabee). (B) Spotted Bullhead Catfish, Ameiurus serracanthus (ANSP 185358; provided by K. Luckenbill). Images show lower branchial elements in ventral view. bb, basibranchial; bh, basihyal; cb, ceratobranchial; ch, ceratohyal. Scale bars = 100 µm.
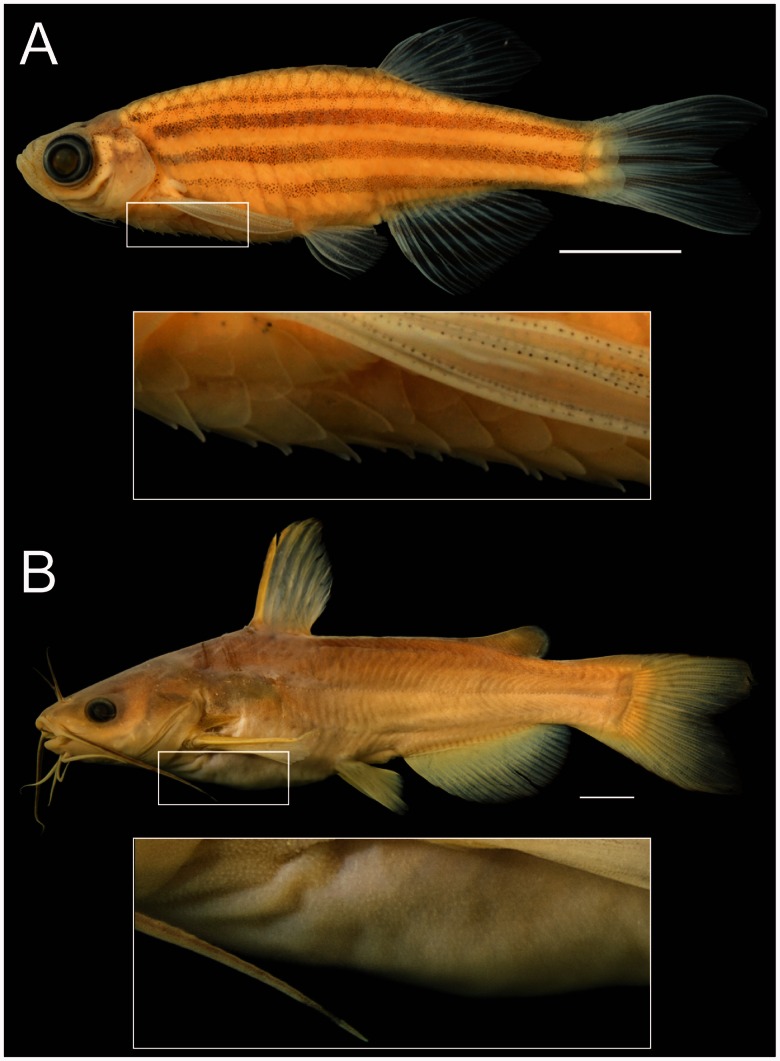
Fig. 3.
Loss of scales in catfishes. Catfishes (Siluriformes) are characterized by the loss of the scales in contrast to relatives, including zebrafish. (A) Zebrafish, Danio rerio (ANSP 189304; provided by K. Luckenbill) in lateral view with close up of scales. (B) White Catfish, Ameiurus catus (ANSP 11678; provided by M. A. Arce-Hernandez) in lateral view with close up of skin. Note that the bumps on the skin visible in the insert are a variety of soft-tissue structures all without bone, likely including externalized taste buds, free-neuromasts, and integumentary glands. Scale bars = 1 cm.
Developmental Morphology and Gene Expression Indicate the Roles of Candidate Genes in Evolutionary Phenotypes
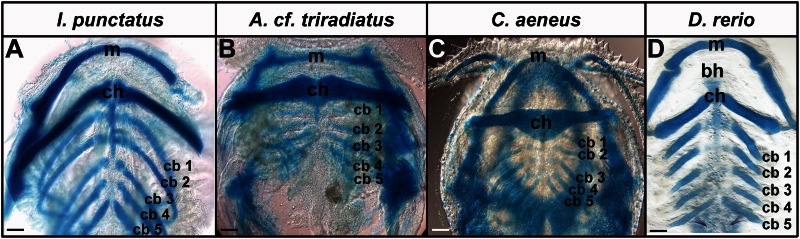
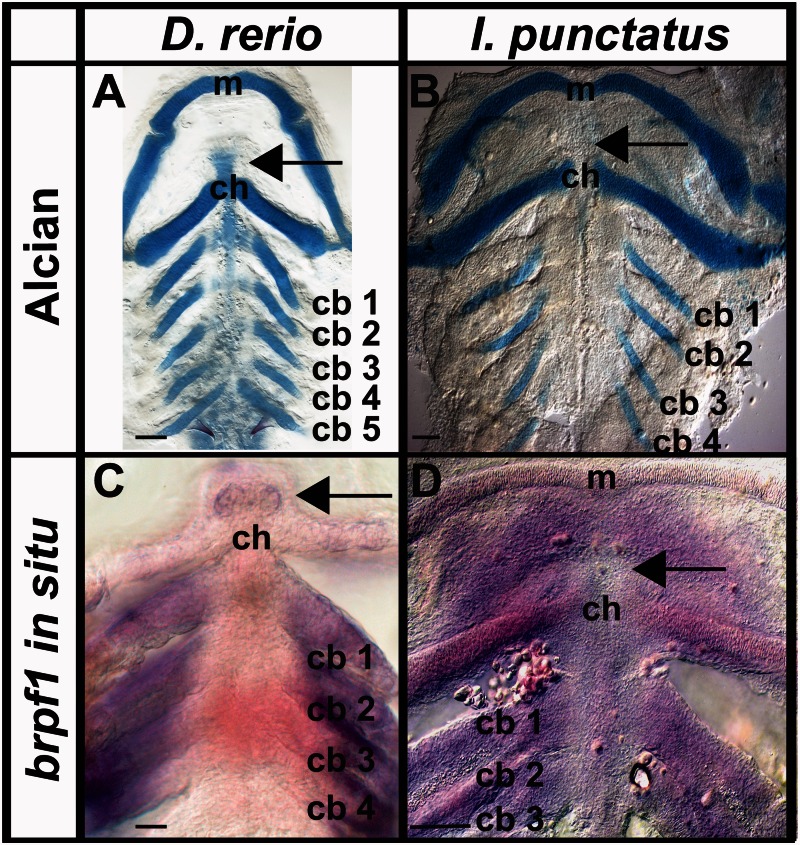
Alcian Blue staining of embryonic I. punctatus, A. cf. triradiatus, and C. aeneus revealed complete chondrification of all arches (mandibular, hyoid, and ceratobranchials 1–5) by 96, 96, and 102 h postfertilization (hpf), respectively (fig. 4).
Fig. 4.
Pharyngeal arches develop later in catfish than in zebrafish. Chondrification of jaw (mandibular arch), hyoid arch, and branchial arches as detected by Alcian Blue staining. Images show lower branchial elements in ventral view. (A) Ictalurus punctatus, 96 hpf; (B) Ancistrus cf. triradiatus, 96 hpf; (C) Corydoras aeneus, 102 hpf; (D) Danio rerio, 72 hpf. m, Meckel’s cartilage; bh, basihyal; ch, ceratohyal cartilage; cb, ceratobranchial cartilages. Scale bars = 100 µm.
Because eda, edar, and brpf1 expression had not previously been investigated in catfishes, we verified the presence of transcripts in all three species by reverse transcription polymerase chain reaction (RT-PCR) across early developmental stages (24–120 hpf; data not shown). Sequencing and ClustalW alignment (Larkin et al. 2007) of RT-PCR products confirmed higher levels of eda and edar nucleotide sequence similarity (i.e., identical nucleotides) among catfish species (97–100% similar) than between catfishes and zebrafish (74–76% similar). Lower levels of nucleotide sequence similarity were observed for brpf1 sequences among catfishes (71–86% similar) than between catfishes and zebrafish (72–73% similar).
We used Alcian Blue and Alcian Green staining to establish 72 and 86 hpf as key time-points in D. rerio and I. punctatus early pharyngeal arch skeletal development (i.e., chondrification of certatobranchials 1–4 and midline cartilages; fig. 5A and B; Eames et al. 2013), respectively. We then targeted these developmental time-points for investigating the loss of basihyal formation as it relates to endogenous brpf1 expression in a representative cypriniform (basihyal present) and siluriform (basihyal absent) species (fig. 5C and D), respectively. Brpf1 expression was observed in all arches of D. rerio by 72 hpf and I. punctatus embryos by 86 hpf, as expected (fig. 5C and D; Laue et al. 2008). However, brpf1 expression was absent in the midline of the hyoid arch (fig. 5D), where the basihyal normally forms in nonsiluriform fishes (e.g., D. rerio, fig. 5C; Cubbage and Mabee 1996; Parichy et al. 2009; Mabee et al. 2011). To ensure that there were actually cells in this region that failed to express brpf1 in I. punctatus, we counterstained embryos after in situ hybridization with Toluidine Blue and removed the pharyngeal arches by dissection. This method confirmed the presence of cells in the anterior hyoid region at these developmental time-points (data not shown). These experiments establish, for the first time, that brpf1 expression is absent in cells immediately anterior to the hyoid arch in I. punctatus during pharyngeal arch development, whereas the basihyal is present and expresses brpf1 in nonsiluriform fishes (e.g., D. rerio; Laue et al. 2008).
Fig. 5.
Catfishes lack brpf1 expression in the region where the basihyal forms in zebrafish. Expression of brpf1 during pharyngeal arch skeletal development in Danio rerio and Ictalurus punctatus, as detected by whole-mount in situ hybridization. Staining of pharyngeal arch cartilages using (A) Alcian Blue in D. rerio (72 hpf) and (B) Alcian Green in I. punctatus (86 hpf). Endogenous brpf1 expression in (C) D. rerio (72 hpf) and (D) I. punctatus (86 hpf). Arrows indicate location of basihyal element in D. rerio (cypriniform) and lack of basihyal element in I. punctatus (siluriform). Images are representative of ≥30 embryos. No labeling was detected in negative controls (data not shown). m, Meckel’s cartilage; ch, ceratohyal cartilage; cb, ceratobranchial cartilages. Scale bars = 100 µm.
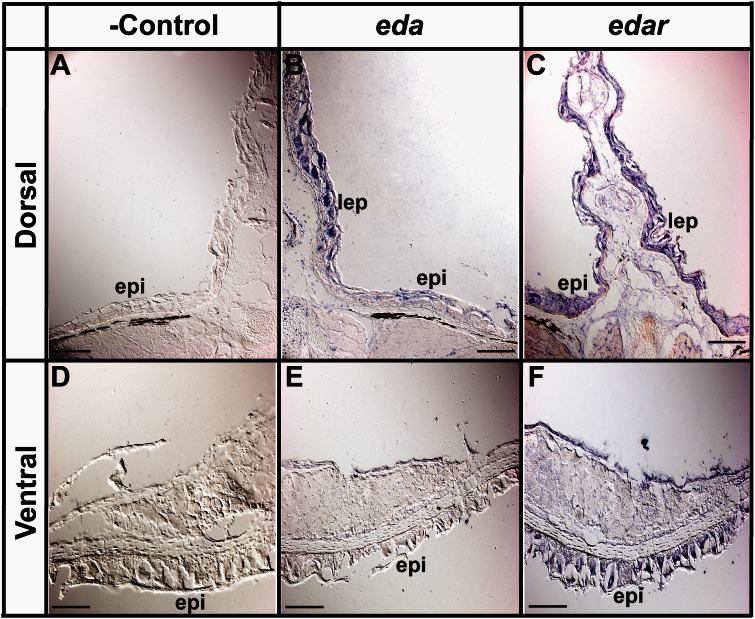
In 4-week-old I. punctatus juveniles, both eda and edar are expressed in dorsal fin lepidotrichia, as expected (fig. 6B and C; Harris et al. 2008). We also detected robust edar expression in the dorsal and ventral epidermis (fig. 6C and F); however, robust eda expression was not detected in the dorsal and ventral epidermis (fig. 6B and E). Because both eda and edar are strongly expressed in zebrafish epidermis (Harris et al. 2008), these in situ hybridization experiments demonstrate that eda expression has been reduced, or potentially silenced, in epidermal cells of juvenile I. punctatus. These results are consistent with the hypothesis that modification of the eda signaling pathway (i.e., loss of ligand expression but not receptor expression) underlies the evolutionary loss of scales in Siluriformes.
Fig. 6.
Epidermis of 4-week-old Ictalurus punctatus lacks eda expression. In situ hybridization using DIG-labeled riboprobes on tissue cryosections was used to determine the patterning of endogenous eda and edar expression (see Materials and Methods). (A, D) Negative hybridization control for dorsal and ventral epidermal tissues, respectively. (B, C) Expression of eda and edar in dorsal fin lepidotrichia as expected (Harris et al. 2008). (E, F) Absent and present endogenous expression of eda and edar in ventral epidermal tissues, respectively. Images are representative of ≥10 cryosectioned juveniles. epi, epidermis; lep, lepidotrichia. Scale bars = 100 µm.
Discussion
A central quest in biology is to understand how the diversity of phenotypes is regulated genetically. Although it has long been possible to comb the literature to find individual candidate genes of interest for a phenotype observed in a different taxon, recent efforts to scale up the capture of both genetic and phenotypic variation (Binder and Zon 2013), and the online accessibility of information from these studies (ZFIN; Ruzicka et al. 2015), provide a strong motivation to develop computational tools that support efficient exploration of causal linkages between genes and phenotypes. Genetic and phenotypic comparisons of phylogenetically disparate model organisms have revealed oftentimes surprisingly deep conservation of the role of genes in developmental processes, as well as an expectation that similar phenotypes (homologous or not) are likely to share genetic pathways and possibly common regulators (e.g., Ritter et al. 2010; Gallant et al. 2014). Techniques for testing developmental genetic hypotheses in nonmodel organisms are becoming increasingly sophisticated, providing experimental investigation of candidate genes for a much wider variety of phenotypes in nature (Abzhanov et al. 2008).
The Phenoscape Knowledgebase, which integrates the peer-reviewed fish morphology literature with zebrafish model organism database data (phenotypes and genotypes), allows evo-devo researchers to develop hypotheses that otherwise would be impossible to generate manually because of the immensity of information (Manda et al. 2015). Currently, morphology data are not present in any other database in a form that can be compared directly with model organism data. The Phenoscape Knowledgebase expedites the process of hypothesis generation and expands it to a scale never before possible. Here, we have demonstrated the viability of such an approach for mining existing genotype–phenotype information computationally to generate candidate genes for phenotypes in a nonmodel organism. The candidate genes brpf1 and eda/edar were predicted by the Phenoscape Knowledgebase to play roles in the evolutionary losses of the basihyal and scales in Siluriformes, respectively. These computational predictions were empirically tested by examining the expression patterns of candidate genes in situ in an experimentally tractable representative of Siluriformes, the channel catfish (I. punctatus).
To investigate the evolutionary losses of the basihyal and scales in Siluriformes, we characterized the timing of pharyngeal arch development in three catfish species (I. punctatus, C. aeneus, and A. cf. triradiatus) as well as the pattern of endogenous brpf1 expression in embryonic and juvenile I. punctatus, and compared these patterns to zebrafish. Although all catfishes lack a basihyal, little else is known about their pharyngeal arch development. Zebrafish develop a basihyal by 72 hpf (Cubbage and Mabee 1996; Parichy et al. 2009; Engeman and Mabee 2012), and our analysis suggested that pharyngeal arch development has reached a similar developmental stage by 78–86 hpf in I. punctatus. Silencing brpf1, which encodes a MOZ histone acetyl transferase complex subunit, prevents basihyal formation and generates a broader and flattened mouth in loss-of-function zebrafish mutants (Laue et al. 2008). Loss of function of brpf1 in medaka (Oryzias latipes) bis mutants results in similar developmental craniofacial skeletal patterning defects, most notably the reduction in basihyal length (Hibiya et al. 2009). We found that I. punctatus lacks brpf1 expression in cells in the location where the basihyal forms in these other species. The absence of brpf1 expression in developing I. punctatus midline cells of the anterior hyoid region and its presence in the pharyngeal arches supports our hypothesis that loss of brpf1 expression (or an upstream regulator) may be mechanistically responsible for the loss of basihyal formation in the catfish lineage. One possible explanation is that brpf1 expression may be repressed in the anterior hyoid region by way of cis- or trans-acting regulators that are evolutionarily unique to Siluriformes. This hypothesis could be tested by examining the endogenous expression of associated Hox genes (Hibiya et al. 2009) in the craniofacial tissues of I. punctatus.
All catfishes also lack true scales and, instead, have a scaleless epidermis (Grizzle et al. 1976; Sire 1993; Konrádsdóttir et al. 2009; Burns et al. 2010). Zebrafish normally form scales, but mutations in the eda or edar genes that encode the ectodysplasin A signaling protein and receptor, respectively, result in failure of scale formation (Harris et al. 2008). Loss of scales was also observed in medaka (O. latipes) following mutational silencing of the rs-3 locus, which encodes the eda receptor protein EDAR (Kondo et al. 2001). Likewise, eda has been associated with variation in scale (postcranial dermal element) number of the three-spine stickleback (Gasterosteus aculeatus) that has undergone repeated selection in freshwater populations worldwide (Colosimo et al. 2005; Bell et al. 2010). We found that although cells of the epidermis in I. punctatus express edar, they fail to express eda (fig. 6).
Given the protein sequence homology for both EDA and EDAR among catfishes, which is consistent with previous observations that the EDA signaling pathway is evolutionarily conserved (Colosimo et al. 2005; Harris et al. 2008), we investigated whether an alteration in the eda signaling pathway underlies evolutionary scale loss in I. punctatus. The eda and edar expression patterns observed in epidermis adjacent to dorsal fin (weak and strong) and ventral epidermis (undetectable and detectable) support our hypothesis that a change in eda signaling pathway balance (i.e., reduction of ligand, but not receptor, expression) underlies this evolutionary difference, respectively. The reduction of eda ligand expression would potentially prevent organization of epithelial cells into signaling centers and dermal placodes and, in doing so, prevent basal epidermis fibroblasts from assembling during early stages of scale formation (Kondo et al. 2001; Colosimo et al. 2005; Bell et al. 2010). This putative mechanism would instead generate the scaleless (i.e., naked) epidermis characteristic of Siluriformes. In contrast, we found expression of edar in epidermal cells in zebrafish, consistent with a previous study (Harris et al. 2008). Presumably, the signaling through EDAR required for scale development is likely to be functionally inhibited by the reduction of the EDA ligand. Changes in the regulation of eda (e.g., transcriptionally or epigenetically) could potentially underlie the undetectable level of expression in juvenile I. punctatus epidermis.
These experimental data demonstrate that in silico hypotheses generated through ontology-driven searches of linked gene-phenotype data in model organisms can be leveraged to discover genes involved in evolutionary changes in morphology. Further development of methods (e.g., artificial intelligence; Gil et al. 2014) to aggregate genetic data from across model organisms and advance the translation of evolutionary morphology into a computable format is thus likely to accelerate our understanding of the genetic changes that underlie evolutionarily diverse phenotypes.
Materials and Methods
Sources of Data
Evolutionary phenotypes in the Phenoscape Knowledgebase are derived from 52 comparative anatomical studies (Fink SV and Fink WL 1981; Sawada 1982; Gardiner 1984; Schaefer 1987, 1991; Siebert 1987; Mayden 1989; Mo 1991; Cavender and Coburn 1992; Coburn and Cavender 1992; Lundberg 1992; Smith 1992; de Pinna 1993, 1996; Johnson and Patterson 1993; Chen 1994; Friel 1994; Bornbusch 1995; Vari 1995; Vari et al. 1995; Poyato-Ariza 1996; Mooi and Johnson 1997; Bockmann 1998; Buckup 1998; Lucena and Menezes 1998; Malabarba 1998; Shibatta 1998; Soares-Porto 1998; Weitzman and Menezes 1998; Arratia 1999; Di Dario 1999; Grande and Poyato-Ariza 1999; Royero 1999; Toledo-Piza 2000; Wiley et al. 2000; Albert 2001; Sanger and McCune 2002; Britto 2003; Chanet 2003; Chang and Maisey 2003; Fang 2003; Armbruster 2004; Grande et al. 2004; Kailola 2004; Otero 2004; Zaragüeta Bagils 2004; Zanata and Vari 2005; de Pinna et al. 2007; Toledo-Piza 2007; Grande T and Grande L 2008; Sidlauskas and Vari 2008; Vigliotta 2008). These are mostly phylogenetic analyses, with characters and states provided by the original authors in a structured and matrix-based format. Character states were annotated with ontology terms according to the Entity–Quality (EQ) formalism (Washington et al. 2009; Mungall et al. 2010) using Phenex software (Balhoff et al. 2010), in which the entity (E) is typically an anatomical structure that bears a phenotype. Here, entity terms are primarily from the Teleost Anatomy Ontology (TAO; Dahdul, Lundberg, et al. 2010) with some drawn from the GO (Blake and Harris 2008) and others from the Spatial Ontology (Dahdul et al. 2014). Quality (Q) terms, which can be applied to any taxon, were drawn from the PATO (Gkoutos, Green, Mallon, Hancock, et al. 2004; Sprague et al. 2008). EQ phenotypes were associated with taxa using a taxonomy ontology (here the Teleost Taxonomy Ontology; Dahdul, Balhoff, et al. 2010; Midford et al. 2013). Each of these associations, termed “taxon phenotype annotations,” represents part of or a full character state for a taxon. In this way, we created 361,346 taxon phenotype annotations for 2,242 taxa.
Associations of EQ phenotypes with genes, termed “gene phenotype annotations,” were obtained from ZFIN. ZFIN records the phenotypes generated by perturbations of genes, either by mutation or by morpholino knockdown, as reported in the developmental biology literature. These phenotypes are also described using the EQ formalism by using anatomical entities from the Zebrafish Anatomy Ontology, which are treated as species-specific subclasses of the multispecies anatomical entities within the TAO (Dahdul, Balhoff, et al. 2010). At the time of analysis, the Phenoscape Knowledgebase included a total of 11,586 gene phenotype annotations curated in this way, relating to genotypes of 2,966 zebrafish genes. Using this unified ontological framework, the Phenoscape Knowledgebase provides a query interface allowing search of both evolutionary and developmental genetic phenotypes using shared anatomical and phenotypic terms.
Experimental Validation of In Silico Candidate Gene Predictions
Embryo Collection
Three species of catfishes (Teleostei: Siluriformes) were used in this study. Ictalurus punctatus (channel catfish) embryos were collected and sent from E.W. Shell Fisheries Research Center (Auburn University, AL) to University of Oregon, Institute of Neuroscience (Eugene, OR). Embryonic suckermouth armored catfish (Ancistrus cf. triradiatus) and armored bronze corydoras catfish (C. aeneus) samples were collected at University of Oregon, Institute of Neuroscience by periodic natural spawning.
Pharyngeal arch developmental timing was established for all three catfishes using samples collected incrementally between 24 and 120 hpf and stained overnight with Alcian Green (Invitrogen, USA). Key time-points in I. punctatus pharyngeal arch development were subsequently targeted for brpf1 in situ hybridization (see below). Endogenous epidermal eda and edar expression patterns were established for I. punctatus using 4-week-old juveniles that originated from the same in vitro fertilization as used for the embryonic time-points. All samples were fixed in 4% paraformaldehyde (PFA) overnight at 4 °C on a rocker, dehydrated using methanol/phosphate-buffered solution (PBS) series and stored at −20 °C in 100% methanol.
Cloning
Total RNA was isolated from all collected developmental stages (24–120 hpf) for all three catfishes using TriZol Reagent (Invitrogen) and RNeasy Spin Columns (Qiagen, USA) as per manufacturer’s instructions. Following isolation, total RNA was reverse transcribed into complementary DNA (cDNA) using SuperScriptIII Reverse Transcriptase and oligo dT20 universal primers (Invitrogen) following manufacturer’s instructions. Bromodomain containing zinc finger 1 (brpf1) and ectodysplasin (eda) were isolated from embryonic cDNA using RT-PCR primers (brpf1-F: TCGGATATGACATGGACGAG and brpf1-R: TAGTGAGGGGCCTCTTGATG; eda-F: GTGCTGCAGGATGGGATGTA and eda-R: CAAGCTGATTGGCTCACGTA) designed from I. punctatus EST sequences ([GH655562] and [FD201975.1]), which are predicted to be orthologs of zebrafish brpf1 and eda ([XP_698063.2] and [XP_001339172.1]), respectively. To isolate ectodysplasin receptor (edar), given that no predicted sequence was available in the I. punctatus EST database, RT-PCR primers (edar-F: AGTGCTGAATACTCGAGCTGT and edar-R: TCCAGCCGCTCGATCTGC) were designed using conserved regions of D. rerio (NP_001108536.1) and O. latipes (NP_001098229.1) nucleotide sequences. Isolated brpf1, eda, and edar RT-PCR products (approximately 750, 450, and 1,300 bp, respectively) were confirmed by sequencing and NCBI BLAST (Basic Local Alignment Search Tool) analyses (Altschul et al. 1990). Verified RT-PCR amplicons were subsequently cloned into PCR4-TOPO vectors according to manufacturer’s instructions (Invitrogen). Clones were sequenced to verify insertion and orientation of target RT-PCR amplicon.
In Situ Hybridization
Endogenous expression patterns for D. rerio and I. punctatus brpf1 were obtained from whole-mount tissue (72 and 86 hpf embryos, respectively) and I. punctatus eda and edar were obtained from cryosectioned tissue (4 weeks postfertilization juveniles). Depending on insert orientation (see above), I. punctatus brpf1, eda, and edar digoxigenin-labeled riboprobes were synthesized in vitro using PCR4-TOPO SP6 or T7 promoter following manufacturer’s instructions (Roche, USA). Danio rerio brpf1 digoxigenin-labeled riborpobes were also synthesized in vitro (Roche) using PCR amplicons (F: ACTGCTACACTGCCTTCCAC; R: TCAGCACCGAGTGTTTCTCC) appended with SP6 or T7 promoter sequence, as detailed elsewhere (Edmunds et al. 2015). Synthesized riboprobes were verified by gel electrophoresis, purified using RNeasy Mini Kit (Qiagen, Inc), quantified by spectrophotometer (Invitrogen, Inc), and diluted in Hyb+ buffer (see below) to a final working concentration of 1 ng/μl.
Brpf1 in situ hybridization was performed on whole-mount tissue using the following protocol modified after Thisse and Thisse (2008): 1) Embryos were collected and fixed in PFA (4%) buffered with 1 × PBS containing 0.1% Tween-20 (PBST) overnight at 4 °C; 2) dehydrated in 100% methanol and transported to Institute of Neuroscience (University of Oregon, Eugene, OR); 3) rehydrated by descending methanol series (75% MeOH/25% PBST, 50% MeOH/50% PBST, 25% MeOH/75% PBST, 100% PBST); 4) dechorionated in PBST; 5) acetone shocked (100% for 10 min at −20 °C); 6) proteinase-K digested (10 μg for 30 min at room temperature); 7) refixed (4% PFA/1 × PBST) at 4 °C overnight; 8) incubated in hybridization solution (Hyb+: 50% formamide, 5× sodium sulfanyl citrate [SSC], 50 µg/ml Heparin, 500 µg/ml yeast ribosomal RNA, 0.1% Tween-20, and citric acid to pH 6.0) for 2 h at 68 °C; 9) incubated in pre-warmed Hyb+ containing 1 µg brpf1 antisense probe overnight at 68 °C; 10) washed with Hyb− solution (Hyb+ without Heparin, yeast ribosomal RNA or DIG-probe) at 68 °C, as follows: 20 min (75% Hyb− in 25% 2× SSC containing 0.1% Tween-20 [SSCT], 50% Hyb− in 50% 2× SSCT and 25% Hyb− in 75% 2× SSCT), 15 min (100% 2× SSCT in PBST) and 2× 30 min (0.2× SSCT in PBST); 11) cooled to room temperature and washed for 10 min each: 75% 0.2× SSCT in 25% PBST, 50% 0.2× SSCT in 50% PBST, 25% 0.2× SSCT in 75% PBST and 100% PBST; 12) incubated in blocking solution (1× PBS, 1 mg bovine serum albumin [BSA], 2.5 ml inactivated sheep serum, 1 ml goat serum, 0.1% Tween-20) for 1 h at room temperature; 13) incubated in anti-DIG antibody (1:5,000 in blocking solution) at 4 °C overnight; and 14) visualized in color buffer (3.75 μg 4-Nitro blue tetrazolium chloride (NBT), 26.3 μg 5-Bromo-4-chloro-3-indolyl phosphate (BCIP), 0.1 M Tris pH 9.5, 0.05 M MgCl2, 0.1 M NaCl, 0.1% Tween-20). Coloration reactions were carried out in the dark at 37 °C until desired degree of labeling.
Similarly, eda and edar in situ hybridizations were performed on cryosectioned tissues using the following protocol modified after Thisse and Thisse (2008): 1) 4-week-old juveniles were collected and fixed in PFA (4%) buffered with 1× PBST at 4 °C overnight, 2) dehydrated in 100% methanol and transported to Institute of Neuroscience (University of Oregon, Eugene, OR), and 3) rehydrated by descending methanol series (75% MeOH/25% PBST, 50% MeOH/50% PBST, 25% MeOH/75% PBST, 100% PBST). Following rehydration, the body trunk region containing the dorsal fin and torso epidermis was isolated by dissection, embedded in agarose (1%, containing sucrose), and incubated at 4 °C overnight. Cryosections (16 μm) were affixed to poly-lysine-treated slides, immersed in hybridization solution (1× salt solution [1.14 g NaCl, 1.4 g Tris–HCl, 0.13 g Tris Base, 0.78 g NaH2PO4·2H20, 0.71 g Na2PO4, 10 ml 0.5 M ethylenediaminetetraacetic acid], 50% formamide, 10% dextran sulfate, 50 mg yeast rRNA, 1× Denhardt’s solution [1% w/v BSA, Ficoll and polyvinylpyrrolidone in water]), and incubated overnight in a humidity chamber (Perspex box with 1× SSC and 50% formamide-wetted Whatman paper). Cryosections were then washed posthybridization for 4× 30 min in wash solution (1× SSC, 50% formamide, 0.1% Tween-20) at 68–70 °C and 3× 30 min in 1× MABT (0.58 g maleic acid, 0.44 g NaCl, 0.1% Tween-20, pH to 7.5 with NaOH), incubated in blocking solution (1× MABT, 2% blocking reagent, 20% heat inactivated sheep serum) for 3 h at room temperature in humidity chamber (Perspex box with 1× PBS-wetted Whatman paper) and then incubated with anti-DIG antibody (1:5,000 in blocking solution) at 4 °C overnight in a humidity chamber (Perspex box with 1× PBS-wetted Whatman paper) before finally being visualized using alkaline phosphate staining buffer (3.75 μg NBT, 26.3 μg BCIP, 0.1 g polyvinyl alcohol, 0.1 M Tris pH 9.5, 0.05 M MgCl2, 0.1 M NaCl, 0.1% Tween-20). Coloration reactions were carried out in the dark at 37 °C until desired degree of labeling.
Toluidine Blue Stock Solution (TBSS) was made by dissolving Toluidine Blue powder (1 g; Allied Chemical Co., New York) in 100 ml 70% ethanol. Sodium Chloride Solution (SCS; 1%) was made by dissolving sodium chloride (0.5 g; Allied Chemical Co.) in 50 ml distilled water and adjusting pH to 2.0–2.5 with 4 M HCl. Toluidine Blue Working Solution contained TBSS (5 ml) and SCS (45 ml) at pH 2.3. Dissected arches were stained for 2–3 min, then rinsed (2×) and washed (3×) in distilled water. Counterstained arches were transferred into glycerol by ascending series (25% glycerol/75% PBST, 50% glycerol/50% PBST, 75% glycerol/25% PBST), slide mounted, and imaged (AxioCam MRc-5; Carl Zeiss Inc., Germany).
Acknowledgments
This material is based upon work supported by the National Science Foundation under Grant Numbers DBI-0641025, DBI-1062404, and DBI-1062542, and supported by the National Evolutionary Synthesis Center under NSF EF-0423641 and NSF EF-0905606. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation. The authors also acknowledge the support of NIH grants HG002659 and HD22486 (to M.W.). They also thank many contributors to the Phenoscape project (http://phenoscape.org/wiki/Acknowledgments). They also thank M. Sabaj for the image of his catfishes used in figure 1 and K. Luckenbill (Academy of Natural Sciences [ANSP], Philadelphia, PA) for his work in developing figure 2.
References
- Abzhanov A, Extavour CG, Groover A, Hodges SA, Hoekstra HE, Kramer EM, Monteiro A. 2008. Are we there yet? Tracking the development of new model systems. Trends Genet. 24:353–360. [DOI] [PubMed] [Google Scholar]
- Albert JS. 2001. Species diversity and phylogenetic systematics of American knifefishes (Gymnotiformes, Teleostei). Misc Publ Mus Zool Univ Mich. 190:1–140. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Armbruster JW. 2004. Phylogenetic relationships of the suckermouth armoured catfishes (Loricariidae) with emphasis on the Hypostominae and the Ancistrinae. Zool J Linn Soc. 141:1–80. [Google Scholar]
- Arratia G. 1999. The monophyly of Teleostei and stem-group teleosts. Consensus and disagreements. In: Arratia G, Schultze H-P, editors. Mesozoic fishes 2—systematics and fossil record. Munchen (Germany): Verlag Dr. F. Pfeil; p. 265–334. [Google Scholar]
- Arratia G, Schultze HP. 1990. The urohyal development and homology within osteichthyans. J Morphol. 203:247–282. [DOI] [PubMed] [Google Scholar]
- Balhoff JP, Dahdul WM, Kothari CR, Lapp H, Lundberg JG, Mabee P, Midford PE, Westerfield M, Vision TJ. 2010. Phenex: ontological annotation of phenotypic diversity. PLoS One 5:e10500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao B, Peatman E, Xu P, Li P, Zeng H, He C, Liu Z. 2006. The catfish liver-expressed antimicrobial peptide 2 (LEAP-2) gene is expressed in a wide range of tissues and developmentally regulated. Mol Immunol. 43:367–377. [DOI] [PubMed] [Google Scholar]
- Baoprasertkul P, Peatman E, Somridhivej B, Liu Z. 2006. Toll-like receptor 3 and TICAM genes in catfish: species-specific expression profiles following infection with Edwardsiella ictaluri. Immunogenetics 58:817–830. [DOI] [PubMed] [Google Scholar]
- Bell MA, Gangavalli AK, Bewick A, Aguirre EW. 2010. Frequency of Ectodysplasin alleles and limited introgression between sympatric threespine stickleback populations. Environ Biol Fishes. 89:189–198. [Google Scholar]
- Binder V, Zon LI. 2013. High throughput in vivo phenotyping: The zebrafish as tool for drug discovery for hematopoietic stem cells and cancer. Drug Discov Today Dis Models. 10:e17–e22. [Google Scholar]
- Blake JA, Harris MA. 2008. The Gene Ontology (GO) project: structured vocabularies for molecular biology and their application to genome and expression analysis. Curr Protoc Bioinformatics. 23:7.2.1–7.2.9. [DOI] [PubMed] [Google Scholar]
- Bockmann FA. 1998. Análise Filogenética da Família Heptapteridae (Teleostei, Ostariophysi, Siluriformes) e Redefenição de seus Gêneros. São Paulo (Brazil): Universidade de São Paulo. [Google Scholar]
- Bornbusch AH. 1995. Phylogenetic relationships within the Eurasian catfish family Siluridae (Pisces: Siluriformes), with comments on generic validities and biogeography. Zool J Linn Soc. 115:1–46. [Google Scholar]
- Britto MR. 2003. Phylogeny of the subfamily Corydoradinae Hoedeman, 1952 (Siluriformes: Callichthyidae), with a definition of its genera. Proc Acad Nat Sci Philadelphia. 153:119–154. [Google Scholar]
- Buckup PA. 1998. Relationships of the Characidiinae and phylogeny of characiform fishes (Teleostei: Ostariophysi). In: Malabarba LR, Reis RE, Vari RP, Lucena ZMS, Lucena CAS, editors. Phylogeny and classification of neotropical fishes. Porto Alegre (Brazil): Edipucrs: p. 123–144. [Google Scholar]
- Burns T, Breathnach S, Cox N, Griffiths C. 2010. Rook’s textbook of dermatology, 4 Volume Set. Chichester, West Sussex, UK; Hoboken, NJ: Wiley-Blackwell. [Google Scholar]
- Cavender TM, Coburn MM. 1992. Phylogenetic relationships of North American Cyprinidae. In: Mayden RL, editor Systematics, historical ecology, and North American freshwater fishes. Stanford (CA): Stanford University Press; p. 293–327. [Google Scholar]
- Chanet B. 2003. Interrelationships of scophthalmid fishes (Pleuronectiformes: Scophthalmidae). Cybium 27:275–286. [Google Scholar]
- Chang MM, Maisey JG. 2003. Redescription of Ellimma branneri and Diplomystus shengliensis, and relationships of some basal Clupeomorphs. Am Mus Novit. 3404:1–35. [Google Scholar]
- Chen F, Lee Y, Jiang Y, et al. 2010. Identification and characterization of full-length cDNAs in channel catfish (Ictalurus punctatus) and blue catfish (Ictalurus furcatus). PLoS One 5:e11546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X-P. 1994. Phylogenetic studies of the amblycipitid catfishes (Teleostei, Siluriformes) with species accounts. Durham (NC): Duke University. [Google Scholar]
- Coburn MM, Cavender TM. 1992. Interrelationships of North American cyprinid fishes. In: Mayden RL, editor. Systematics, historical ecology, and North American freshwater fishes. Stanford (CA): Stanford University Press; p. 328–373. [Google Scholar]
- Colosimo PF, Hosemann KE, Balabhadra S, et al. 2005. Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science 307:1928–1933. [DOI] [PubMed] [Google Scholar]
- Cubbage CC, Mabee PM. 1996. Development of the cranium and paired fins in the zebrafish Danio rerio (Ostariophysi, Cyprinidae). J Morphol. 229:121–160. [DOI] [PubMed] [Google Scholar]
- Dahdul WM, Balhoff JP, Engeman J, et al. 2010. Evolutionary characters, phenotypes and ontologies: curating data from the systematic biology literature. PLoS One 5:e10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahdul WM, Cui H, Mabee PM, Mungall CJ, Osumi-Sutherland D, Walls RL, Haendel MA. 2014. Nose to tail, roots to shoots: spatial descriptors for phenotypic diversity in the Biological Spatial Ontology. J Biomed Semantics. 5:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahdul WM, Lundberg JG, Midford PE, Balhoff JP, Lapp H, Vision TJ, Haendel MA, Westerfield M, Mabee PM. 2010. The teleost anatomy ontology: anatomical representation for the genomics age. Syst Biol. 59:369–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pinna MCC. 1993. Higher-level phylogeny of Siluriformes (Teleostei, Ostariophysi), with a new classification of the order. New York: City University of New York. [Google Scholar]
- de Pinna MCC. 1996. A phylogenetic analysis of the Asian catfish families Sisoridae, Akysidae, and Amblycipitidae, with a hypothesis on the relationships of the neotropical Aspredinidae (Teleostei, Ostariophysi). Fieldiana Zool. 84:i–iv + 1-83. [Google Scholar]
- de Pinna MCC. 1998. Phylogenetic relationships of Neotropical Siluriformes (Teleostei: Ostariophysi): historical overview and synthesis of hypotheses. In: Malabarba LR, Reis RE, Vari RP, Lucena ZMS, Lucena CAS, editors. Phylogeny and classification of neotropical fishes. Porto Alegre (Brazil): Edipucrs; p. 279–330. [Google Scholar]
- de Pinna MCC, Ferraris CJJ, Vari RP. 2007. A phylogenetic study of the neotropical catfish family Cetopsidae (Osteichthys, Ostariophysi, Siluriformes), with a new classification. Zool J Linn Soc. 150:755–813. [Google Scholar]
- Deans AR, Lewis SE, Huala E, et al. 2015. Finding our way through phenotypes. PLoS Biol. 13.1:e1002033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan BM. 2010. The rise of genomics sheds light on the dawn of animals. Evol Dev. 12:425–427. [DOI] [PubMed] [Google Scholar]
- Di Dario F. 1999. Filogenia de Pristigasteroidea (Teleostei, Clupeomorpha). São Paulo (Brazil): Universidade de São Paulo. [Google Scholar]
- Eames BF, DeLaurier A, Ullmann B, Huycke TR, Nichols JT, Dowd J, McFadden M, Sasaki MM, Kimmel CB. 2013. FishFace: interactive atlas of zebrafish craniofacial development at cellular resolution. BMC Dev Biol. 13:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmunds RC, Gill JA, Baldwin DH, Linbo TL, French BL, Brown TL, Esbaugh AJ, Mager EM, Stieglitz J, Hoenig R, et al. 2015. Corresponding morphological and molecular indicators of crude oil toxicity to the developing hearts of mahi mahi. Sci Rep. Advance Access published December 10, 2015; doi:10.1038/srep17326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engeman JM, Mabee PM. 2012. Segmentation and fusion on the midline: basibranchial homologies in cypriniform fishes. J Morphol. 273:725–736. [DOI] [PubMed] [Google Scholar]
- Fang F. 2003. Phylogenetic analysis of the Asian Cyprinid genus Danio (Teleostei, Cyprinidae). Copeia 2003:714–728. [Google Scholar]
- Fink SV, Fink WL. 1981. Interrelationships of the ostariophysan fishes (Teleostei). Zool J Linn Soc. 72:297–353. [Google Scholar]
- Fink SV, Fink WL. 1996. Interrelationships of Ostariophysan fishes (Teleostei). In: Stiassny MLJ, Parenti LR, Johnson GD, editors. Interrelationships of fishes. San Diego (CA): Academic Press; p. 209–249. [Google Scholar]
- Friel JP. 1994. A phylogenetic study of the Neotropical banjo catfishes (Teleostei: Siluriformes: Aspredinidae). Durham (CA): Duke University. [Google Scholar]
- Gallant JR, Traeger LL, Volkening JD, et al. 2014. Nonhuman genetics. Genomic basis for the convergent evolution of electric organs. Science 344:1522–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner BG. 1984. The relationships of the palaeoniscid fishes, a review based on new specimens of Mimia and Moythomasia from the Upper Devonian of Western Australia. Bull Br Mus Nat Hist. 37:173–428. [Google Scholar]
- Geerinckx T, Adriaens D. 2007. Ontogeny of the intermandibular and hyoid musculature in the suckermouth armoured catfish Ancistrus cf. triradiatus (Loricariidae, Siluriformes). Anim Biol Leiden Neth. 57:339–358. [Google Scholar]
- Geerinckx T, Adriaens D. 2008. Ontogeny of the suspensorial and opercular musculature in the suckermouth armoured catfish Ancistrus cf. triradiatus (Loricariidae, Siluriformes). Zoomorphology 127:83–95. [Google Scholar]
- Geerinckx T, Brunain M, Adriaens D. 2005. Development of the chondrocranium in the suckermouth armored catfish Ancistrus cf. triradiatus (Loricariidae, Siluriformes). J Morphol. 266:331–355. [DOI] [PubMed] [Google Scholar]
- Geerinckx T, Brunain M, Adriaens D. 2007. Development of the osteocranium in the suckermouth armored catfish Ancistrus cf. triradiatus (Loricariidae, siluriformes). J Morphol. 268:254–274. [DOI] [PubMed] [Google Scholar]
- Gil Y, Greaves M, Hendler J, Hirsh H. 2014. Amplify scientific discovery with artificial intelligence. Science 346:171–172. [DOI] [PubMed] [Google Scholar]
- Gilbert SF. 2009. The adequacy of model systems for evo-devo; modeling the formation of organisms. In: Barberousse A, Morange M, Pradeu T, editors. Mapping the future of biology, Boston Studies in the Philosophy of Science. Springer. p. 57–68. [Google Scholar]
- Gkoutos GV, Green ECJ, Mallon A-M, Blake A, Greenaway S, Hancock JM, Davidson D. 2004. Ontologies for the description of mouse phenotypes. Comp Funct Genomics. 5:545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkoutos GV, Green EC, Mallon AM, Hancock JM, Davidson D. 2004. Using ontologies to describe mouse phenotypes. Genome Biol. 6:R8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grande T, Grande L. 2008. Reevaluation of the gonorynchiform genera Ramallichthys, Judeichthys and Notogoneus, with comments on the families Charitosomidae and Gonorynchidae. In: Arratia G, Schultze H-P, Wilson MVH, editors. Mesozoic fishes 4—homology and phylogeny. München (Germany): Verlag Dr. Friedrich Pfeil; p. 295–310. [Google Scholar]
- Grande T, Laten H, Lopez JA. 2004. Phylogenetic relationships of extant esocid species (Teleostei: Salmoniformes) based on morphological and molecular characters. Copeia 2004:743–757. [Google Scholar]
- Grande T, Poyato-Ariza FJ. 1999. Phylogenetic relationships of fossil and recent gonorynchiform fishes (Teleostei: Ostariophysi). Zool J Linn Soc. 125:197–238. [Google Scholar]
- Grizzle JM, Rogers WA, Station AUAE, Service USNMF. 1976. Anatomy and histology of the channel catfish. Auburn (AL): Auburn University, Agricultural Experiment Station. [Google Scholar]
- Haendel MA, Ballhoff JP, Bastian FB, et al. 2014. Uberon: unification of multi-species vertebrate anatomy ontologies for comparative biology. J Biomed Semantics. 5:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MP, Rohner N, Schwarz H, Perathoner S, Konstantinidis P, Nüsslein-Volhard C. 2008. Zebrafish eda and edar mutants reveal conserved and ancestral roles of ectodysplasin signaling in vertebrates. PLoS Genet. 4:e1000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibiya K, Katsumoto T, Kondo T, Kitabayashi I, Kudo A. 2009. Brpf1, a subunit of the MOZ histone acetyl transferase complex, maintains expression of anterior and posterior Hox genes for proper patterning of craniofacial and caudal skeletons. Dev Biol. 329:176–190. [DOI] [PubMed] [Google Scholar]
- Hoehndorf R, Schofield PN, Gkoutos GV. 2011. PhenomeNET: a whole-phenome approach to disease gene discovery. Nucleic Acids Res. 39:e119–e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huysentruyt F, Brunain M, Adriaens D. 2008. Ontogeny of the chondrocranium in Corydoras aeneus (Gill, 1858) (Callichthyidae, Siluriformes). J Morphol. 269:522–532. [DOI] [PubMed] [Google Scholar]
- Huysentruyt F, Brunain M, Adriaens D. 2009. Ontogeny of the cranial musculature in Corydoras aeneus Callichthyidae, Siluriformes. J Fish Biol. 75:1601–1614. [DOI] [PubMed] [Google Scholar]
- Huysentruyt F, Geerinckx T, Adriaens D. 2007. A descriptive myology of Corydoras aeneus (Gill, 1858) (Siluriformes: Callichthyidae), with a brief discussion on adductor mandibulae homologies. Anim Biol Leiden Neth. 57:433–452. [Google Scholar]
- Huysentruyt F, Geerinckx T, Brunain M, Adriaens D. 2011. Development of the osteocranium in Corydoras aeneus (Gill, 1858) Callichthyidae, Siluriformes. J Morphol. 272:573–582. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Gao X, Liu S, Zhang Y, Liu H, Sun F, Bao L, Waldbieser G, Liu Z. 2013. Whole genome comparative analysis of channel catfish (Ictalurus punctatus) with four model fish species. BMC Genomics 14:780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DG, Patterson C. 1993. Percomorph phylogeny: a survey of acanthomorphs and a new proposal. Bull Mar Sci. 52:554–626. [Google Scholar]
- Kailola PJ. 2004. A phylogenetic exploration of the catfish family Ariidae (Otophysi; Siluriformes). Beagle (Darwin) 20:87-166. [Google Scholar]
- Kocabas AM, Li P, Cao D, Karsi A, He C, Patterson A, Ju Z, Dunham RA, Liu Z. 2002. Expression profile of the channel catfish spleen: analysis of genes involved in immune functions. Mar Biotechnol (NY). 4:526–536. [DOI] [PubMed] [Google Scholar]
- Kondo S, Kuwahara Y, Kondo M, et al. 2001. The medaka rs-3 locus required for scale development encodes ectodysplasin-A receptor. Curr Biol. 11:1202–1206. [DOI] [PubMed] [Google Scholar]
- Konrádsdóttir F, Loftsson T, Sigfússon SD. 2009. Fish skin as a model membrane: structure and characteristics. J Pharm Pharmacol. 61:121–124. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. ClustalW and ClustalX version 2. Bioinformatics 23(21):2947–2948. [DOI] [PubMed] [Google Scholar]
- Laue K, Daujat S, Crump JG, Plaster N, Roehl HH, Kimmel CB, Schneider R, Hammerschmidt M. 2008. The multidomain protein Brpf1 binds histones and is required for Hox gene expression and segmental identity. Development 135:1935–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Peatman E, Wang S, et al. 2007. Towards the ictalurid catfish transcriptome: generation and analysis of 31,215 catfish ESTs. BMC Genomics 8:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li RW, Waldbieser GC. 2006. Production and utilization of a high-density oligonucleotide microarray in channel catfish, Ictalurus punctatus. BMC Genomics 7:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Zhou Z, Lu J, et al. 2011. Generation of genome-scale gene-associated SNPs in catfish for the construction of a high-density SNP array. BMC Genomics 12:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. 2011. Development of genomic resources in support of sequencing, assembly, and annotation of the catfish genome. Comp Biochem Physiol Part D Genomics Proteomics. 6:11–17. [DOI] [PubMed] [Google Scholar]
- Lu J, Peatman E, Yang Q, Wang S, Hu Z, Reecy J, Kucuktas H, Liu Z. 2011. The catfish genome database cBARBEL: an informatic platform for genome biology of ictalurid catfish. Nucleic Acids Res. 39:D815–D821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucena CAS, Menezes NA. 1998. A phylogenetic analysis of Roestes Günther and Gilbertolus Eigenmann with a hypothesis on the relationships of the Cynodontidae and Acestrorhynchidae (Teleostei, Ostariophysi, Characiformes). In: Malabarba LR, Reis RE, Vari RP, Lucena ZMS, Lucena CAS, editors. Phylogeny and classification of neotropical fishes. Porto Alegre (Brazil): Edipucrs; p. 261–278. [Google Scholar]
- Lundberg JG. 1992. The phylogeny of ictalurid catfishes: a synthesis of recent work. In: Mayden RL, editor. Systematics, historical ecology, & North American freshwater fishes. Stanford (CA): Stanford University Press; p. 392–420. [Google Scholar]
- Mabee PM, Arratia G, Coburn M, Haendel M, Hilton EJ, Lundberg JG, Mayden RL, Rios N, Westerfield M. 2007. Connecting evolutionary morphology to genomics using ontologies: a case study from Cypriniformes including zebrafish. J Exp Zool B Mol Dev Evol. 308B:655–668. [DOI] [PubMed] [Google Scholar]
- Mabee PM, Ashburner M, Cronk Q, Gkoutos GV, Haendel M, Segerdell E, Mungall C, Westerfield M. 2007. Phenotype ontologies: the bridge between genomics and evolution. Trends Ecol Evol. 22:345–350. [DOI] [PubMed] [Google Scholar]
- Mabee PM, Grey E, Arratia G, Bogutskaya NG, Boron A, Coburn MM, Conway KW, He S, Naseka A, Rios N, et al. 2011. Gill arch and hyoid arch diversity and cypriniform phylogeny: distributed integration of morphology and web-based tools. Zootaxa 2877:1–40. [Google Scholar]
- Malabarba LR. 1998. Monophyly of the Cheirodontinae, characters and major clades (Ostariophysi: Characidae). In: Malabarba LR, Reis RE, Vari RP, Lucena ZMS, Lucena CAS, editors. Phylogeny and classification of neotropical fishes. Porto Alegre (Brazil): Edipucrs; p. 193–234. [Google Scholar]
- Manda P, Balhoff JP, Lapp H, Mabee P, Vision TJ. 2015. Using the Phenoscape Knowledgebase to relate genetic perturbations to phenotypic evolution. Genesis http://dx.doi.org/10.1002/dvg.22878. [DOI] [PubMed] [Google Scholar]
- Mayden RL. 1989. Phylogenetic studies of North American minnows, with emphasis on the genus Cyprinella (Teleostei: Cypriniformes). Misc Publ Univ Kans Mus Nat Hist. 80:1–189. [Google Scholar]
- Midford PE, Dececchi T, Balhoff JP, et al. 2013. The vertebrate taxonomy ontology: a framework for reasoning across model organism and species phenotypes. J Biomed Semantics. 4:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CT, Yelon D, Stainier DY, Kimmel CB. 2003. Two endothelin 1 effectors, hand2 and bapx1, pattern ventral pharyngeal cartilage and the jaw joint. Development 130:1353–1365. [DOI] [PubMed] [Google Scholar]
- Mo T. 1991. Anatomy, relationships and systematics of the Bagridae (Teleostei: Siluroidei) with a hypothesis of siluroid phylogeny. Koenigstein (Germany): Koeltz. [Google Scholar]
- Mooi RD, Johnson GD. 1997. Dismantling the Trachinoidei: evidence of a scorpaenoid relationship for the Champsodontidae. Ichthyol Res. 44:143–176. [Google Scholar]
- Mungall CJ, Gkoutos GV, Smith CL, Haendel MA, Lewis SE, Ashburner M. 2010. Integrating phenotype ontologies across multiple species. Genome Biol. 11:R2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhauss SC, Solnica-Krezel L, Schier AF, Zwartkruis F, Stemple DL, Malicki J, Abdelilah S, Stainier DY, Driever W. 1996. Mutations affecting craniofacial development in zebrafish. Development 123:357–367. [DOI] [PubMed] [Google Scholar]
- Ninwichian P, Peatman E, Liu H, et al. 2012. Second-generation genetic linkage map of catfish and its integration with the BAC-based physical map. G3 2:1233–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oellrich A, Hoehndorf R, Gkoutos GV, Rebholz-Schuhmann D. 2012. Improving disease gene prioritization by comparing the semantic similarity of phenotypes in mice with those of human diseases. PLoS One 7:e38937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero O. 2004. Anatomy, systematics and phylogeny of both recent and fossil latid fishes (Teleostei, Perciformes, Latidae). Zool J Linn Soc. 141:81–133. [Google Scholar]
- Parichy DM, Elizondo MR, Mills MG, Gordon TN, Engeszer RE. 2009. Normal table of postembryonic zebrafish development: staging by externally visible anatomy of the living fish. Dev Dyn. 238:2975–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peatman E, Baoprasertkul P, Terhune J, et al. 2007. Expression analysis of the acute phase response in channel catfish (Ictalurus punctatus) after infection with a Gram-negative bacterium. Dev Comp Immunol. 31:1183–1196. [DOI] [PubMed] [Google Scholar]
- Poyato-Ariza FJ. 1996. A revision of the ostariophysan fish family Chanidae, with special reference to the Mesozoic forms. Palaeo Ichthyol. 6:1–52. [Google Scholar]
- Ritter DI, Li Q, Kostka D, Pollard KS, Guo S, Chuang JH. 2010. The importance of being cis: evolution of orthologous fish and mammalian enhancer activity. Mol Biol Evol. 27:2322–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan BA, Weigel D, Koenig D. 2011. Developmental genetics and new sequencing technologies: the rise of nonmodel organisms. Dev Cell. 21:65–76. [DOI] [PubMed] [Google Scholar]
- Royero R. 1999. Studies on the systematics and phylogeny of the catfish family Auchenipteridae (Teleostei: Siluriformes). Bristol (United Kingdom): University of Bristol. [Google Scholar]
- Ruzicka L, Bradford YM, Frazer K, Howe DG, Paddock H, Ramachandran S, Singer A, Toro S, Van Slyke CE, Eagle AE, et al. 2015. ZFIN, The zebrafish model organism database: Updates and new directions. Genesis 53(8):498–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger TJ, McCune AR. 2002. Comparative osteology of the Danio (Cyprinidae: Ostariophysi) axial skeleton with comments on Danio relationships based on molecules and morphology. Zool J Linn Soc. 135:529–546. [Google Scholar]
- Sawada Y. 1982. Phylogeny and zoogeography of superfamily Cobitoidea (Cyprinoidei, Cypriniformes). Memoirs of the Faculty of Fisheries, Hokkaido University. 28:65–223. [Google Scholar]
- Schaefer SA. 1987. Osteology of Hypostomus plecostomus (Linnaeus), with a phylogenetic analysis of the loricariid subfamilies (Pisces: Siluroidei). Contr Sci Mus Nat His Los Angeles. 1–31. [Google Scholar]
- Schaefer SA. 1991. Phylogenetic analysis of the loricariid subfamily Hypoptopomatinae (Pisces: Siluroidei: Loricariidae), with comments on generic diagnoses and geographic distribution. Zool J Linn Soc. 102:1–41. [Google Scholar]
- Schilling TF, Piotrowski T, Grandel H, et al. 1996. Jaw and branchial arch mutants in zebrafish I: branchial arches. Development 123:329–344. [DOI] [PubMed] [Google Scholar]
- Schwend T, Ahlgren SC. 2009. Zebrafish con/disp1 reveals multiple spatiotemporal requirements for Hedgehog-signaling in craniofacial development. BMC Dev Biol. 9:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibatta OA. 1998. Sistemática e Evolução da Família Pseudopimelodidae (Ostariophysi, Siluriformes), com a Revisão Taxonômica de Gênero Pseudopimelodus. [Ph.D. Dissertation]. [São Carlos (Brasil)]: Universidade Federal São Carlos. [Google Scholar]
- Sidlauskas BL, Vari RP. 2008. Phylogenetic relationships within the South American fish family Anostomidae (Teleostei, Ostariophysi, Characiformes). Zool J Linn Soc. 154:70–210. [Google Scholar]
- Siebert DJ. 1987. Interrelationships among families of the order Cypriniformes (Teleostei) [Ph.D. Dissertation]. [New York]: City University of New York. [Google Scholar]
- Sire J-Y, Akimenko M-A. 2004. Scale development in fish: a review, with description of sonic hedgehog (shh) expression in the zebrafish (Danio rerio). Int J Dev Biol. 48:233–248. [DOI] [PubMed] [Google Scholar]
- Sire J-Y, Huysseune A. 2003. Formation of dermal skeletal and dental tissues in fish: a comparative and evolutionary approach. Biol Rev. 78:219–249. [DOI] [PubMed] [Google Scholar]
- Sire JY. 1993. Development and fine structure of the bony scutes in Corydoras arcuatus (Siluriformes, Callichthyidae). J Morphol. 215:225–244. [DOI] [PubMed] [Google Scholar]
- Smith GR. 1992. Phylogeny and biogeography of the Catostomidae, freshwater fishes of North America and Asia. In: Mayden RL, editor. Systematics, historical ecology, and North American freshwater fishes. Stanford (CA): Stanford University Press; p. 778–826. [Google Scholar]
- Soares-Porto LM. 1998. Monophyly and interrelationships of the Centromochlinae (Siluriformes: Auchenipteridae). In: Malabarba LR, Reis RE, Vari RP, Lucena ZMS, Lucena CAS, editors. Phylogeny and classification of neotropical fishes. Porto Alegre (Brazil): Edipucrs; p. 331–350. [Google Scholar]
- Sprague J, Bayraktaroglu L, Bradford Y, et al. 2008. The Zebrafish Information Network: the zebrafish model organism database provides expanded support for genotypes and phenotypes. Nucleic Acids Res. 36:D768–D772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinke D, Salzburger W, Meyer A. 2006. Novel relationships among ten fish model species revealed based on a phylogenomic analysis using ESTs. J Mol Evol. 62:772–784. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B. 2008. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 3:59–69. [DOI] [PubMed] [Google Scholar]
- Toledo-Piza M. 2000. The neotropical fish subfamily Cynodontinae (Teleostei: Ostariophysi: Characiformes): a phylogenetic study and revision of Cynodon and Rhaphiodon. Am Mus Novit. 3286:1–88. [Google Scholar]
- Toledo-Piza M. 2007. Phylogenetic relationships among Acestrorhynchus species (Ostariophysi: Characiformes: Acestrorhynchidae). Zool J Linn Soc. 151:691–757. [Google Scholar]
- USDA. 2005. Catfish Production Report. Washington (DC): National Agricultural Statistics Service USDA; (July 23, 2005). [Google Scholar]
- Vari RP. 1995. The Neotropical fish family Ctenoluciidae (Teleostei: Ostariophysi: Characiformes): supra and intrafamilial phylogenetic relationships, with a revisionary study. Smithson Contrib Zool. 564:1–97. [Google Scholar]
- Vari RP, Castro RMC, Raredon SJ. 1995. The Neotropical fish family Chilodontidae (Teleostei: Characiformes): a phylogenetic study and a revision of Caenotropus Günther. Smithson Contrib Zool. 577:32. [Google Scholar]
- Vigliotta TR. 2008. A phylogenetic study of the African catfish family Mochokidae (Osteichthyes, Ostariophysi, Siluriformes), with a key to its genera. Proc Acad Nat Sci Philadelphia. 157:73–136. [Google Scholar]
- Waldbieser GC, Bosworth BG, Nonneman DJ, Wolters WR. 2001. A microsatellite-based genetic linkage map for channel catfish, Ictalurus punctatus. Genetics 158:727–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Peatman E, Abernathy J, et al. 2010. Assembly of 500,000 inter-specific catfish expressed sequence tags and large scale gene-associated marker development for whole genome association studies. Genome Biol. 11:R8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington NL, Haendel MA, Mungall CJ, Ashburner M, Westerfield M, Lewis SE. 2009. Linking human diseases to animal models using ontology-based phenotype annotation. PLoS Biol. 7:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzman SH, Menezes NA. 1998. Relationships of the tribes and genera of the Glandulocaudiinae (Ostariophysi: Characiformes: Characidae), with a description of a new genus. In: Malabarba LR, Reis RE, Vari RP, Lucena ZMS, Lucena CAS, editors. Phylogeny and classification of Neotropical fishes. Porto Alegre (Brazil): Edipucrs; p. 171–192. [Google Scholar]
- Wiley E, David Johnson G, Wheaton Dimmick W. 2000. The interrelationships of acanthomorph fishes: a total evidence approach using molecular and morphological data. Biochem Syst Ecol. 28:319–350. [DOI] [PubMed] [Google Scholar]
- Wood JD, Bonath F, Kumar S, Ross CA, Cunliffe VT. 2009. Disrupted-in-schizophrenia 1 and neuregulin 1 are required for the specification of oligodendrocytes and neurones in the zebrafish brain. Hum Mol Genet. 18:391–404. [DOI] [PubMed] [Google Scholar]
- Xu P, Wang S, Liu L, Peatman E, Somridhivej B, Thimmapuram J, Gong G, Liu Z. 2006. Channel catfish BAC-end sequences for marker development and assessment of syntenic conservation with other fish species. Anim Genet. 37:321–326. [DOI] [PubMed] [Google Scholar]
- Yan YL, Miller CT, Nissen RM, et al. 2002. A zebrafish sox9 gene required for cartilage morphogenesis. Development 129:5065–5079. [DOI] [PubMed] [Google Scholar]
- Zanata AM, Vari RP. 2005. The family Alestidae (Ostariophysi, Characiformes): a phylogenetic analysis of a trans-Atlantic clade. Zool J Linn Soc. 145:1–144. [Google Scholar]
- Zaragüeta Bagils R. 2004. Basal clupeomorphs and ellimmichthyiform phylogeny. In: Arratia G, Tintori A, editors. Mesozoic fishes 3—systematics, paleoenvironments and biodiversity. Munich (Germany): Verlag Dr. F. Pfeil; p. 391–404. [Google Scholar]
- Zhang Y, Liu S, Lu J, Jiang Y, Gao X, Ninwichian P, Li C, Waldbieser G, Liu Z. 2013. Comparative genomic analysis of catfish linkage group 8 reveals two homologous chromosomes in zebrafish and other teleosts with extensive inter-chromosomal rearrangements. BMC Genomics 14:387. [DOI] [PMC free article] [PubMed] [Google Scholar]