Abstract
The temporal change of phenotypes during the adaptive process remains largely unexplored, as do the genetic changes that affect these phenotypic changes. Here we focused on three mutations that rose to high frequency in the early stages of adaptation within 12 Escherichia coli populations subjected to thermal stress (42 °C). All the mutations were in the rpoB gene, which encodes the RNA polymerase beta subunit. For each mutation, we measured the growth curves and gene expression (mRNAseq) of clones at 42 °C. We also compared growth and gene expression with their ancestor under unstressed (37 °C) and stressed conditions (42 °C). Each of the three mutations changed the expression of hundreds of genes and conferred large fitness advantages, apparently through the restoration of global gene expression from the stressed toward the prestressed state. These three mutations had a similar effect on gene expression as another single mutation in a distinct domain of the rpoB protein. Finally, we compared the phenotypic characteristics of one mutant, I572L, with two high-temperature adapted clones that have this mutation plus additional background mutations. The background mutations increased fitness, but they did not substantially change gene expression. We conclude that early mutations in a global transcriptional regulator cause extensive changes in gene expression, many of which are likely under positive selection for their effect in restoring the prestress physiology.
Keywords: experimental evolution, bacterial adaptation, gene expression
Introduction
Organisms are often exposed to stressful environments. To cope, they have evolved responses based on the duration of the stress. For example, bacteria that are exposed to increased temperatures display a transient heat shock response, which involves upregulation of genes encoding heat stress proteins (Richter et al. 2010), followed by a period of phenotypic acclimation (Gunasekera et al. 2008). If the environmental stress is sustained over a long period of time, individuals may eventually accumulate mutations that result in long-term adaptation of the population. Although acute responses to stress have been well characterized (Gunasekera et al. 2008), the mechanisms of stress acclimation and adaptation are understood less well (Riehle et al. 2003; Gunasekera et al. 2008).
One possible mechanism for adaptation to stressful conditions is genetic change that produces novel traits or new physiological functions, such as antibiotic resistance or the ability to use new metabolic pathways (Blount et al. 2012; Quandt et al. 2014). Another mechanism is genetic change that confers restoration of cellular functions to a prestressed state. Rather than creating new functions, restorative mutations revert some aspect of the individuals’ altered physiological state back to an unstressed or “normal” state (Carroll and Marx 2013). This pattern of restoration has been recently observed during metabolic perturbation and high-temperature adaptation (Fong et al. 2005; Carroll and Marx 2013; Sandberg et al. 2014). Although both novelty and restoration likely drive long-term adaptation, it is not yet clear which is more frequent.
Another issue that remains largely unexplored is the temporal pace of phenotypic change during adaptation to stress. After an environment becomes stressful, the acclimated state becomes the initial phenotypic state upon which natural selection acts. Thereafter, each adaptive mutation moves the population toward a phenotypic optimum (i.e., to a phenotype that best fits the stressful environment; Fisher 1930; Orr 2005). For historical and methodological reasons (Orr 2005), most evolutionary studies have focused on the end product of adaptation, leaving the early and intermediate steps of adaptation largely unexplored.
Fortunately, studies on experimental evolution may provide insight into the sequential magnitude of fitness changes during adaptation. To date, these studies have shown that the first beneficial mutation, or the “first-step mutation,” generally confers a large fitness advantage, perhaps because early, large-effect mutations outcompete other beneficial mutations in the population. In contrast, subsequent mutations confer more moderate fitness gains (Chou et al. 2011; Khan et al. 2011), in part due to diminishing returns epistasis (Kryazhimskiy et al. 2014).
Although our knowledge about the temporal trajectory of fitness continues to grow, few studies have examined shifts in phenotypes during the adaptive process. One exception is the study of Fong et al. (2005), which used microarrays to follow the phenotype of gene expression (GE) after a shift in growth from glucose to lactate and from glucose to glycerol (Fong et al. 2005). Fong et al. (2005) observed that 39% of genes were differentially expressed during the process of acclimation to glycerol medium; however, the number of differentially expressed genes (DEGs) decreased at an intermediate point of adaptation and also at the endpoint of the experiment. Most of the GE changes during adaptation were restorative (Fong et al. 2005), that is, they restored GE to normal, prestress levels. Unfortunately, however, the genetic bases of these phenotypic changes were not resolved; it was unclear if the changes in GE during the intermediate point of adaptation were caused by an early adaptive mutation or were caused by combinations of mutations that accumulated during the experiment. Thus, many questions regarding phenotypic adaptation remain unresolved, such as the following: What are the molecular effects underlying the large fitness advantage conferred by first-step mutations? And, what is the phenotypic contribution of the first-step mutation compared with later steps in an adaptive walk?
We have decided to explore these questions based on our recent, large-scale evolution experiment (Tenaillon et al. 2012). In this experiment, a strain of Escherichia coli B was evolved at 42 °C in 114 replicate populations. After 2,000 generations, we isolated single clones from each evolved population and identified the genetic changes that had accumulated during the experiment. Overall, the rpoB gene, which encodes the β subunit of RNA polymerase (RNAP), accumulated most mutations, with a total of 87 mutations in the 114 genomes. Three rpoB mutations were especially interesting because they resulted in amino acid substitutions in the active site of RNAP and conferred rifampicin resistance (Rodríguez-Verdugo et al. 2013). These mutations, all of which are located in codon 572, were driven to high frequency in 12 populations and were also beneficial at high temperature in the low glucose medium (Rodríguez-Verdugo et al. 2013). Furthermore, these mutations typically appeared early in the evolution experiment. In one population, for example, the rpoB I572L mutation swept to fixation before 100 generations; it was furthermore shown to confer a large fitness benefit of about 20% (Rodríguez-Verdugo et al. 2013). Because these rpoB mutations are located in the contact region between the downstream DNA duplex and RNAP (proximal active site), they could play key roles in modulating the enzyme’s activity in all three stages of transcription: Initiation, elongation, and termination (Ederth et al. 2006).
Yet, some questions remain. First, we have not identified the mechanistic bases of their fitness advantage. Second, although we know that these parallel mutations converge on having a fitness advantage at 42 °C (Rodríguez-Verdugo et al. 2013), with disadvantages at low temperatures (Rodríguez-Verdugo et al. 2014), we have yet to explore if they converged on other phenotypes. That is, it is still unclear if these mutations confer the same phenotypic effects as rpoB mutations found in other regions of RNAP that were also selected in some lineages during the thermal adaptation experiment. In this study, we address all these questions. Solving these questions is relevant for understanding the adaptive mechanisms prevalent in our experimental conditions and also in other evolutionary experiments, because mutations that affect transcriptional regulators (such as RNAP and the Rho termination factor) typically appear in the early stages of stress adaptation (Applebee et al. 2008; Goodarzi et al. 2009; Kishimoto et al. 2010). Numerous observations point to the possibility that highly pleiotropic mutations in transcriptional regulators could be the first step of a general mechanism of adaptation (Fong et al. 2005).
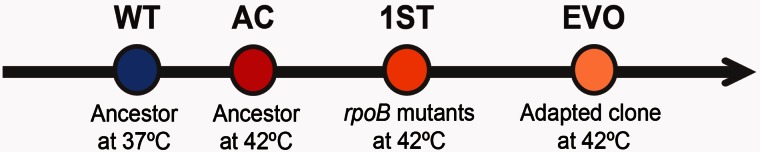
With these considerations in mind, we have investigated the timeline of adaptive change during our evolution experiment (fig. 1). To do so, we first describe growth characteristics and GE during acclimation to thermal stress at 42 °C. We then explore first-step mutations in terms of their effect on growth and GE relative to the ancestral strain. More specifically, we explore whether three amino acid substitutions in the RNAP active site converge on the same expression phenotype and also whether their effects are similar to another rpoB mutation that affects an amino acid substitution far from the RNAP active site. Finally, we evaluated the phenotypic contribution of one of these first-step mutations relative to the end product of adaptation, as represented by two clones from the end of the 2,000 generation experiment.
Fig. 1.
Thermal stress adaptation timeline. We characterized four states during thermal stress adaptation: A wild-type (WT) state, which corresponds to the ancestor grown at 37 °C; an acclimatized (AC) state, which corresponds to the ancestor acclimatized at 42 °C; a first-step adaptive (1ST) state, which corresponds to the rpoB single mutants; and an evolved (EVO) state, which corresponds to the high-temperature adapted clones.
Results
Acclimation to Thermal Stress Involved Many Changes in GE
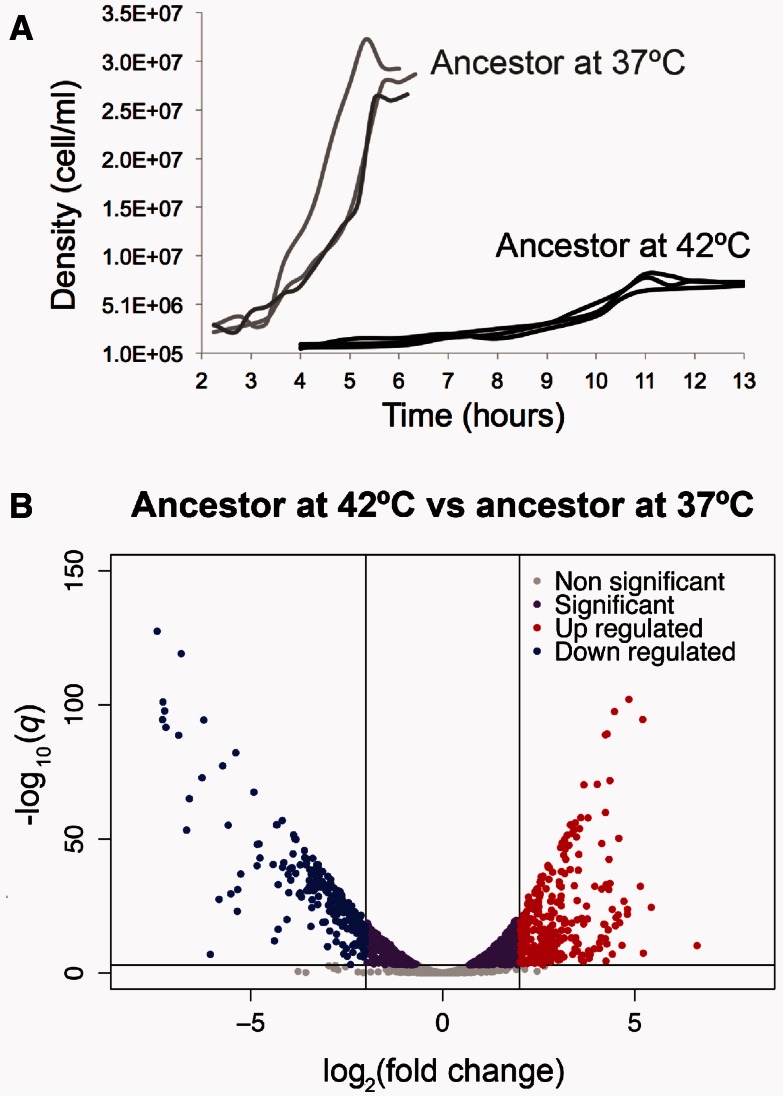
To explore the phenotypic effect of high temperature on the ancestor, we first characterized the ancestor’s growth at 37 °C, which we consider to be the prestress condition, and at 42 °C, the stress condition. The ancestor had a significantly longer effective lag phase and a significantly lower maximum growth rate and final yield when grown at 42 °C than when grown at 37 °C (fig. 2A and table 1), reflecting the fact that 42 °C is a stressful temperature.
Fig. 2.
Phenotypic characterization of the ancestor at 37 °C and 42 °C. (A) Growth curves of the ancestor grown at 37 °C and 42 °C (three replicates at each temperature). (B) Volcano plot showing the global differential expression of genes (represented as dots) between the ancestor grown at 42 °C compared with the ancestor grown at 37 °C. Colors represent status with respect to 2-fold expression difference, represented by two vertical lines, and a significance at q = 0.001, represented by an horizontal line.
Table 1.
Growth Parameters of the First-Step Mutants and the Ancestor.
| Strain | µmaxa 37 °C | µmaxa 42 °C | Pb | τc 37 °C | τc 42 °C | Pb | Yieldd 37 °C | Yieldd 42 °C | Pb |
|---|---|---|---|---|---|---|---|---|---|
| AncestorSE | 0.756 (0.018) | 0.418 (0.033) | 0.002 | 3.4 (0.1) | 6.7 (0.7) | 0.035 | 2.79 × 107 (8.15 × 105) | 7.21 × 106 (1.46 × 105) | 0.001 |
| I572FSE | 0.876 (0.085) | 0.734 (0.021) | 0.233 | 3.4 (0.4) | 6.8 (0.4) | 0.004 | 2.44 × 107 (1.08 × 106) | 1.72 × 107 (6.44 × 105) | 0.008 |
| Pe | 0.293 | 0.002 | 1.000 | 0.846 | 0.063 | 0.003 | |||
| I572LSE | 0.898 (0.090) | 0.809 (0.092) | 0.526 | 3.2 (0.4) | 6.8 (0.2) | 0.005 | 2.72 × 107 (9.86 × 105) | 1.57 × 107 (4.24 × 105) | 0.003 |
| Pe | 0.252 | 0.039 | 0.713 | 0.829 | 0.615 | 0.001 | |||
| I572NSE | 1.124 (0.219) | 0.622 (0.067) | 0.140 | 3.4 (0.2) | 8.5 (0.0) | 0.001 | 3.05 × 107 (1.76 × 106) | 1.28 × 107 (5.95 × 105) | 0.005 |
| Pe | 0.235 | 0.074 | 0.908 | 0.111 | 0.274 | 0.008 |
aMaximum growth rate with standard error in parenthesis.
bSignificance value from a two-sample t-test. The null hypothesis is that the mean at 37 °C and the mean at 42 °C are equal.
cDuration of the effective lag phase with standard error in parenthesis.
dFinal yield (cells/ml) with standard error in parenthesis.
eSignificance value from a two-sample t-test. The null hypothesis is that the mean of the ancestor and the mean of the mutant are equal. Numbers in bold correspond to P < 0.05.
Next, we explored the global GE profile after acclimation to 42 °C by obtaining RNA-seq data from three replicates of the ancestor at the midexponential growth phase at both 37 °C and 42 °C. Even under a stringent criterion of significance (q < 0.001), we identified 1,737 genes that were differentially expressed at 42 °C relative to 37 °C (fig. 2B and supplementary data set S1, Supplementary Material online). Of these DEGs, 240 genes were highly downregulated (log2-fold change < −2; blue dots in fig. 2B) and 293 genes were highly upregulated (log2-fold change > 2; red dots in fig. 2B) at 42 °C relative to 37 °C. Based on an enrichment analysis of gene ontology (GO) functional categories, we identified significant downregulation of genes involved in translation during heat acclimation (GO: 0006412; P = 4.55 × 10−29). Of the genes involved in translation, 54 genes transcribed products that were structural constituents of ribosomes (GO: 0003735), including rpl, rpm, and rps genes (supplementary table S1, Supplementary Material online). Other significantly downregulated biological processes involved the following: 1) Amino acid biosynthesis (GO: 0008652; P = 2.17 × 10−15), particularly genes involved in the biosynthesis of methionine (met genes); 2) biosynthesis of ribonucleosides (GO: 0042455; P = 3.12 × 10−7), including purines and pyrimidine (pur and pyr genes); and 3) bacterial-type flagellum-dependent cell motility (GO: 0071973; P = 5.83 × 10−3), including flg and flh genes (supplementary table S1, Supplementary Material online). Surprisingly, the heat shock inducible genes (Nonaka et al. 2006) were mostly downregulated during heat acclimation, as were the subunits of the core RNAP (rpoA, rpoB, rpoC, and rpoZ; supplementary table S2, Supplementary Material online).
Among the upregulated genes, we identified a significant enrichment of genes involved in the following: 1) Amino acid degradation (GO: 0009063; P = 6.55 × 10−4) such as degradation of arginine (ast genes); 2) alcohol degradation (GO: 0046164; P = 5.44 × 10−4); 3) glycerol metabolic process (GO: 0015794; P = 1.73 × 10−2); 4) monosaccharide transport (GO: 0015749; P = 1.11 × 10−2); and 5) cellular response to stress (GO: 0033554; P = 4.60 × 10−2). Of the 66 genes that had been identified previously as a component of the general stress response (Weber et al. 2005), 57 (86%) were significantly upregulated at 42 °C in our experiment (supplementary table S2, Supplementary Material online).
Overall patterns of GE suggest that the ancestor at 42 °C induced an early stationary phase. To be sure that this GE profile did not result from sampling at the wrong phase of the growth curve, we repeated the experiment. We again sampled three replicates at midexponential, repeated RNA sampling, and transcriptome analyses. The results were qualitatively similar between experiments, implying both that the ancestor at 42 °C cannot unleash full growth and that the population expands at a low rate in this stressed physiological state.
First-Step Adaptive Mutations Changed GE at 42 °C Dramatically
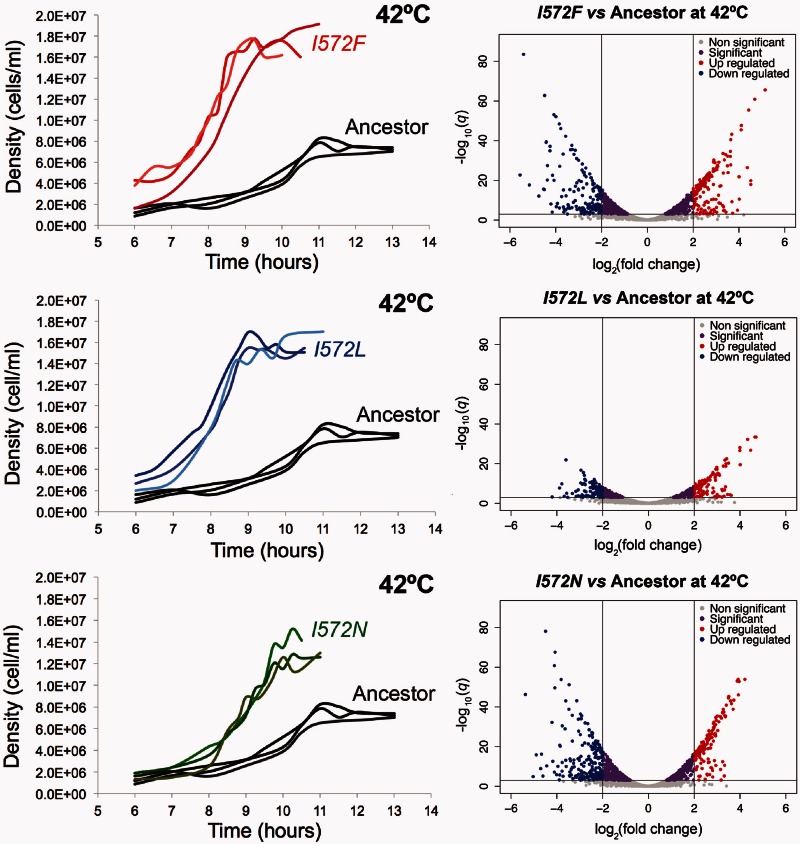
Given insights into the acclimation response at 42 °C, we then investigated the effect of three potential first-step mutations—rpoB I572F, I572L, and I572N—on cell growth and GE. Previous estimates of relative fitness indicated that these mutations are advantageous at 42 °C (Rodríguez-Verdugo et al. 2013). We complemented this observation by estimating two components of fitness—the duration of the lag phase and the maximum growth rate. We also calculated the final yield, which is not a component of fitness (Vasi et al. 1994). These life-history traits were estimated from the mutant’s growth curves at both 42 °C and 37 °C. The mutants had a significantly longer effective lag phase (τ) and lower final yield at 42 °C compared with their growth at 37 °C (table 1). We note, however, that an alternative definition of lag phase, which was interpolated from the maximum growth rate (see Materials and Methods), reflects a shorter but not significantly different lag phase in the mutants (supplementary table S3, Supplementary Material online). Nevertheless, the mutants had a significantly higher maximum growth rate compared with the ancestor grown at 42 °C, reflecting improved fitness under thermal stress (table 1 and fig. 3).
Fig. 3.
Phenotypic characterization of the mutants compared with the ancestor at 42 °C. The left side of the figure shows the growth curves of the ancestor and the mutants grown at 42 °C (three replicates per each genotype). The right side of the figure shows the volcano plots showing the global differential expression of genes (represented as dots) between the mutants grown at 42 °C and the ancestor grown at 42 °C. Colors represent status with respect to 2-fold expression difference, represented by two vertical lines, and a significance at q = 0.001, represented by an horizontal line.
We explored two hypotheses about the molecular mechanisms that may underlie the growth improvement of the mutants at 42 °C. Our first hypothesis was the mutants grew better at 42 °C, because the mutated RNAP was more efficient than the ancestral RNAP at transcribing DNA to RNA (Jin et al. 1992; Reynolds 2000). Our second, nonexclusive hypothesis was that the mutated RNAP led directly or indirectly to altered transcription of a set of genes and this differential expression underlies growth improvement (Conrad et al. 2010).
Regarding the first hypothesis, we predicted that the rpoB mutations slowed the RNAP complex (Rodríguez-Verdugo et al. 2014), which is otherwise accelerated at high temperatures (Ryals et al. 1982; Mejia et al. 2008). In turn, we reasoned that the reduced speed of RNAP increases both transcription fidelity and termination efficiency (Jin et al. 1988; Zhou et al. 2013), resulting in an overall higher transcription efficiency (Jin et al. 1992). We measured the transcription efficiency of the mutants and the ancestor at 42 °C. In our experiment, transcription efficiency captures both the speed and accuracy of all events during transcription, including initiation, elongation, and termination (Reynolds 2000). To measure the transcription efficiency we measured the relative abundance of an inducible fluorescent gene (YFP) inserted in our strains, at different points post induction (see Materials and Methods). Using this method (Reynolds 2000; Brandis et al. 2012), we found that two of the three mutants (I572L and I572F) yielded higher slopes than the ancestor at 42 °C, consistent with our hypothesis of higher efficiency. However, the assay has wide confidence intervals, and there were therefore no statistical differences in the transcription efficiency of the mutants relative to the ancestor at 42 °C (supplementary fig. S1, Supplementary Material online). Within the limits of our experiment, we cannot conclude that the growth improvement of the mutants at 42 °C is caused by higher RNAP efficiency.
We also addressed our second hypothesis that the rpoB mutations lead to changes in GE with consequent growth improvements at 42 °C. To test this hypothesis, we obtained RNAseq data from each mutant, based on two replicates per mutant at midexponential growth at 42 °C. We then contrasted the GE profile from each mutant against that of the ancestor grown at 42 °C.
All three mutations displayed hundreds to thousands of DEGs (q < 0.001; fig. 3). The mutant I572F had 1,332 DEGs at 42 °C with 195 highly downregulated (log2-fold change<−2) and 182 highly upregulated (log2-fold change > 2) genes relative to the 42 °C ancestor. The mutant I572L had 598 DEGs with 126 highly downregulated and 126 highly upregulated genes. Finally, the mutant I572N had 1,360 DEGs with 233 highly downregulated and 164 highly upregulated genes. Within the confines of this experiment, we cannot differentiate as to whether these changes in GE were caused by direct effects, such as modified binding of the RNAP, or indirect, downstream effects. Nonetheless, it is clear that single mutations within codon 572 of rpoB lead to numerous, pleiotropic shifts in GE.
rpoB Mutations in Codon 572 Restored GE Back Toward the Ancestral State
To investigate the general trend in GE changes from the ancestor to acclimation and then from acclimation to first-step mutations (fig. 1), we used two approaches. The first was Principal Component Analysis, which simultaneously considered the expression of all genes and all clones. The resultant plot of the first and second components exhibited differences among clones and also clearly indicated that the clones with first-step mutations shifted their GE profiles toward the 37 °C ancestor (supplementary fig. S2, Supplementary Material online).
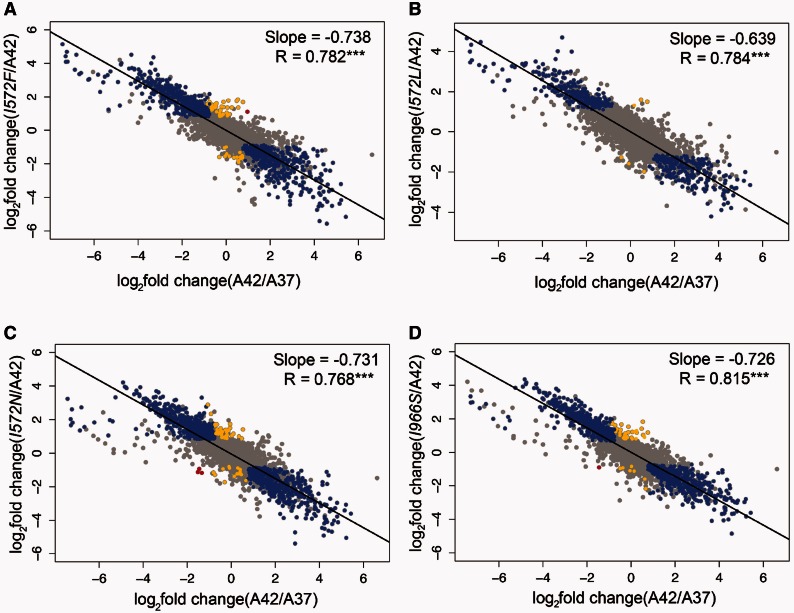
Our second approach was to plot the log2-fold expression change during the acclimation response (ancestor grown at 42 °C vs. ancestor grown at 37 °C, x-axis) against the log2-fold GE change during the adaptive response (mutant grown at 42 °C vs. ancestor grown at 42 °C, y-axis; fig. 4). If there were no relationship between shifts in GE during acclimation and during the adaptive response, we expected a regression slope of 0; alternatively, if all the genes of a mutant were restored by an adaptive mutation—such that the expression of each genes changed from a stressed state back to a prestressed state—the slope of the fitted regression would approach −1.0. For the three mutants we observed a highly significant negative correlation, with slopes of −0.738 for I572F, −0.639 for I572L, and −0.731 for I572N (fig. 4A–C). All three slopes were highly significant (P < 0.001) based on permutation tests (see Material and Methods), indicating that the main phenotypic effect of the first-step mutations was to restore global GE toward the prestressed state.
Fig. 4.
Global changes in GE during the acclimation and adaptive response. In all the graphs, the x-axis represents the acclimation changes (ancestor grown at 42 °C vs. ancestor grown at 37 °C). The y-axis represents the changes at different steps of the adaptive walk: First-step adaptive mutations (I572F, I572L, I572N) and a single mutant (I966S) vs. ancestor grown at 42 °C. Changes in expression were categorized and colored as follows: Restored (blue), reinforced (red), and novel (yellow). Both unrestored and uninformative genes are colored in grey. The black line represents the linear regression fitted to the dots in each graph.
We next examined the GE changes in more detail by characterizing the expression of individual genes into one of the four patterns of change that denote the direction of the effect of the mutated RNAP (see Materials and Methods and supplementary table S4, Supplementary Material online). First, the expression of an individual gene could be “restored” back toward the prestressed state. Second, the expression of a gene could be “reinforced” into an exaggeration of the acclimated state, such that the mutated RNAP produced more transcripts (in the case of upregulated genes) or fewer transcripts (in the case of downregulated genes) than the acclimated state. Third, a gene was “unrestored” if the mutated RNAP did not change GE significantly relative to the ancestral RNAP at 42 °C. Finally, GE was “novel” if it was not differentially expressed during the acclimation response, but was expressed significantly differently during the adaptive response. For these definitions, we investigated genes that differed in expression either during acclimation or between acclimation and first-mutation clones (supplementary table S4, Supplementary Material online); as a result, we categorized directional shifts of roughly half of 4,202 E. coli genes.
Following this categorization, we observed that most (63%) of the genes that were differentially expressed during acclimation were restored by mutant I572F (blue dots in fig. 4A; table 2), as expected from the strong overall negative correlation in figure 4A. The same was true for mutant I572N, for which most (67%) genes were restored (table 2). For mutant I572L, we found most genes to be unrestored (68%), but a substantial proportion of genes (32%) had restored expression (table 2). In contrast to restoration, only a small proportion (∼3% or less) of characterized genes exhibited novel expression in the rpoB mutants (table 2).
Table 2.
Classification of the Genes into Four Patterns of Expression Change.
| Category | I572F | I572L | I572N | I966S |
|---|---|---|---|---|
| 1. Restored | 1,092 | 563 | 1,165 | 1,308 |
| 2. Reinforced | 1 | 0 | 3 | 1 |
| 3. Unrestored | 644 | 1,174 | 569 | 428 |
| 4. Novel | 59 | 6 | 53 | 39 |
Finally, we based our analyses on two replicates of each of our single mutant clones. However, the broad similarity in results across the 3 codon 572 mutations suggest that the 3 clones can be treated as pseudoreplicates. Accordingly, we combined the six replicates (two from each of the three single mutations) and reran GE analyses to examine the effect of increased sampling. Using all 6 replicates, we identified 867 genes that were differentially expressed relative to the 42 °C ancestor (q < 0.001), which is within the range of 598 to 1,360 genes based on analyses of separate mutations (fig. 3). Importantly, the directional information from this combined analysis was similar to the analyses based on single mutants, because we estimated that 45% of genes that were differentially expressed during acclimation had restored GE. Thus, although the statistical power to detect differences likely varies among clones, the combined data reinforce the observation that a large proportion of genes were restored in GE by codon 572 rpoB mutants.
An rpoB Mutation away from the Active Site of the RNAP also Restores GE
Given that rpoB mutations in codon 572 converged in phenotype toward restorative GE, we sought to know if additional mutations in rpoB had similar effects. To address this question, we constructed a single mutant, rpoB I966S, which alters one of the two parallel α-helices of the Eco flap domain of RNAP (Opalka et al. 2010). The I966S mutation was found in 15 of our high-temperature adapted clones (Tenaillon et al. 2012), and its fitness advantage at 42 °C was confirmed by competition experiments (Rodríguez-Verdugo et al. 2014).
We obtained RNAseq data from two replicates of this mutant at 42 °C and performed the same GE analyses. We observed a global pattern of GE change similar to that for the I572F, I572L, and I572N mutants. That is, the I966S mutation tended to restore GE from the acclimated state toward the prestressed state (fig. 4D).
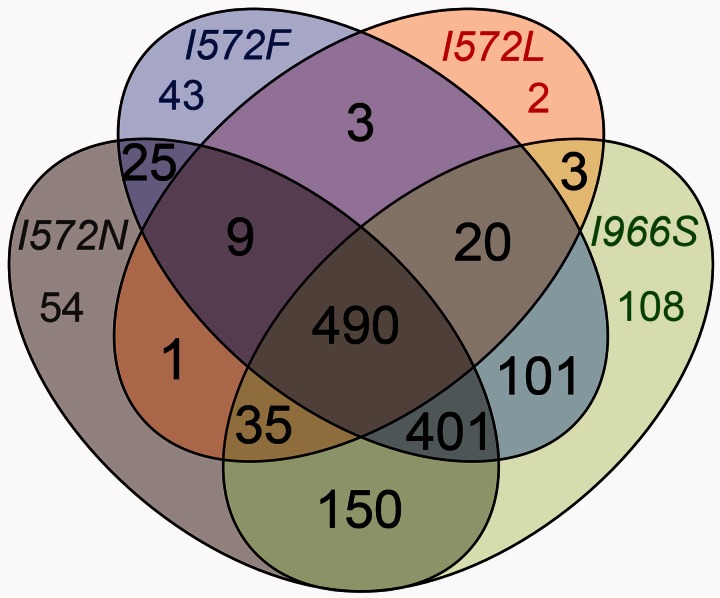
To explore the convergent effects of the I966S, I572F, I572L, and I572N mutations further, we determined the number of genes with restored GE shared among the four mutants. A third of the restored genes were shared between the four mutants, and 86% of the genes were share by at least two mutants (fig. 5), indicating a high level of expression convergence. This high level of phenotypic convergence was also highlighted by the pairwise comparisons of differential expression between the mutants (supplementary fig. S3, Supplementary Material online). We performed an enrichment analysis of GO assignments for the 490 restored genes that were shared among the 4 mutants. Not surprisingly, given the overall pattern of restoration (fig. 4), the restored genes represented the same sets of genes that were enriched for the acclimation response. For example, significantly downregulated genes during the acclimation response, such as the genes involved in translation (rpl, rpm, and rps), were significantly upregulated in the four mutants. This suggests that the different RNAP mutants restored GE toward a pattern of growth.
Fig. 5.
Convergence of genes with restored expression in single mutants. Number of shared genes with restored expression.
We also determined how many genes of novel GE were shared between the four single mutants: I572F, I572L, I572N, and I966S. Only 2% of the genes with novel GE were shared between the four mutants, and 33% of the genes were shared by at least two mutants (supplementary fig. S4, Supplementary Material online), indicating lower expression convergence for genes with novel GE than for restored genes. The complete set of genes with differential GE shared and unique among the four single mutant clones is provided in supplementary data set S2, Supplementary Material online.
In conclusion, the phenotypic convergence observed between the mutants I572F, I572L, I572N, and I966S suggests the following: 1) Restoration of the altered physiological state back toward a prestressed state, 2) that much of that restoration was achieved by single, highly pleiotropic mutations, and 3) similar effects result from different amino acid mutations at the same site (codon 572 for mutants I572F, I572L, and I572N) or at a different site (codon 966 for mutant I966S).
Mutations Fixed during Adaptation Contributed Few Changes in GE
Following the temporal schematic of this study (fig. 1), our last step was to contrast the GE phenotype of one first-step mutation (I572L) to end products of our adaptation experiment (i.e., clones evolved 2,000 generations at 42 °C; fig. 6A). As end products, we chose two high-temperature adapted clones: Clone 27 and clone 97 (clone numbers correspond to reference Tenaillon et al. 2012), which were isolated from two populations in which the mutation I572L swept to fixation before 400 generations (Rodríguez-Verdugo et al. 2013). Although sharing the same rpoB mutation, these two high-temperature adapted clones differed in their genetic backgrounds (supplementary tables S5 and S6, Supplementary Material online). Clone 27 had 2 large deletions of 2,896 and 71,416 bp affecting > 65 genes, a 138-bp deletion disrupting the tRNA-Met gene, and 7-point mutations in 7 genes. In contrast, clone 97 had only one 4-bp small deletion, an insertion sequence (IS), and 8-point mutations in eight different genes (Tenaillon et al. 2012).
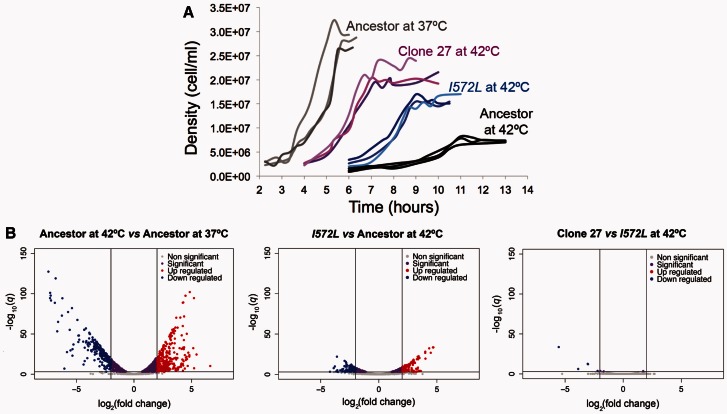
Fig. 6.
Phenotypic changes during thermal stress adaptation of clone 27. (A) Growth improvements during the heat stress adaptive walk. (B) Changes in GE during the acclimation and adaptive response.
We again characterized the clones’ growth curves at 42 °C and compared them with the ancestor grown at 37 °C, the ancestor grown at 42 °C, and the mutant I572L grown at 42 °C (table 3, fig. 6A, and supplementary fig. S5, Supplementary Material online). The high-temperature adapted clones 27 and 97 had significantly shorter effective lag phases compared with I572L mutant, and an alternative measure of lag phase revealed qualitatively similar results but with less statistical significance (supplementary table S7, Supplementary Material online). In addition, clone 27 had a significantly higher final yield than I572L mutant (table 3). It thus appears that the mutations accumulated after the first-step mutations contribute to better growth at 42 °C. We note, however, that none of the adapted clones grow as well as the ancestor at 37 °C, which had a significantly shorter effective lag phase and a higher final yield (table 3 and fig. 6A).
Table 3.
Growth Parameters of the High-Temperature Adapted Clones 27 and 97 at 42 °C Compared with the Mutant I572L and the Ancestor at 37 °C and 42 °C (values in table 1).
| Strain | µmaxa | τb | Yieldc |
|---|---|---|---|
| 27 | 0.769 (0.041) | 4.7 (0.3) | 2.09 × 107 (1.22 × 106) |
| 97 | 0.794 (0.063) | 4.7 (0.3) | 1.87 × 107 (2.20 × 106) |
| Comparison | Significanced | ||
| 27 vs. ancestor at 42 °C | 0.003 | 0.076 | 0.007 |
| 97 vs. ancestor at 42 °C | 0.013 | 0.076 | 0.034 |
| 27 vs. I572L at 42 °C | 0.716 | 0.011 | 0.039 |
| 97 vs. I572L at 42 °C | 0.651 | 0.011 | 0.302 |
| 27 vs. ancestor at 37 °C | 0.801 | 0.049 | 0.011 |
| 97 vs. ancestor at 37 °C | 0.613 | 0.049 | 0.040 |
Note.—Numbers in bold correspond to P < 0.05.
aMaximum growth rate with standard error in parenthesis.
bDuration of the effective lag phase with standard error in parenthesis.
cFinal yield (cells/ml) with standard error in parenthesis.
dSignificance value from a two-sample t-test. The null hypothesis is that the means are equal.
Knowing that the evolved clones grew more quickly and to higher yields than the single mutant I572L at 42 °C, we sought to identify GE changes that could explain their improved growth. We obtained RNA-seq data from each high-temperature adapted clone (two replicates per clone) grown at 42 °C. We then contrasted the GE from each clone against the GE of the first-step mutation I572L at 42 °C. To our surprise, we observed very few genes that were differentially expressed (q < 0.001): 7 for clone 27 in addition to 56 genes with no measurable GE (fig. 6B and supplementary table S5, Supplementary Material online) and 16 for clone 97 (supplementary fig. S5, Supplementary Material online). Furthermore, the high-temperature adapted clones maintained the general pattern of restoration back to the ancestral physiological state previously observed for the mutant I572L (slope of−0.654 for clone 27 and slope of −0.645 for clone 97; supplementary fig. S6, Supplementary Material online).
These observations suggest that the mutations accumulated in later steps of thermal stress adaptation did not substantially change the expression profile caused by the first-step mutation; that is, most detectable GE shifts were caused by the I572L mutation and were associated with the restoration of the growth profile. To emphasize this point, we plotted the log2-fold expression change during the first-step adaptive response (mutant I572L grown at 42 °C vs. ancestor grown at 42 °C; supplementary fig. S7, Supplementary Material online) against the log2-fold expression change during the complete adaptive response (high-temperature adapted clone grown at 42 °C vs. ancestor grown at 42 °C; supplementary fig. S7, Supplementary Material online). We observed a highly significant positive correlation (slope of 0.977 for clone 27 and slope of 1.004 for clone 97; supplementary fig. S7A and B, Supplementary Material online), confirming high similarity in the overall GE pattern between the mutant I572L and the high-temperature adapted clones. Finally, when we contrasted the GE from the two high-temperature adapted clones at 42 °C (clone 97 vs. clone 27), there were only 15 DEGs (q < 0.001; supplementary figs. S3 and S7C, Supplementary Material online), beyond the 52 genes that were part of the 2 large deletions in clone 27 and did not have any measureable GE. Therefore, the GE profiles for the two high-temperature adapted clones were nearly identical within the limitations of our experiment, despite their differences in genetic background. These observations confirm that the first-step mutation I572L contributed to most of the changes in GE during high-temperature adaptation.
Discussion
Two aspects of adaptation that have been largely unexplored are the temporal change of phenotypes during the adaptive process and the genetic changes underlying these changes. Here we have focused on the phenotypic effects of first-step mutations during adaptation of E. coli to high temperature (42 °C). A significant finding of our study is that single mutations in RNAP led to either the direct or indirect alteration of GE for thousands of genes—most of which were differentially expressed during acclimation to 42 °C—and conferred large fitness advantages. A major phenotypic effect of these first-step mutations was to restore global GE back toward the prestressed state. The GE profile of the ancestor at 42 °C revealed that the cells were physiologically stressed, but the first-step mutations restored GE toward an efficient growth profile. Subsequent mutations also increased fitness but did not substantially change GE.
Heat Stress Acclimation Involves a Balance between Energy Conservation and Stress Resistance
The acute response to thermal stress, known as the heat shock response, occurs in diverse organisms (Richter et al. 2010). In E. coli, the heat shock response is transient (in the order of magnitude of minutes) and is characterized by the induction of stress-related proteins mediated by the alternative σ32 factor (Nonaka et al. 2006). The σ32 regulon encodes the following: 1) Molecular chaperons (i.e., ClpB, DnaK, DnaJ, IbpB, GrpE, GroEL, GroES) that promote protein folding; 2) cytosolic proteases (i.e., ClpP, ClpX) that clear misfolded and aggregated proteins; 3) metabolic enzymes; 4) DNA/RNA repair enzymes; 5) regulatory proteins; 6) proteins involved in maintaining cellular integrity; and 7) proteins involved in transport and detoxification (Nonaka et al. 2006; Richter et al. 2010). Although the heat shock response has been studied widely, the expression of heat shock genes after hours or days of thermal stress (i.e., thermal acclimation) is an open question.
We have found that most heat shock induced genes (Nonaka et al. 2006; Richter et al. 2010) were not differentially expressed during acclimation and were, in fact, downregulated. For example, most of the heat shock genes encoding chaperones, such as clpB, dnaJ, dnaK, groEL, and groES, were downregulated during the acclimation response (supplementary table S2, Supplementary Material online). However, one exception is spy, which encodes a periplasmic chaperone and was upregulated during acclimation (log2-fold change = 5.0, q < 0.001; supplementary data set S1, Supplementary Material online). Previous studies on Saccharomyces cerevisiae and E. coli have reported a decrease in the expression of molecular chaperones after ∼15 min of heat stress induction (Eisen et al. 1998; Zhao et al. 2005; Jozefczuk et al. 2010; Richter et al. 2010). Therefore, it is possible that the heat shock genes were initially expressed during our experiment, immediately after transfer to 42 °C, but were subsequently downregulated.
Nevertheless, two previous studies have explored the physiological acclimation of E. coli to high temperature and reported upregulation of heat shock genes (Gunasekera et al. 2008; Sandberg et al. 2014). The discrepancy among studies may be explained by differences in genetic backgrounds (E. coli K-12 strain vs. E. coli B strain), by differences in temperature (42 °C vs. 43 °C in Gunasekera et al. 2008), by differences in culture conditions (ours favor microaerophiles), and by differences in sampling. For example, in our study bacterial clones were allowed to acclimate at 42 °C for 1 day before being sampled the next day during midexponential phase at 42 °C. Thus, the bacteria spent one complete cycle of growth (lag, exponential, and stationary phase) at 42 °C, in addition to the approximately half cycle of growth (lag and half exponential phase) at 42 °C (in total ∼1.5 days). For other studies, the time that the bacteria spent at high temperature before the RNA extraction is unclear (Sandberg et al. 2014). Similarly, we have carefully documented the growth curve of the ancestor at 42 °C to help ensure that our RNAseq samples originated from the exponential growth phase (supplementary fig. S8, Supplementary Material online). Other studies report to have sampled during midexponential phase but have not reported growth curves. Under normal conditions growth curves may not be necessary, but the exponential phase can be very short under extreme stress. It is thus possible that previous studies have reported GE on slightly different phases of the growth cycle.
Micro-organisms often resist stressful conditions by modulating GE to limit growth (López-Maury et al. 2008). As a consequence, genes with growth-related functions, which are energy demanding, are downregulated, allowing a redistribution of resources and energy to the expression of genes related to stress resistance (López-Maury et al. 2008; Jozefczuk et al. 2010; Jin et al. 2012). Surprisingly, we have observed a downregulation of genes encoding different subunits of RNAP during the acclimation response. Assuming that lower expression also reflects protein abundance, this observation implies that the ancestor contains fewer RNAP molecules when grown at 42 °C than when grown at 37 °C.
The reduction of RNAP molecules can have important physiological consequences, given that RNAP is limiting for genome-wide transcription (Ishihama 2000). A reduction in the total number of RNAP would limit the transcription rate and favors our hypothesis that mutated RNAP have higher transcriptional efficiency at 42 °C (Rodríguez-Verdugo et al. 2014). Accordingly, we have examined RNAP efficiency and found a slight trend toward higher rates of transcriptional efficiency for two of the three mutations in codon 572 of rpoB. However, none of these differences were supported statistically (supplementary fig. S1, Supplementary Material online). A limitation in RNAP molecules could also indirectly affect bacterial growth (Jin et al. 2012). For example, when growth conditions are unfavorable, RNAP molecules are released from the rRNA operons, thereby reducing rRNA synthesis (i.e., reducing growth) so that more RNAP molecules become free and available for genome-wide transcription (Jin et al. 2012). Therefore, the reduction in growth at 42 °C may be explained in part by the downregulation of genes involved in translation and ribosome biogenesis but also by a potential limitation of RNAP.
Other downregulated genes involved in energy demanding processes include genes associated with the biosynthesis of amino acids, nucleotides, ribonucleotides and constituents of the flagella (supplementary table S1, Supplementary Material online). A similar pattern of expression has been observed during acute exposure to thermal stress (Jozefczuk et al. 2010). In addition, metabolomic studies have reported a sharp decline in the levels of nucleotides in E. coli cultures exposed to heat stress (Jozefczuk et al. 2010; Ye et al. 2012). Therefore, our study suggests that the pattern of energy conservation mediated through downregulation of energy demanding processes not only occurs during the acute response to stress but also occurs during the acclimation response.
We hypothesize that some of the resources and energy are redistributed to express genes controlled by the sigma S factor or σS, the master regulator of the stress response (Battesti et al. 2011). Using previous observations (Weber et al. 2005; Keseler et al. 2013), we generated a list of 66 genes expressed under several stress conditions (including high temperature) and under regulation of σS. Of these, 62 genes (94%) were significantly upregulated during acclimation to 42 °C (supplementary table S2, Supplementary Material online). σS induced genes were related to: 1) The synthesis molecules responsible to deal with the detrimental effects of stress, 2) transport systems, and 3) the production of metabolic enzymes, mostly involved in the central energy metabolism (Weber et al. 2005).
In conclusion, the ancestor acclimates to thermal stress by launching alternative GE programs (López-Maury et al. 2008). One involves the downregulation of growth-related pathways, potentially resulting in energy conservation. The other involves the upregulation of stress genes that are involved in repair and metabolic adjustments to high temperature.
Restoration of the Ancestral Physiological State Is Advantageous
This study has shown that first-step mutations dramatically altered GE from the acclimated state and acted primarily to restore GE toward the prestressed state (fig. 3). Previous studies have reported that small deletions in RNAP (Conrad et al. 2010) and mutations in other RNAP subunits (Carroll et al. 2015) change GE of hundreds of genes. Taken together, these observations suggest that rpoB mutations have important downstream effects that affect complex networks of interacting genes and their products (i.e., global regulators of GE or “hubs”; Barabasi and Oltvai 2004). Our study also adds to previous observations that global regulators are a primary target of natural selection in microbial evolution experiments (Philippe et al. 2007; Hindre et al. 2012; Carroll et al. 2015).
At least two pieces of evidence suggest that the restoration of GE was advantageous in our experiment, leading to the rapid fixation of rpoB mutations in evolved populations (Rodríguez-Verdugo et al. 2013). First, restoration in GE may be advantageous because it “reactivates” some of the growth-related functions that were downregulated during the acclimation response. The GO categories related to growth, such as translation and ribosomal biogenesis, were significantly enriched in the four mutants relative to the acclimated state. The upregulation of growth-related genes may explain why the mutants have higher maximum growth rates and higher final yields than the ancestor (fig. 3), as well as higher relative fitness values (Rodríguez-Verdugo et al. 2013, 2014). Second, the general trend of restoration occurred in parallel in all four rpoB mutants (I572F, I572L, I572N, and I966S); such phenotypic convergence is commonly interpreted as a sign of adaptive evolution (Christin et al. 2010).
Not only did we observe convergence in global GE (fig. 4), but we also observed substantial overlap of genes with restored expression: 86% of the affected genes were shared by at least two mutations (fig. 5). Surprisingly, these observations extend not only to three different mutations in the rpoB codon (572) but also to a mutation (I966S) that is not in the RNAP active site (figs. 4D and 5). It is somewhat surprising to us that the I966S mutation produces similar effects as those in codon 572. We note, however, that our RNAP mutations differ from those found in other evolution experiments, such as the rpoB R562V mutation found in adaptation to glycerol (Herring et al. 2006), rpoA mutations found in adaptation to methanol (Carroll et al. 2015), and rpoC mutations uncovered during evolution to heat stress (Tenaillon et al. 2012; Sandberg et al 2014). Taken together, these observations suggest that RNAP can be fine-tuned by mutations in a number of different subunits and residues (Tenaillon et al. 2012). We conjecture that many of these changes may be nearly equivalent with respect to their tendency to move GE toward restoration (Carroll et al. 2015).
It is unclear whether the effects of these RNAP mutants are direct or indirect. For a direct effect, the mutated RNAPs would have had new intrinsic binding affinities for promoters throughout the genome. Alternatively, because RNAP is a sensor of the physiological state of the cell (Ishihama 2000), it may induce different GE programs. For example, under amino acid starvation RNAP modifies recruitment of sigma factors through its interaction with the alarmone ppGpp, and it consequently induces the stringent response program (Chatterji and Ojha 2001). The fact that mutations with similar effects are not limited to the active site of the RNAP favors the possibility of indirect effects, but further experiments are needed to test this hypothesis.
It is worth noting that restoration to the nonstress phenotype is not complete, because first-step mutations differ markedly from the 37 °C ancestor in GE dynamics. For example, each of the 4 rpoB mutations differ from the ancestor in the expression of a minimum of 392 genes (I572L) and as many as 688 genes (I572N). The important point is that first-step mutations restore hundreds of genes toward nonstress expression dynamics but substantial differences remain relative to the wild-type phenotype. These differences may explain, in part, differences in growth characteristics between first-step mutations and the ancestor at 37 °C (supplementary fig. S9, Supplementary Material online).
In contrast to restorative changes in GE, we find that few novel changes in GE occurred in parallel among the four rpoB mutants (supplementary fig. S4, Supplementary Material online). These observations add to a growing literature that suggests restorative changes in GE are a general trend in laboratory adaptation experiments, but novel (and/or reinforced) changes are less common (Fong et al. 2005; Carroll and Marx 2013; Sandberg et al. 2014; although see Szamecz et al. 2014).
Pleiotropy and Compensation
The four rpoB mutations (I572F, I572L, I572N, and I966S) were advantageous in the conditions of our experiment, but they also had pleiotropic effects, such as fitness trade-offs at low temperatures (Rodríguez-Verdugo et al. 2014). Previous studies have shown that mutations in global regulators often have maladaptive side effects (Hindre et al. 2012; Carroll et al. 2015). We hypothesized, therefore, that later adaptive mutations compensate for maladaptive side effects of highly pleiotropic first-step mutations (Hindre et al. 2012).
To contrast first-step mutations to the end products of our adaptation experiment (fig. 1), we compared the I572L rpoB mutation with two clones (97 and 27) that included the I572L mutation. Each of the two clones differed from the single-mutant background by ∼10 mutations; these mutations included both point mutations and more complex genetic changes, such as a large deletion. Based on these contrasts, we have found that the first-step mutation I572L contributed most of the GE variation during thermal stress adaptation (987 DEGs), while later mutations contributed fewer changes in GE (63 and 16 DEGs in clones 27 and 97, respectively; fig. 6 and supplementary fig. S5, Supplementary Material online). Interestingly, these few changes in GE may contribute to significant changes in growth parameters (table 3). Therefore, the number of DEGs was not proportional to fitness gains.
This “disconnect” between the number of genes differentially expressed and the magnitude of the fitness advantage might be caused by compensatory changes to the pleiotropic side effects of mutation I572L. Under this model, we presume that some of the hundreds of DEGs in I572L have beneficial effects, while others have deleterious effects, netting an overall beneficial change in GE. If true, it is likely that subsequent mutations change the expression of only few genes, but most of these changes are beneficial. For example, the large deletion in clone 27 contained genes involved in iron acquisition (fep and ent operons; Crosa and Walsh 2002), and copper and silver efflux system (cus operons; Long et al. 2010). Costly, nonfunctional pathways are often downregulated in order to save energy that would be otherwise used to produce unnecessary proteins and metabolisms (Cooper et al. 2001; Lewis et al. 2010). Therefore the large deletion of 71 kb in length may be an energetic benefit of large phenotypic effect (Hug and Gaut 2015), explaining its occurrence in 35 high-temperature adapted clones (Tenaillon et al. 2012).
Clone 97, which lacked the large deletions of clone 27, displayed fewer changes in GE than clone 27. In this clone, one of the significantly downregulated genes (q < 0.001) was the rmf gene, which encodes the ribosome modulation factor (RMF). RMF has been associated with decreased translation activity and is expressed during slow growth conditions, such as stationary phase (Polikanov et al. 2012). Therefore, the downregulation of rmf, occurring in parallel in clone 27 and 97, might increase protein synthesis and thus growth.
Finally, we observed that the genes involved in flagellum-dependent cell motility (e.g., flg genes) were highly downregulated during the acclimation response (log2-fold <−3 and q < 0.001) but were again upregulated in the single mutants I572F and I572L (q < 0.001; supplementary data set S1, Supplementary Material online). In the conditions of our evolution experiment—a well-mixed environment lacking physical structure—motility seems unnecessary. Given that the biosynthesis of flagella is costly (Soutourina and Bertin 2003), reducing the expression of flg genes might be beneficial (Cooper et al. 2003; Fong et al. 2005). We therefore posit that the restoration in GE of flg genes might be costly and have deleterious effects. Interestingly, some of the flg genes were downregulated (q < 0.05) in clone 97 when compared with the first-step mutant I572L (supplementary table S8, Supplementary Material online). Therefore, later adaptive mutations might contribute to the fine-tuning of GE by compensating the side effects of restoration. We note, however, that we have not yet identified the mutation that causes the downregulation of flg genes in clone 97. That being said, the upregulation of flagellar genes after restorative shifts in GE and the fixation of subsequent mutations that downregulate them has been observed previously (Sandberg et al. 2014).
Concluding Remarks
Mutations in global regulators of GE are observed recurrently in laboratory evolution experiments (Applebee et al. 2008; Goodarzi et al. 2009; Kishimoto et al. 2010; Carroll et al. 2015). It is not always clear if these mutations represent the first step of an adaptive walk or a later step, but at least in the case of rpoB and rpoC mutations it seems they are often first-step mutations (Herring et al. 2006). Therefore, the pattern that we have observed in our study may not be specific to our system but instead a more general phenomenon. Moreover, these results tend to support the view that evolution to stressful environments is driven, at least initially, by shifts in regulatory proteins (Wilson 1976).
Based on our results, we propose a general, two-step adaptive process. First, a mutation affecting global transcriptional regulator appears in the population and changes global GE. This expression change is mostly restorative, so that the stressed state moves toward a prestressed (ancestral) state. These changes in GE confer a high advantage, promoting the rapid fixation of the mutation in the population. Once the cell recovers its normal homeostatic state, other mutations accumulate and contribute to novel functions (fine-tuning of adaptive traits) or/and compensate for the side effects of the first-step pleiotropic mutation. Future directions to confirm this general pattern may require time course studies of GE (including heat shock and acclimation response, as well as all the intermediate steps of adaptation) coupled with genomic data across a diverse set of environmental treatments.
Materials and Methods
Growth Conditions
Unless otherwise noted, the culture conditions used for the physiological assays (growth curves, transcription efficiency, and RNA-seq assays) were the same used during the high-temperature evolution experiment (Tenaillon et al. 2012). Briefly, strains were revived in Luria-Bertani (LB) broth and incubated at 37 °C with constant shaking (120 rpm). Overnight cultures were diluted 104-fold into 10 ml Davis minimal medium supplemented with 25 µg/ml glucose (DM25) and incubated for 1 day at 37 °C to allow the strains to acclimate to the culture conditions. The following day, we transferred 100 µl of the overnight culture in 9.9 ml of fresh DM25 (100-fold dilution) and we incubated them at 42 °C for 1 day to allow the strains to acclimate to high temperature.
Growth Curves
Acclimated strains were grown at the assay temperature (either 37 °C or 42 °C) and the densities were measured approximately every hour during the lag phase and approximately every 30 minutes during the exponential phase. Population densities were measured using an electronic particle counter (Coulter Counter model Multisizer 3 equipped with a 30-µm diameter aperture tube). To measure density, 50 µl of the culture was diluted in 9.9 mL Isoton II diluent (Beckman Coulter), and 50 µl of the resulting dilution was counted electronically. To estimate the maximum growth rate, µmax, we fitted a linear regression to the natural logarithm of the cell density over time during the exponential phase (i.e., linear part) using the lm function in R version 3.0.2 (R Development Core Team 2013; supplementary fig. S8, Supplementary Material online). Three estimates of µmax were obtained for each genotype. The duration of the effective lag phase, τ, was defined as the time elapsed before the maximal growth rate was reached. Because this definition measure may include some of the initial period of exponential growth, we also explored an alternative definition to estimate the end of the lag phase as the intersection point between the initial cell density and the linear regression fitted to the data measured in exponential phase. The final yield was estimated from the cell counts at the end of the exponential phase.
RNA Extraction and Preparation for Sequencing
To investigate the GE profile prior to thermal stress, we grew the ancestor (REL1206), previously acclimated to the growth conditions, at 37 °C until it reached midexponential phase (three replicates). To investigate the GE profile during acclimation, we grew the ancestor at 42°, previously acclimated to 42 °C (two replicates), until midexponential phase. The remaining strains (single first-step mutants and high-temperature adapted clones) were grown at 42 °C until midexponential phase, with two replicates for each.
Briefly, bacterial cultures were grown in DM25 medium until they reached midexponential phase, which we determined by electronic counts. Eighty milliliter of the culture was filtered through a 0.2 µm cellulose membrane (Life Science, Germany). Cells, concentrated in the filter, were stabilized with Qiagen RNA-protect Bacteria Reagent and pellet for storage at −80 °C prior to RNA extraction. Total RNA was extracted using RNeasy Mini Kit (Qiagen). Total RNA was DNase treated using Turbo DNA-free kit (Ambion) and ribosomal RNA (rRNA) was depleted using the Ribo-Zero rRNA Removal kit for Gram-Negative Bacteria (Epicentre Biotechnologies, Medion, WI). cDNA library was constructed using TruSeq RNA v2 kit (Illumina, San Diego, CA). Libraries were multiplexed 8-fold and sequenced on an Illumina Hiseq 2000 platform. A total of 100-bp single-end reads were generated.
mRNAseq Data Analyses
Reads were mapped to the E. coli B REL606 genome reference (CP000819.1) using bwa 0.6.2 (Li and Durbin 2009), using default parameters (http://bio-bwa.sourceforge.net/bwa.shtml). Only unique, perfectly matching reads to the 4,204 annotated coding regions were retained for further analyses (supplementary table S9, Supplementary Material online). Differential expression analysis was performed using the DESeq R package (Anders and Huber 2010). We used the P values adjusted by the Benjamini and Hochberg approach (q values), which controls for false discovery rate. Genes with q less than 0.001 were considered significantly differentially expressed. For the GO term enrichment analyses, we used the Enrichment analysis tool from http://geneontology.org/page/go-enrichment-analysis.
DEGs were classified in one of the four categories (restored, reinforced, unrestored, and novel) based on the contrasts for which they were significant and the direction and value of their fold change (supplementary table S4, Supplementary Material online). We also examined the overall trends in GE shifts by plotting two ratios that share an underlying parameter (fig. 4 and supplementary fig. S6, Supplementary Material online), which creates an inherent negative bias for correlation values. We therefore tested the strength of correlation by randomization; we chose the three independent parameters at random from the data set, constructed ratios for the observed number of genes, estimated a correlation coefficient, and repeated the process 1,000 times to achieve a distribution of correlation values that reflect the inherent bias.
Construction of the Fluorescently Labeled Strains for the Transcription Efficiency Assay
We generated an ∼4-kb long linear DNA fragment carrying the CAT, tetR, and YFP genes from an E. coli strain carrying the CAT:tetR:YFP genomic cassette (Fehér et al. 2012) kindly provided by Csaba Pál (Biological Research Center, Szeged, Hungary). We amplified the CAT:tetR:YFP cassette by PCR using the primers ARV19 and ARV20 (supplementary table S10, Supplementary Material online) and Pfu DNA polymerase (Promega). The purified PCR product was integrated into the ancestral strain REL1206 carrying the pKD46 recombineering plasmid as previously described by Datsenko and Wanner (2000). Briefly, the ancestral strain carrying the pKD46 plasmid was grown overnight at 30 °C in 5 ml of LB with 100 µg/ml of ampicillin. The overnight culture was 100-fold diluted in 100 ml of LB with ampicillin and 1 mM l-arabinose (Sigma) and grown at 30 °C to an optical density (OD600) of 0.6. Electrocompetent cells were made by washing the culture five times with ice-cold water. Approximately 200 ng of linear DNA was electroporated into 25 µl of cells. After electroporation, 1 ml of LB was added, and the cells were incubated at 30 °C for 2 h with shaking, then plated 100 µl on LB agar plates with chloramphenicol (20 µg/ml). We selected a single colony and purified it in LB agar plate containing chloramphenicol. Correct integration was verified by PCR using primers ARV34 and ARV35 (supplementary table S10, Supplementary Material online).
Transcription Efficiency Assay
To measure the transcription efficiency of the ancestor and the mutants, we performed a quantitative reverse transcription polymerase chain reaction (RT-PCR) assay (Reynolds 2000; Brandis et al. 2012). In brief, we grew the fluorescently labeled strains in the same culture conditions previously described except that we supplemented the DM medium with 100 µg/ml glucose (DM100). We confirmed the advantage of these mutations to high temperature despite the higher amount of glucose based on growth curves of the ancestor and the mutants at 42 °C in DM100. Acclimated cultures were grown at 42.2 °C until they reached midexponential phase. We took 1 ml of uninduced cells (sample T0) and stabilized them in RNAprotect Bacteria Reagent (Qiagen). Immediately after, we induced cells by adding 10 µl of anhydotetracycline (66 µg/ml) to the medium. Samples of 1 ml of culture were taken at 1, 2, 3, and 4 min after induction (samples T1, T2, T3, and T4) and stabilized with RNA protect Bacteria Reagent and pelleted for storage at −80 °C prior to RNA extraction. Total RNA was prepared using RNeasy Mini Kit (Qiagen). RNA was DNase treated using Turbo DNA-free kit (Ambion). We used 300 ng of DNA-free RNA to produce cDNA with the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). From each reverse transcribed product, we quantified the abundance of cDNA of the gene reference gst (Gst, glutathione transferase; Pfaffl 2001) and the target gene YFP (Yellow fluorescent protein) using Fast SYBR Green Master Mix (Applied Biosystems) quantified on a Stratagene MX3005P QPCR System (Agilent Technologies). For each PCR reaction, we used 0.625 µM forward primer (ARV48 for gst or ARV50 for YFP; supplementary table S10, Supplementary Material online), 0.625 µM reverse primer (ARV49 for gst or ARV51 for YFP; supplementary table S10, Supplementary Material online), 3 µl cDNA template, 4.5 µl RNase-free water, and 10 µl of Fast SYBR Green Master Mix to have a final reaction volume of 20 µl. To control for the intra-assay variation (repeatability), we prepared three replicates of each reaction. The PCR thermal cycling conditions were 95 °C for 20 s followed by 40 cycles of 95 °C for 3 s and 60 °C for 30 s.
The efficiency of the amplifications for each pair of primers was determined from a standard curve using the formula E = 10[−1/s], where s is the slope of the standard curve (Pfaffl 2001). To calculate the “relative expression ratio (i.e., the relative change in GE of the target gene YFP normalized to the reference gene gst and relative to the uninduced control sample T0), we used the mathematical model for relative quantification in real-time RT-PCR developed by Pfaffl (Pfaffl 2001; Brandis et al. 2012). Transcription efficiency was calculated as the slope of the fitted linear regression between the relative expression ratio against time, based on three replicates.
Supplementary Material
Supplementary data set S1 and S2, figures S1 and S2, and tables S1–S10 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank P. McDonald and R. Gaut for technical assistance. This work was supported by the National Science Foundation grant DEB-0748903 to B.G. O.T. was supported by European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013/ERC Grant 310944). A.R.V. was supported by the University of California Institute for Mexico and the United States-Consejo Nacional de Ciencia y Technología (Mexico) Fellowship and by the Chateaubriand Fellowship in Science, Technology Engineering, and Mathematics (STEM).
References
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol. 11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applebee MK, Herrgard MJ, Palsson BO. 2008. Impact of individual mutations on increased fitness in adaptively evolved strains of Escherichia coli. J Bacteriol. 190:5087–5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabasi AL, Oltvai ZN. 2004. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 5:101–113. [DOI] [PubMed] [Google Scholar]
- Battesti A, Majdalani N, Gottesman S. 2011. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol. 65:189–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount ZD, Barrick JE, Davidson CJ, Lenski RE. 2012. Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature 489:513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandis G, Wrande M, Liljas L, Hughes D. 2012. Fitness-compensatory mutations in rifampicin-resistant RNA polymerase. Mol Microbiol. 85:142–151. [DOI] [PubMed] [Google Scholar]
- Carroll SM, Chubiz LM, Agashe D, Marx CJ. 2015. Parallel and divergent evolutionary solutions for the optimization of an engineered central metabolism in Methylobacterium extorquens AM1. Microorganisms 3:152–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SM, Marx CJ. 2013. Evolution after introduction of a novel metabolic pathway consistently leads to restoration of wild-type physiology. PLoS Genet. 9:e1003427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterji D, Ojha AK. 2001. Revisiting the stringent response, ppGpp and starvation signaling. Curr Opin Microbiol. 4:160–165. [DOI] [PubMed] [Google Scholar]
- Chou HH, Chiu HC, Delaney NF, Segre D, Marx CJ. 2011. Diminishing returns epistasis among beneficial mutations decelerates adaptation. Science 332:1190–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin PA, Weinreich DM, Besnard G. 2010. Causes and evolutionary significance of genetic convergence. Trends Genet. 26:400–405. [DOI] [PubMed] [Google Scholar]
- Conrad TM, Frazier M, Joyce AR, Cho BK, Knight EM, Lewis NE, Landick R, Palsson BO. 2010. RNA polymerase mutants found through adaptive evolution reprogram Escherichia coli for optimal growth in minimal media. Proc Natl Acad Sci U S A. 107:20500–20505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TF, Rozen DE, Lenski RE. 2003. Parallel changes in gene expression after 20,000 generations of evolution in Escherichia coli. Proc Natl Acad Sci U S A. 100:1072–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper VS, Schneider D, Blot M, Lenski RE. 2001. Mechanisms causing rapid and parallel losses of ribose catabolism in evolving populations of Escherichia coli B. J Bacteriol. 183:2834–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosa JH, Walsh CT. 2002. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol Mol Biol Rev. 66:223–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 97:6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ederth J, Mooney RA, Isaksson LA, Landick R. 2006. Functional interplay between the jaw domain of bacterial RNA polymerase and allele-specific residues in the product RNA-binding pocket. J Mol Biol. 356:1163–1179. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. 1998. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 95:14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehér T, Bogos B, Méhi O, Fekete G, Csörgő B, Kovács K, Pósfai G, Papp B, Hurst LD, Pál C. 2012. Competition between transposable elements and mutator genes in bacteria. Mol Biol Evol. 29:3153–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. 1930. The genetical theory of natural selection. Oxford: Oxford University Press. [Google Scholar]
- Fong SS, Joyce AR, Palsson BØ. 2005. Parallel adaptive evolution cultures of Escherichia coli lead to convergent growth phenotypes with different gene expression states. Genome Res. 15:1365–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi H, Hottes AK, Tavazoie S. 2009. Global discovery of adaptive mutations. Nat Methods. 6:581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekera TS, Csonka LN, Paliy O. 2008. Genome-wide transcriptional responses of Escherichia coli K-12 to continuous osmotic and heat stresses. J Bacteriol. 190:3712–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herring CD, Raghunathan A, Honisch C, Patel T, Applebee MK, Joyce AR, Albert TJ, Blattner FR, Van den Boom D, Cantor CR. 2006. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat Genet. 38:1406–1412. [DOI] [PubMed] [Google Scholar]
- Hindre T, Knibbe C, Beslon G, Schneider D. 2012. New insights into bacterial adaptation through in vivo and in silico experimental evolution. Nat Rev Microbiol. 10:352–365. [DOI] [PubMed] [Google Scholar]
- Hug SM, Gaut BS. 2015. The phenotypic signature of adaptation to thermal stress in Escherichia coli. BMC Evol Biol. 15:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama A. 2000. Functional modulation of Escherichia coli RNA polymerase. Annu Rev Microbiol. 54:499–518. [DOI] [PubMed] [Google Scholar]
- Jin DJ, Burgess RR, Richardson JP, Gross CA. 1992. Termination efficiency at rho-dependent terminators depends on kinetic coupling between RNA polymerase and rho. Proc Natl Acad Sci U S A. 89:1453–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin DJ, Cagliero C, Zhou YN. 2012. Growth rate regulation in Escherichia coli. FEMS Microbiol Rev. 36:269–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin DJ, Walter WA, Gross CA. 1988. Characterization of the termination phenotype of rifampicin-resistant mutants. J Mol Biol. 202:245–253. [DOI] [PubMed] [Google Scholar]
- Jozefczuk S, Klie S, Catchpole G, Szymanski J, Cuadros-Inostroza A, Steinhauser D, Selbig J, Willmitzer L. 2010. Metabolomic and transcriptomic stress response of Escherichia coli. Mol Syst Biol. 6:364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keseler IM, Mackie A, Peralta-Gil M, Santos-Zavaleta A, Gama-Castro S, Bonavides-Martínez C, Fulcher C, Huerta AM, Kothari A, Krummenacker M. 2013. EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res. 41:D605–D612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AI, Dinh DM, Schneider D, Lenski RE, Cooper TF. 2011. Negative epistasis between beneficial mutations in an evolving bacterial population. Science 332:1193–1196. [DOI] [PubMed] [Google Scholar]
- Kishimoto T, Iijima L, Tatsumi M, Ono N, Oyake A, Hashimoto T, Matsuo M, Okubo M, Suzuki S, Mori K. 2010. Transition from positive to neutral in mutation fixation along with continuing rising fitness in thermal adaptive evolution. PLoS Genet. 6:e1001164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryazhimskiy S, Rice DP, Jerison ER, Desai MM. 2014. Global epistasis makes adaptation predictable despite sequence-level stochasticity. Science 344:1519–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis NE, Hixson KK, Conrad TM, Lerman JA, Charusanti P, Polpitiya AD, Adkins JN, Schramm G, Purvine SO, Lopez-Ferrer D. 2010. Omic data from evolved E. coli are consistent with computed optimal growth from genome-scale models. Mol Syst Biol. 6:390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F, Su CC, Zimmermann MT, Boyken SE, Rajashankar KR, Jernigan RL, Edward WY. 2010. Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport. Nature 467:484–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Maury L, Marguerat S, Bähler J. 2008. Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nat Rev Genet. 9:583–593. [DOI] [PubMed] [Google Scholar]
- Mejia YX, Mao HB, Forde NR, Bustamante C. 2008. Thermal probing of E-coli RNA polymerase off-pathway mechanisms. J Mol Biol. 382:628–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka G, Blankschien M, Herman C, Gross CA, Rhodius VA. 2006. Regulon and promoter analysis of the E. coli heat-shock factor, σ32, reveals a multifaceted cellular response to heat stress. Genes Dev. 20:1776–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opalka N, Brown J, Lane WJ, Twist K-AF, Landick R, Asturias FJ, Darst SA. 2010. Complete structural model of Escherichia coli RNA polymerase from a hybrid approach. PLoS Biol. 8:e1000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA. 2005. The genetic theory of adaptation: a brief history. Nat Rev Genet. 6:119–127. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe N, Crozat E, Lenski RE, Schneider D. 2007. Evolution of global regulatory networks during a long‐term experiment with Escherichia coli . BioEssays 29:846–860. [DOI] [PubMed] [Google Scholar]
- Polikanov YS, Blaha GM, Steitz TA. 2012. How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science 336:915–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt EM, Deatherage DE, Ellington AD, Georgiou G, Barrick JE. 2014. Recursive genomewide recombination and sequencing reveals a key refinement step in the evolution of a metabolic innovation in Escherichia coli. Proc Natl Acad Sci U S A. 111:2217–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. 2013. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing. [Google Scholar]
- Reynolds MG. 2000. Compensatory evolution in rifampin-resistant Escherichia coli. Genetics 156:1471–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter K, Haslbeck M, Buchner J. 2010. The heat shock response: life on the verge of death. Mol Cell. 40:253–266. [DOI] [PubMed] [Google Scholar]
- Riehle MM, Bennett AF, Lenski RE, Long AD. 2003. Evolutionary changes in heat-inducible gene expression in lines of Escherichia coli adapted to high temperature. Physiol Genomics. 14:47–58. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Verdugo A, Carrillo-Cisneros D, González-González A, Gaut BS, Bennett AF. 2014. Different tradeoffs result from alternate genetic adaptations to a common environment. Proc Natl Acad Sci U S A. 111:12121–12126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Verdugo A, Gaut BS, Tenaillon O. 2013. Evolution of Escherichia coli rifampicin resistance in an antibiotic-free environment during thermal stress. BMC Evol Biol. 13:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals J, Little R, Bremer H. 1982. Temperature dependence of RNA synthesis in Escherichia coli. J Bacteriol. 151:879–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg TE, Pedersen M, LaCroix RA, Ebrahim A, Bonde M, Herrgard MJ, Palsson BO, Sommer M, Feist AM. 2014. Evolution of Escherichia coli to 42° C and subsequent genetic engineering reveals adaptive mechanisms and novel mutations. Mol Biol Evol. 31:2647–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutourina OA, Bertin PN. 2003. Regulation cascade of flagellar expression in Gram‐negative bacteria. FEMS Microbiol Rev. 27:505–523. [DOI] [PubMed] [Google Scholar]
- Szamecz B, Boross G, Kalapis D, Kovács K, Fekete G, Farkas Z, Lázár V, Hrtyan M, Kemmeren P, Koerkamp MJAG, et al. 2014. The genomic landscape of compensatory evolution. PLoS Biol. 12:e1001935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon O, Rodriguez-Verdugo A, Gaut RL, McDonald P, Bennett AF, Long AD, Gaut BS. 2012. The molecular diversity of adaptive convergence. Science 335:457–461. [DOI] [PubMed] [Google Scholar]
- Vasi F, Travisano M, Lenski RE. 1994. Long-term experimental evolution in Escherichia coli. II. Changes in life-history traits during adaptation to a seasonal environment. Am Nat. 144:432–456. [Google Scholar]
- Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J Bacteriol. 187:1591–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AC. 1976. Gene regulation in evolution. In: Ayala FJ, editor. Molecular evolution. Sunderland (MA): Sinauer Associates; p. 225–234. [Google Scholar]
- Ye Y, Zhang L, Hao F, Zhang J, Wang Y, Tang H. 2012. Global metabolomic responses of Escherichia coli to heat stress. J Proteome Res. 11:2559–2566. [DOI] [PubMed] [Google Scholar]
- Zhao K, Liu M, Burgess RR. 2005. The global transcriptional response of Escherichia coli to induced σ32 protein involves σ32 regulon activation followed by inactivation and degradation of σ32 in vivo. J Biol Chem. 280:17758–17768. [DOI] [PubMed] [Google Scholar]
- Zhou YN, Lubkowska L, Hui M, Chen S, Strathern J, Jin DJ, Kashlev M. 2013. Isolation and characterization of RNA Polymerase rpoB mutations that alter transcription slippage during elongation in Escherichia coli. J Biol Chem. 288:2700–2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.