Abstract
The opportunistic pathogen Pseudomonas aeruginosa is capable of establishing severe and persistent infections in various eukaryotic hosts. It encodes a wide array of virulence factors and employs several strategies to evade immune detection. In the present study, we screened the Harvard Medical School transposon mutant library of P. aeruginosa PA14 for bacterial factors that modulate interleukin-8 responses in A549 human airway epithelial cells. We found that in addition to the previously identified alkaline protease AprA, the elastase LasB is capable of degrading exogenous flagellin under calcium-replete conditions and prevents flagellin-mediated immune recognition. Our results indicate that the production of two proteases with anti-flagellin activity provides a failsafe mechanism for P. aeruginosa to ensure the maintenance of protease-dependent immune-modulating functions.
INTRODUCTION
Pathogenic bacteria possess an extensive arsenal of virulence factors that allow them to survive in the host and cause disease. Among such virulence factors are secreted extracellular proteases. They are known to facilitate bacterial colonization by inducing damage to host tissue and actively subverting immune responses (1). Pseudomonas aeruginosa produces several extracellular proteases that function for this purpose. Two of these, the 50-kDa alkaline protease AprA and the 33-kDa elastase LasB, often are implicated in P. aeruginosa infections. They can be detected under in vitro culture conditions and in vivo at various sites of infection (2–5). Both AprA and LasB are zinc metalloproteases that require calcium ions for stability (6, 7). They have been demonstrated to possess proteolytic activity against several different types of substrates (8) and to combat host immune responses. For instance, LasB has been reported to degrade mucins and surfactant proteins which collectively function to promote bacterial clearance (9–11). Similarly, AprA also has been shown to impede bacterial clearance by degrading the C2 component of the complement system, thereby preventing complement-mediated phagocytosis (12). In addition to acting against responding host factors, the immune-modulating capabilities of AprA also include the ability to degrade flagellin, a known activator of proinflammatory responses via toll-like receptor 5 (TLR5) recognition (13, 14).
In the context of host-pathogen interactions, the importance of flagella, their constituents, and the motility they confer have been well established (15–19). Flagellin in particular has become a major focus of study due to its ability to elicit robust responses from both the innate and adaptive arms of the defense systems in eukaryotes (20–22). It is interesting that, despite this, flagellin remains conserved across several bacterial species. This indicates a functional significance that outweighs its immunogenic consequences. Interestingly, as a countermeasure, many bacteria employ various strategies to compromise or even prevent flagellin-mediated recognition. For instance, Campylobacter jejuni, Helicobacter pylori, and Bartonella bacilliformis have been demonstrated to be capable of producing altered flagellin that cannot be recognized by TLR5 (23). Modification of flagellin by glycosylation also has been observed in bacteria, although its implications in the context of host immune recognition are less clear (24, 25). In P. aeruginosa, in addition to the above-mentioned flagellin degradation by AprA, a more widely known mechanism involves the repression of flagellar biosynthesis in response to environmental cues as part of a global response observed in chronic infections (26–28).
Both flagellin and extracellular proteases are considered indicators of virulence, which in P. aeruginosa is largely regulated by quorum sensing (QS). In QS, cells communicate in a cell population-dependent manner via the production and detection of diffusible signal molecules, ultimately prompting necessary changes in the execution of various cell functions (29). QS also is implicated in the formation of biofilms which are frequently associated with persistent Pseudomonas infections. Three major QS circuits exist in P. aeruginosa, the Las, Rhl, and PQS systems. The Las system governs the production of the extracellular proteases AprA and LasB.
In this study, we investigated the contributions of P. aeruginosa proteases in immune evasion and demonstrate that in addition to the previously identified alkaline protease AprA, the elastase LasB is capable of degrading exogenous flagellin under calcium-replete conditions and therefore ensures effective immune evasion. By degrading free flagellin as the ligand for immune receptors, the pathogen retains its motility phenotype but escapes flagellin-dependent immune recognition.
MATERIALS AND METHODS
Bacterial strains and plasmids.
All bacterial strains, plasmids, and primers used in this study are listed in Table 1. Bacteria were grown in broth or plate cultures in either Luria-Bertani (LB) or brain heart infusion (BHI) medium at 37°C under aerobic conditions. DNA manipulations were carried out in accordance with standard molecular biology procedures or by following the kit manufacturer's protocols whenever applicable. In-frame deletion mutants were constructed by following protocols previously described by Shi et al. (30). Using overlap extension PCR (31), upstream and downstream regions of the gene of interest first were separately replicated and, following a second round of PCR, allowed to anneal to generate fragments in which only the first and last 30 bp of the target gene are retained. The resulting fragment was ligated into the pEX18Ap vector and transformed into Escherichia coli WM3064, which then was used to chromosomally integrate the mutagenesis construct into P. aeruginosa via conjugation. Transconjugants carrying the construct were isolated by the use of carbenicillin as a selecting agent, and resulting clones were plated simultaneously onto LB supplemented with either 400 μg/ml carbenicillin or 10% sucrose. Clones demonstrating a carbenicillin-resistant sucrose-susceptible phenotype then were grown in LB liquid medium for approximately 20 passages and replated onto LB medium with 10% sucrose. Mutants were selected by picking clones displaying a sucrose-resistant carbenicillin-susceptible phenotype and were verified via PCR and sequencing.
TABLE 1.
Bacterial strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Relevant feature(s) or sequencea | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | Strain used for standard cloning experiments | 55 |
| WM3064 | Donor strain for conjugation | 56 |
| P. aeruginosa | ||
| PA14 WT | WT reference strain | 39 |
| PA14 aprA::Tn | aprA transposon mutant from the PA14 transposon mutant library, Gmr | 39 |
| PA14 ΔaprA | aprA mutant of PA14 WT | This study |
| PA14 ΔlasB | lasB mutant of PA14 WT | This study |
| PA14 ΔlasR | lasR mutant of PA14 WT | S. Brouwer, unpublished |
| PA14 ΔaprA ΔlasB | aprA and lasB double mutant of PA14 WT | This study |
| P. putida | ||
| KT2440 WT | WT reference strain | 57 |
| KT2440 fleQ::Tn | fleQ transposon mutant of KT2440 WT, Kmr | 58 |
| Plasmid | ||
| pEX18Ap | oriT+ sacB+, gene replacement vector with MCS from pUC18, Apr | 59 |
| Primers | ||
| AprAUF | CCCAAGCTTCGAACTTCGTTTCCGGCGAG | |
| AprAUR | CGCCTGGCCACCTTTCAATGCAAGAGAATTGC | |
| AprADF | TTGAAAGGTACCCAGGCCGACATCGTC | |
| AprADR | GCTCTAGACGTGAGGCAGGCGGTATCG | |
| LasBUF | CCCAAGCTTGAAGCTGGTGCTGAAAAGCG | |
| LasBUR | CACGCCGACCAACAGGTCAAGCGTAGAAACC | |
| LasBDF | GACCTGTTGGTCGGCGTGACCTGCC | |
| LasBDR | GCTCTAGATGGACCCCGGCGACC | |
| AprA_F | GTGTCAGTTTGGACTTCGGCC | |
| AprA_R | CTTCGCTCCTGTGGAAGACAC | |
| LasR_F | AACGTGCCGGATATCGGGTG | |
| LasR_R | CGAGAACCTGCCCTTCCCTA |
Apr, ampicillin resistance; Cbr, carbenicillin resistance; Gmr, gentamicin resistance; Kmr, kanamycin resistance; Tcr, tetracycline resistance; MCS, multiple cloning site. Primer sequences are oriented 5′ to 3′.
Collection of cell-free supernatant samples.
Cell-free supernatant samples were collected from bacterial cultures grown for 18 h in either LB or BHI, with or without the addition of 1 mM CaCl2. Cells were pelleted via centrifugation at 4,000 rpm for 20 min, after which supernatants were passed through 0.2-μm membrane filters to exclude any remaining bacteria. Supernatant aliquots were stored at −20°C until use, and proteolytic activity was assessed semiqualitatively via skim milk agar assays.
Eukaryotic cell cultures.
For all in vitro infection assays, we used A549-Gluc cells that previously were generated from the A549 human alveolar adenocarcinoma cell line via lentiviral transduction to possess a Gaussia luciferase viability reporter (32, 33). Cells were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum, 100 U/ml penicillin streptomycin (Life Technologies), 1× MEM nonessential amino acids (Life Technologies), and 2 mM l-glutamine (here referred to as DC medium). DC medium additionally was supplemented with 10 μg/ml blasticidin for the maintenance of A549-Gluc cells (DC-Bla). Cell cultures were grown and maintained at 37°C in the presence of 5% CO2. Host cell viability was monitored during live bacterial infections and flagellin incubation assays and were performed directly on collected A549-Gluc cell supernatants as previously described (34, 35). Percent viability was calculated as the relative light unit (RLU) ratio between infected and uninfected controls. Samples with viability readouts falling below the range set by the PA14 wild-type (WT) controls for each assay were excluded from data analysis.
Flagellin isolation and Western blot analysis.
Monomeric flagellin was isolated from PA14 WT by following a protocol modified from Bardoel et al. (13). PA14 WT was grown in LB for 6 h, and cells were collected by centrifugation at 4,000 rpm for 30 min. Cell pellets were resuspended in PBS and sheared by vortexing with 5-mm glass beads for 5 min. The sheared cell suspension was centrifuged at 4,000 rpm for 20 min, filtered to eliminate cells and debris, and then centrifuged at 25,000 rpm for 1 h. The resulting pellet containing flagella was resuspended in PBS and heated at 70°C for 20 min to obtain monomeric flagellin. The flagellin concentration was determined by Bradford assay, and the purity of flagellin preparations was assessed by SDS-PAGE. The specific detection of flagellin was achieved by immunoblotting using a polyclonal rabbit antibody against P. aeruginosa type b flagellin (kindly provided by L. Wiehlmann, Hannover Medical School). Following SDS-PAGE, proteins were transferred to an Immobilon-P membrane (Merck Millipore), blocked for 1 h with 10% skim milk in Tris-buffered saline with 0.05% Tween 20 (TBST), incubated with anti-FliC antibody for 1 h, washed, and subsequently incubated with horseradish peroxidase (HRP)-linked goat anti-rabbit IgG for 1 h (Cell Signaling Technology). Bands were visualized using the ECL Western blotting detection system (GE Healthcare Life Sciences).
Bacterial infection assays.
Bacterial inocula were prepared from liquid cultures that had been grown in LB or BHI for 6 h. Cell suspensions were prepared in DC medium without penicillin streptomycin (DC-P) and adjusted to a multiplicity of infection (MOI) of 10. A549-Gluc cells were grown in 96-well plates to 80 to 100% confluence. Cells were washed once with PBS prior to the addition of bacterial inocula. Plates were centrifuged for 5 min at 500 × g to facilitate contact between bacteria and eukaryotic cells and then incubated for 3 h at 37°C in the presence of 5% CO2. Cell supernatant samples were collected following plate centrifugation for 20 min at 4,000 rpm to pellet bacteria and cell debris and stored at −80°C until use for Gaussia luciferase assays and interleukin-8 (IL-8) enzyme-linked immunosorbent assay (ELISA) measurements.
Exposure of A549-Gluc cells to flagellin.
Flagellin digestion was carried out by coincubating 1 ng/μl flagellin with either 1 ng/μl purified P. aeruginosa elastase (Merck Millipore) or with cell-free supernatants obtained from cultures grown for 18 h in LB or BHI and then diluted to 10 ng/μl total protein concentration in a 250-μl reaction mixture for 1 h at 37°C. Polymyxin B was added to this reaction mixture at a concentration of 10 ng/μl to inactivate bacterial lipopolysaccharide (LPS). Reaction mixtures then were used to treat A549-Gluc cells at a 1:10 dilution in DC-P medium to limit cytotoxicity due to cell starvation. Preparations of A549-Gluc and assay conditions were identical to those of bacterial infections, except plates were not centrifuged prior to the collection of cell culture supernatants at the end of each assay.
Detection of IL-8.
Cell supernatant samples collected from A549-Gluc cells were tested for IL-8 levels using the human CXCL8/IL-8 DuoSet kit (R&D Systems) by following the manufacturer's protocols.
Skim milk plate assays.
Live bacteria were assessed for proteolytic activity by growing cultures on LB or BHI agar plates supplemented with 1% skim milk. Casein hydrolysis, seen as zones of clearing surrounding bacteria, indicated positive proteolytic activity. Cell-free supernatant samples from LB or BHI cultures, to which 1 mM CaCl2 was added either prior to bacterial inoculation or after sample collection, were tested for proteolytic activity in a similar manner, but 1% agarose gels supplemented with 1% skim milk were used instead.
Sequential PAGE separation of proteins.
LB or BHI cell-free culture supernatants first were resolved by native PAGE using the Tris-tricine system as described by Haider et al. (36), with minor modifications: 100 mM Tris-tricine running buffer was adjusted to pH 8.8 instead of 8.25. Electrophoresis was carried out at 4°C for 2 h, after which gels were either stained with PageBlue protein staining solution (Thermo Scientific) to visualize bands or overlain onto LB milk agar plates and incubated for 30 to 60 min at 37°C to detect proteolytically active bands. Upon visual confirmation of casein hydrolysis, native PAGE gel sections overlaying clearing zones were carefully excised, cut into pieces, placed into SDS sample buffer, and then heated at 95°C for 10 min to denature proteins. Samples then were resolved by SDS-PAGE using standard procedures.
Protein identification by MS.
Following SDS-PAGE, bands on the polyacrylamide gel containing proteins of interest were excised. Proteins were digested with trypsin and subjected to mass spectrometry (MS) analysis by following the protocols of Olling and colleagues (37), with some modifications. MS and tandem MS (MS-MS) spectra were acquired in positive ion mode with a scan range of m/z 300 to 1600 using a top10 method and collision-induced dissociation (CID) fragmentation. Acquired MS data were processed with the Proteome Discoverer 1.3 software package (Thermo Fisher Scientific) and searched against a strain-specific protein database (generated by combining both locally obtained data from the Institute for Molecular Bacteriology and P. aeruginosa PA14 entries available at the UniProt online database [38]) using the Mascot 2.1 search engine (Matrix Science).
RESULTS
Flagella are major elicitors of host reactions.
To identify bacterial factors that could be involved in preventing the immune recognition of P. aeruginosa, we screened the Harvard Medical School PA14 transposon mutant library (39) for mutants eliciting an altered immune reaction. Screening was performed in vitro using A549-Gluc cells and interluleukin-8 (IL-8) as the readout for immune stimulation. IL-8 production by human epithelial cells upon P. aeruginosa recognition is well characterized and readily observable (25, 40). We tested approximately 5,800 mutants. Mutants that exhibited an altered cytotoxicity were excluded by determining Gaussia luciferase activity in cell culture supernatants of exposed A549-Gluc cells. After multiple rounds of screening, we identified 113 mutants that elicited either a decreased or increased IL-8 response compared to that of the PA14 WT control strain (see Table S1 in the supplemental material). Not surprisingly, many of these candidates harbored transposon insertions within genes involved in flagellar biosynthesis.
In follow-up experiments, we compared the motility as well as the IL-8 responses induced by P. aeruginosa PA14 and the soil bacterium Pseudomonas putida KT2440 (Fig. 1). P. putida has rarely been reported to cause human infections, likely owing to its lack of an innate virulence potential (41, 42). Of note, P. aeruginosa expresses a single polar flagellum, while P. putida displays multiple polar flagella (43, 44). The comparison of the IL-8 induction between the two WT strains showed a remarkable difference. PA14 exposure resulted in a severely reduced response compared to that of KT2440. In contrast, the fleQ mutants of both strains induced similar low levels of IL-8 (Fig. 1B). FleQ is the primary transcriptional regulator responsible for initiating flagellar biosynthesis in both P. aeruginosa and P. putida (45). The drop in IL-8 observed for both flagellum loss mutants of the two pseudomonads lays further emphasis on the dominance of flagellum-mediated immune recognition of P. aeruginosa.
FIG 1.
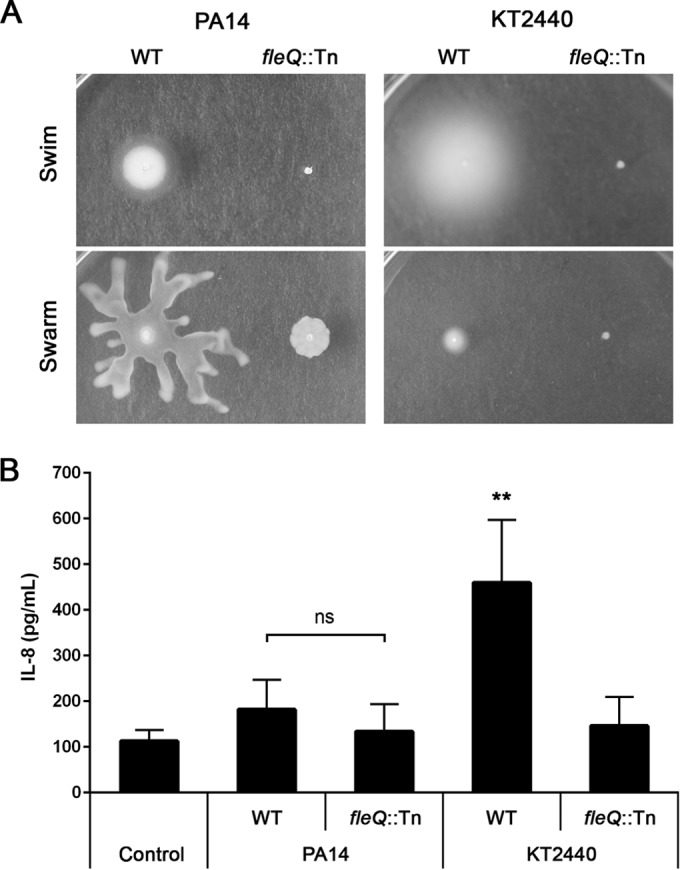
Bacterial motility and IL-8 production by A549-Gluc cells upon exposure to P. aeruginosa and P. putida. (A) Motility plate assays showing swimming and swarming phenotypes of P. aeruginosa PA14 and P. putida KT2440 wild types (WT) and respective fleQ mutants. (B) IL-8 response of A549-Gluc cells exposed to P. aeruginosa and P. putida WT strains and their respective fleQ mutants. The control represents A549-Gluc cells treated with PBS. Data are presented as means from five experiments ± standard deviations (SD). Significance was evaluated by Mann-Whitney U test. **, P < 0.01 relative to the PA14 WT control; ns, not significant.
P. aeruginosa produces more than one protease with anti-flagellin activity.
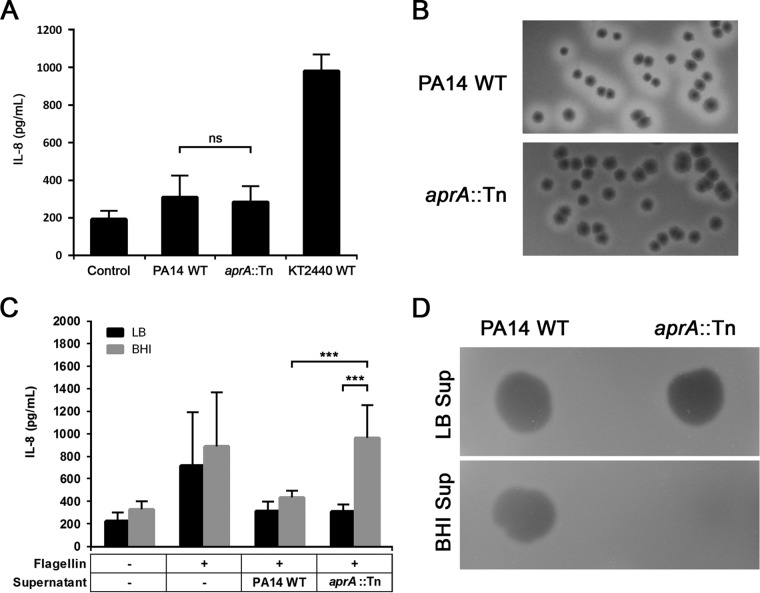
In a previous study, Bardoel and colleagues proposed a mechanism to explain how P. aeruginosa evades host immune recognition by using the secreted alkaline protease AprA to degrade exogenous flagellin (13). However, in our screen the PA14 transposon mutant of aprA did not elicit the expected high IL-8 response. Instead, we detected a response similar to that of the PA14 WT strain (Fig. 2A). Qualitative assessment of proteolytic activity in LB agar supplemented with 1% skim milk revealed visibly decreased clearing zones surrounding the aprA mutant compared to those of PA14 WT. Still, a residual proteolytic activity was detectable for the aprA mutant (Fig. 2B). This indicates that the production of other extracellular proteases in the mutant remains unaffected.
FIG 2.
Characterization of proteolytic activities of PA14 WT and aprA::Tn. (A) IL-8 response of A549-Gluc cells at an MOI of 10. The control represents A549-Gluc cells treated with PBS. (B) Qualitative analysis of protease activity using milk plate assays (LB agar supplemented with 1% skim milk). Zones of clearing surrounding colony growth indicate positive casein (a primary milk protein) hydrolysis. (C) Coincubation of A549-Gluc cells with PA14 WT flagellin pretreated with cell-free supernatant (Sup) samples obtained from cultures grown for 18 h in either LB or BHI. Data are presented as means from at least six experiments ± SD. Significance was evaluated by Wilcoxon test. ***, P < 0.001; ns, not significant. (D) Qualitative analysis of protease activity of cell-free supernatant samples obtained from cultures grown for 18 h in either LB or BHI, using skim milk assays (1% agarose gel supplemented with 1% skim milk).
For confirmation, flagellin purified from PA14 WT was pretreated with cell-free supernatant of the aprA mutant that had been cultured in LB for 18 h. When A549-Gluc cells were incubated with this preparation, we observed a low IL-8 response similar to that of flagellin pretreated with PA14 WT supernatant (Fig. 2C). These results strongly hint that other protease(s) with antiflagellin activity are produced by the aprA mutant.
We additionally investigated the effect of a different culture medium (BHI medium) on extracellular protease activity. Interestingly, after being grown in liquid BHI medium for 18 h, proteolytic activity could no longer be detected in the cell-free culture supernatant of the aprA mutant. In contrast, PA14 WT supernatant retained its protease activity (Fig. 2D). In accordance, antiflagellin activity also was lost in the aprA mutant BHI supernatant (Fig. 2C). Taken together, these results indicate that P. aeruginosa can produce at least one alternative protease that exerts immune modulation by means of an antiflagellin activity. However, this activity cannot be detected in BHI-grown cultures.
Production of proteases with antiflagellin activity in quorum-sensing-deficient P. aeruginosa strains.
It is well established that virulence factors such as extracellular proteases are produced by P. aeruginosa during later stages of growth and that their production is largely controlled by quorum sensing (29, 46). Therefore, we set out to investigate the possible quorum-sensing regulatory pathways involved in the production of the alternative protease with antiflagellin activity. To achieve this, we tested the PA14 ΔlasR, rhlR::Tn, and pqsA::Tn quorum-sensing mutants, which have nonfunctional Las, Rhl, and Pqs systems, respectively. All three mutants exhibited proteolytic activity on skim milk agar plates (Fig. 3A). However, unlike the BHI culture supernatants of the rhlR and pqsA mutants, supernatant of the lasR mutant did not show any detectable proteolytic activity (Fig. 3B). We then treated PA14 flagellin with cell-free supernatants of 18-h-old LB and BHI cultures of the three QS mutants and tested for IL-8 response. As previously observed with the aprA mutant, flagellin pretreated with the BHI supernatant of the lasR mutant induced a high IL-8 response in A549-Gluc cells, whereas the others did not (Fig. 3C). These results suggest a strong influence of the Las system on the production of the antiflagellin protease AprA. The effect appears to be much less on the alternative protease, whose activity remains evident under LB growth conditions.
FIG 3.
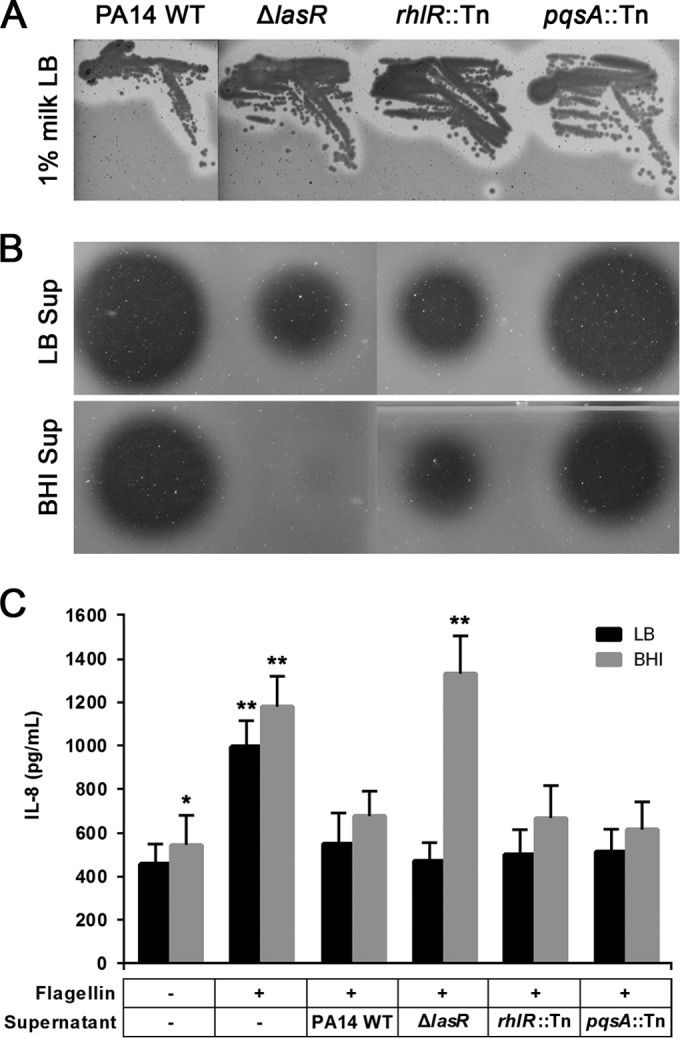
Characterization of proteolytic activities of quorum-sensing mutants. (A) Qualitative analysis of protease activity using milk plate assays; (B) Qualitative analysis of protease activity of cell-free supernatant samples, after growth in LB or BHI for 18 h, using skim milk. (C) Coincubation of A549-Gluc cells with PA14 WT flagellin and flagellin pretreated with cell-free supernatant samples obtained from cultures grown in LB or BHI for 18 h. Data are presented as means from eight experiments ± SD. Significance was evaluated by Wilcoxon test. P < 0.05 (*) and P < 0.01 (**) relative to results for the corresponding PA14 WT controls.
LasB has antiflagellin activity that can prevent flagellin-mediated immune recognition in A549-Gluc cells.
To identify the alternative protease with antiflagellin activity that is active under LB culture conditions, we used native PAGE and SDS-PAGE to separate the proteins of cell-free supernatants of PA14 WT and aprA::Tn grown in LB and BHI for 18 h. Postrun native PAGE gels were overlaid onto 1% skim milk LB agar to detect bands with proteolytic activity that were present in the PA14 WT under both culture conditions but only in the LB culture for the aprA::Tn mutant. Indeed, a predicted proteolytic pattern could be detected (see Fig. S1 in the supplemental material). We then excised native polyacrylamide gel pieces and confirmed their proteolytic activity by coincubation with purified flagellin (data not shown). The excised native PAGE gel samples then were subjected to protein separation by SDS-PAGE. Comparison of protein bands between total supernatant and excised native PAGE samples revealed a 33-kDa band in both LB- and BHI-grown PA14 WT samples as well as in the LB-grown aprA::Tn sample. This band was absent from the BHI-grown aprA::Tn sample (see Fig. S2). Since this protein could account for the antiflagellin activity, we excised and analyzed the 33-kDa band using mass spectrometry (MS). Peptide identification using MS-MS spectra revealed the protein to be the extracellular elastase LasB.
To validate this result, we coincubated commercial P. aeruginosa elastase with flagellin and monitored the degradation of flagellin via immunoblotting. Coincubation of flagellin with elastase resulted in flagellin degradation (Fig. 4A). Similarly, pretreatment of flagellin with commercial elastase efficiently abolished flagellin-mediated immune recognition, resulting in low levels of induced IL-8 in A549-Gluc cells (Fig. 4B).
FIG 4.
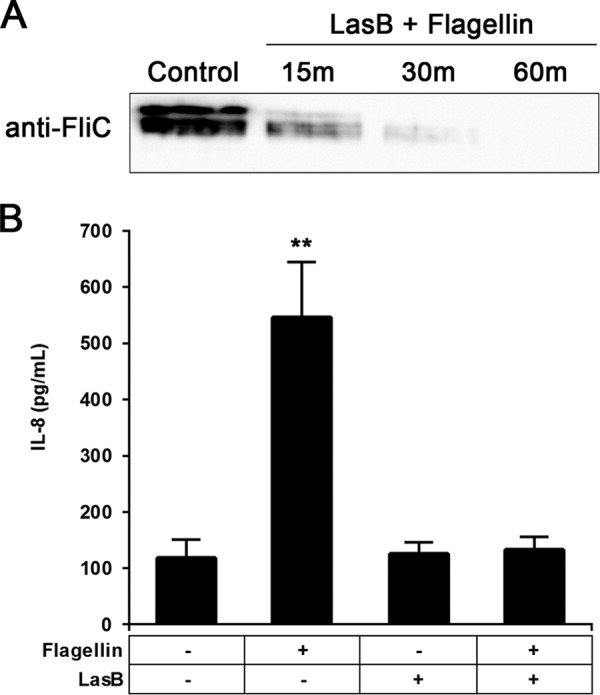
Antiflagellin activity of commercially available LasB. Flagellin degradation by LasB was directly monitored over time (15, 30, and 60 min of coincubation) by the use of an antiflagellin antibody visualized by Western blotting (A) and indirectly by measuring the IL-8 response of A549-Gluc cells pretreated with flagellin (B). Data are presented as means from six experiments ± SD. Significance was evaluated by Wilcoxon test. **, P < 0.01.
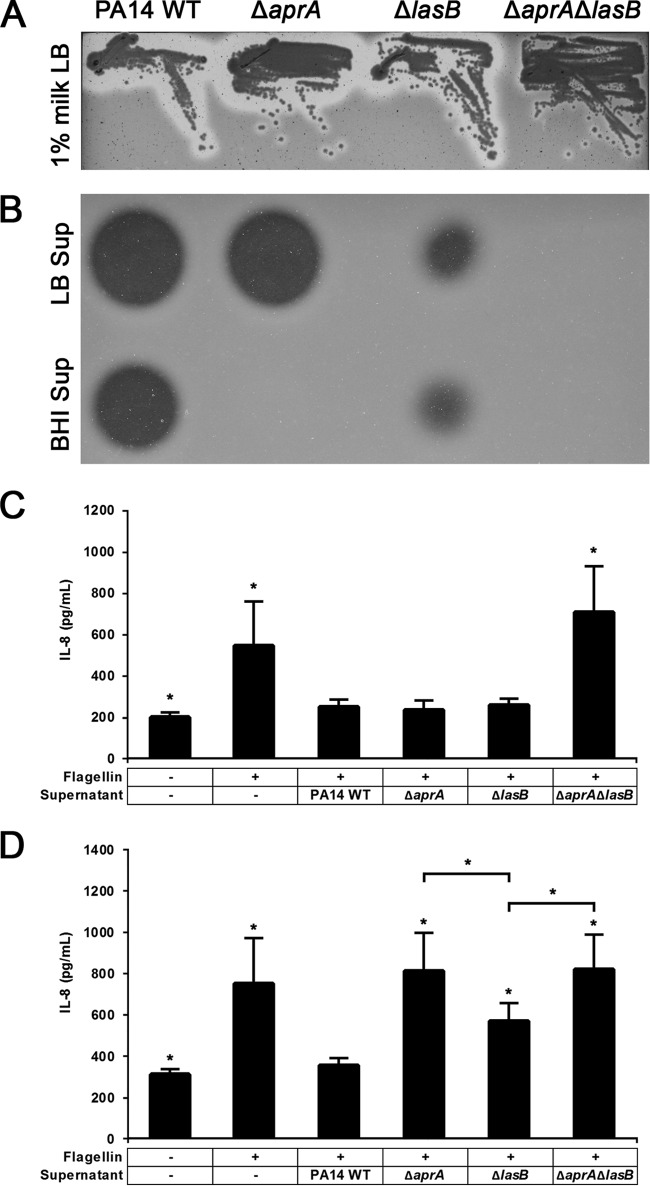
The ΔaprA ΔlasB double mutant exhibits no detectable flagellin protease activity.
To further examine the contribution of LasB to the anti-inflammatory mechanisms of P. aeruginosa, we constructed single- and double-knockout mutants, the ΔaprA, ΔlasB, and ΔaprA ΔlasB mutants, and evaluated their protease activities (Fig. 5A). Interestingly, cell-free supernatants of the ΔaprA ΔlasB double-knockout mutant showed no proteolytic activity in LB or BHI cultures that had been grown for 18 h (Fig. 5B). In contrast, the ΔlasB mutant exhibited reduced overall proteolytic activity, and this reduction appears to be unaffected by the culture conditions. We then exposed A549-Gluc cells to flagellin pretreated with culture supernatants collected from 18-h-old cultures and found that both LB- and BHI-grown supernatants of the double mutant failed to eliminate flagellin, which resulted in the induction of a high-level IL-8 response (Fig. 5C and D).
FIG 5.
Characterization of proteolytic activities of ΔaprA, ΔlasB and ΔaprA ΔlasB mutants. (A) Qualitative analysis of protease activity using milk plate assays. (B) Qualitative analysis of protease activity of cell-free supernatant samples, treated with LB or BHI for 18 h, using skim milk assays. Incubation of A549-Gluc cells were incubated with with flagellin pretreated cell-free supernatant samples grown in LB (C) or BHI (D) for 18 h. Data are presented as means from six experiments ± SD. Significance was evaluated by Wilcoxon test. *, P < 0.05 relative to results for the PA14 WT control.
LasB activity is dependent on the presence of calcium.
Our initial results showed a strong WT-like proteolytic activity for the ΔaprA mutant when grown in LB medium that was lost upon switching to BHI medium. Previous studies have described BHI medium as calcium deficient (47, 48). Since both LasB and AprA proteases require calcium for stability (6, 7, 49), we tested whether the addition of calcium to BHI could restore the proteolytic activity of the single-knockout mutants. Indeed, supplementing BHI medium with as little as 1 mM CaCl2 was sufficient to restore LasB activity in BHI medium such that the ΔaprA mutant displayed WT levels of proteolytic activity in milk plate assays (Fig. 6A). The proteolytic activity of the ΔlasB mutant supernatant also could be enhanced, albeit to a lesser extent. Interestingly, the addition of CaCl2 to filtered BHI supernatant samples did not restore proteolytic activity for the ΔaprA mutant (Fig. 6B). Based on these results, it is apparent that while both proteases require calcium for full functionality, LasB is more sensitive to calcium depletion. However, despite its sensitivity to calcium concentrations, it appears that under calcium-sufficient conditions, LasB is either produced in greater quantities or acts with higher efficiency. Thus, the AprA activity could provide a failsafe mechanism for P. aeruginosa in the event of calcium starvation. These conditions may ensure the maintenance of protease-dependent immune modulating functions.
FIG 6.
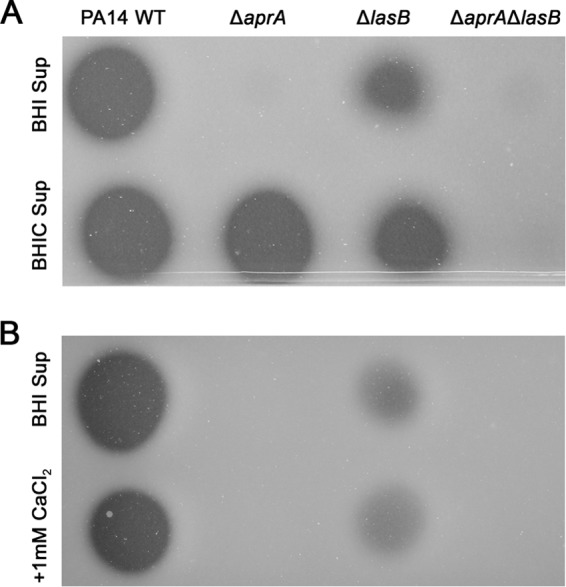
Effect of calcium supplementation on protease activity. Qualitative analysis of protease activity of BHI supernatant samples using skim milk assays. (A) Addition of 1 mM CaCl2 to BHI medium prior to bacterial inoculation (BHIC) restores WT protease activity in the ΔaprA mutant and increases the proteolytic activity of the ΔlasB mutant. (B) Addition of 1 mM CaCl2 after collection of filtered supernatants (plus 1 mM CaCl2) did not restore WT protease activity. Images shown are representative of three independent experiments.
DISCUSSION
The role of flagella in eliciting host immune responses during bacterial infections has been widely investigated over the years. Studies have shown flagellar components and motility to be key elements implicated in host immune recognition (15–19). We observed that the WT strains of P. aeruginosa and P. putida induced strikingly different levels of IL-8 in A549-Gluc cells. The latter showed a significantly higher IL-8 response. In contrast, both of their nonflagellated, nonmotile fleQ mutant counterparts showed very similar phenotypes of low-level IL-8 response. Thus, expressing flagella was sufficient to induce a strong, robust immune response by P. putida, whereas P. aeruginosa appears to manage to successfully obstruct flagellum-mediated host immune recognition.
An earlier study by Bardoel and colleagues demonstrated that the alkaline protease AprA of P. aeruginosa can efficiently remove flagellin from its immediate surroundings and therefore escape flagellin-mediated immune recognition (13). In the present study, we found that P. aeruginosa produces yet another protease with antiflagellin activity besides AprA. We identified this protease to be the elastase LasB.
LasB, also known as pseudolysin, is a highly potent extracellular protease often implicated in tissue damage in P. aeruginosa infections. Both LasB and AprA are virulence factors regulated by the Las quorum system, although the former also has been shown to be regulated by the Rhl quorum system (3, 50). It is known that both AprA and LasB proteases require calcium for stability and function (49, 51). In accordance with this, we observed a loss of detectable proteolytic activity in the ΔaprA mutant grown in BHI. This was reversible upon the addition of calcium to the medium, but only when cells were allowed to grow in the presence of calcium and not when calcium was added to the supernatant after the cells had been removed. A similar calcium dependence could not be detected for AprA. Only minor proteolytic activity was recovered when the BHI medium for the ΔlasB mutant was supplemented with calcium. This indicates that LasB is more sensitive to calcium depletion. There are differences in the protein structure between the two proteases. AprA has been described previously to contain eight calcium ion binding sites, whereas LasB has only one (6, 7). In addition, LasB is produced as a precursor protein that requires autocatalytic processing to ensure proper folding as it is exported across the outer membrane (48, 52). Our observation of LasB activity when cultures were grown in calcium-supplemented medium and not after the addition of calcium to conditioned medium indicates that calcium is needed either for efficient transcription of lasB or during the process of folding and export, as calcium plays a role in the structural integrity of LasB.
The importance of cations for bacterial growth has been well established. During infections, high concentrations of extracellular DNA found in the extracellular matrix of P. aeruginosa biofilms have been shown to efficiently chelate cations such as calcium, resulting in a cation-limited environment (53, 54). It is interesting that under these conditions where P. aeruginosa moves toward persistence, strains often become less virulent and frequently acquire inactivating mutations within LasR, which governs the production of LasB.
Under our experimental conditions, proteolytic activity was observed in the quorum-sensing ΔlasR, rhlR::Tn, and pqsA::Tn mutants when cultures were grown in LB. This proteolytic activity was lost when the ΔlasR variant was grown in BHI medium. LasR governs the production of both AprA and LasB in a cell density-dependent manner and thus also should direct the production of both proteases in LB. Interestingly, a previous study on proteomes of quorum-sensing mutants by Nouwens and colleagues reported that the inactivation of LasR, independent of RhlR activity, resulted in the loss of AprA production. In contrast, the loss of LasB was observed only when both LasR and RhlR were inactivated (3). Thus, it is possible that the induction of LasB production is increased by the combined regulatory activities of the Las and Rhl systems in a manner not applicable to AprA.
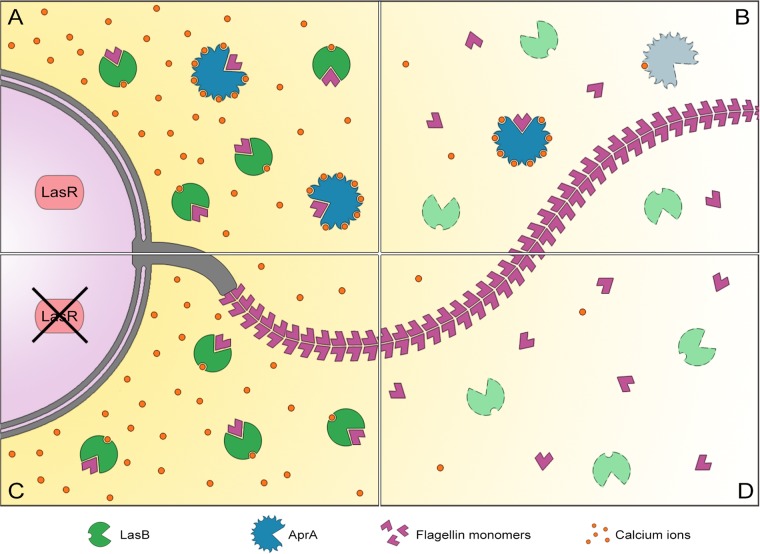
It is interesting that despite the functional redundancy between LasB and AprA, the apparent differences indicate a cooperative relationship. Thus, not only is the efficiency of providing immune-modulating functions enhanced but also the preservation of this function under changing environments is assured. Both proteases act in concert to prevent immune recognition by clearing immunogenic flagellin. This proves advantageous in at least two aspects: first, differences in regulation between the two proteases provide a level of protection against mutations in regulatory elements (Fig. 7A and C). Second, differences in calcium requirements add a level of functional resilience against certain kinds of environmental stress (Fig. 7B and D). Ultimately, it would take the simultaneous inactivation of both proteases to fully induce flagellin-mediated immunogenicity.
FIG 7.
LasB and AprA act in concert to prevent flagellin-mediated immune recognition in P. aeruginosa. Differences between the two proteases provide means for conservation of protease-dependent immune-modulatory functions. (A) Under optimal growth conditions, both LasB and AprA are produced and can act to clear flagellin. (B) Low availability of calcium ions lead to the loss of LasB activity; activity of AprA is less sensitive to the calcium shortage and acts as an anti-flagellin agent. (C) Loss of LasR abolishes AprA production. In the presence of sufficient amounts of calcium, LasB activity clears flagellin. (D) Loss of LasR results in the loss of AprA, and low availability of calcium ions renders LasB nonfunctional. The inactivation of both proteases leads to increased immunogenicity due to flagellin-mediated immune recognition.
Supplementary Material
ACKNOWLEDGMENTS
We extend our gratitude to Juan-Luis Ramos for providing the P. putida KT2440 fleQ::Tn mutant strain, Lutz Wiehlmann for providing the polyclonal antibody against P. aeruginosa FliC, Stephan Brouwer for providing the PA14 ΔlasR strain, Andreas Pich for granting us the use of the MS laboratory facilities, and Thomas Pietschmann and Kathrin Hüging for providing the A549-Gluc cell line.
Funding Statement
The European Research Council (ERC) provided a starter grant to Susanne Häussler under the number Resistome 260276. Further funding was provided by the Ministry for for Science and Culture of Lower Saxony via the Georg Lichtenberg Scholarship.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00939-15.
REFERENCES
- 1.Potempa J, Pike RN. 2009. Corruption of innate immunity by bacterial proteases. J Innate Immun 1:70–87. doi: 10.1159/000181144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tingpej P, Smith L, Rose B, Zhu H, Conibear T, Al Nassafi K, Manos J, Elkins M, Bye P, Willcox M, Bell S, Wainwright C, Harbour C. 2007. Phenotypic characterization of clonal and nonclonal Pseudomonas aeruginosa strains isolated from lungs of adults with cystic fibrosis. J Clin Microbiol 45:1697–1704. doi: 10.1128/JCM.02364-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nouwens AS, Beatson SA, Whitchurch CB, Walsh BJ, Schweizer HP, Mattick JS, Cordwell SJ. 2003. Proteome analysis of extracellular proteins regulated by the las and rhl quorum sensing systems in Pseudomonas aeruginosa PAO1. Microbiology 149:1311–1322. doi: 10.1099/mic.0.25967-0. [DOI] [PubMed] [Google Scholar]
- 4.Louis D, Sorlier P, Wallach J. 1998. Quantitation and enzymatic activity of the alkaline protease from Pseudomonas aeruginosa in culture supernatants from clinical strains. Clin Chem Lab Med 36:295–298. [DOI] [PubMed] [Google Scholar]
- 5.Upritchard HG, Cordwell SJ, Lamont IL. 2008. Immunoproteomics to examine cystic fibrosis host interactions with extracellular Pseudomonas aeruginosa proteins. Infect Immun 76:4624–4632. doi: 10.1128/IAI.01707-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thayer MM, Flaherty KM, McKay DB. 1991. Three-dimensional structure of the elastase of Pseudomonas aeruginosa at 1.5-A resolution. J Biol Chem 266:2864–2871. [DOI] [PubMed] [Google Scholar]
- 7.Baumann U, Wu S, Flaherty KM, McKay DB. 1993. Three-dimensional structure of the alkaline protease of Pseudomonas aeruginosa: a two-domain protein with a calcium binding parallel beta roll motif. EMBO J 12:3357–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andrejko M, Zdybicka-Barabas A, Janczarek M, Cytrynska M. 2013. Three Pseudomonas aeruginosa strains with different protease profiles. Acta Biochim Pol 60:83–90. [PubMed] [Google Scholar]
- 9.Mun JJ, Tam C, Kowbel D, Hawgood S, Barnett MJ, Evans DJ, Fleiszig SM. 2009. Clearance of Pseudomonas aeruginosa from a healthy ocular surface involves surfactant protein D and is compromised by bacterial elastase in a murine null-infection model. Infect Immun 77:2392–2398. doi: 10.1128/IAI.00173-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuang Z, Hao Y, Walling BE, Jeffries JL, Ohman DE, Lau GW. 2011. Pseudomonas aeruginosa elastase provides an escape from phagocytosis by degrading the pulmonary surfactant protein-A. PLoS One 6:e27091. doi: 10.1371/journal.pone.0027091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alcorn JF, Wright JR. 2004. Degradation of pulmonary surfactant protein D by Pseudomonas aeruginosa elastase abrogates innate immune function. J Biol Chem 279:30871–30879. doi: 10.1074/jbc.M400796200. [DOI] [PubMed] [Google Scholar]
- 12.Laarman AJ, Bardoel BW, Ruyken M, Fernie J, Milder FJ, van Strijp JA, Rooijakkers SH. 2012. Pseudomonas aeruginosa alkaline protease blocks complement activation via the classical and lectin pathways. J Immunol 188:386–393. doi: 10.4049/jimmunol.1102162. [DOI] [PubMed] [Google Scholar]
- 13.Bardoel BW, van der Ent S, Pel MJ, Tommassen J, Pieterse CM, van Kessel KP, van Strijp JA. 2011. Pseudomonas evades immune recognition of flagellin in both mammals and plants. PLoS Pathog 7:e1002206. doi: 10.1371/journal.ppat.1002206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pel MJ, Van Dijken AJ, Bardoel BW, Seidl MF, Van der Ent S, Van Strijp JA, Pieterse C. 2014. Pseudomonas syringae evades host immunity by degrading flagellin monomers with alkaline protease AprA. Mol Plant Microbe Interact 27:603–610. doi: 10.1094/MPMI-02-14-0032-R. [DOI] [PubMed] [Google Scholar]
- 15.Hayashi F, Smith KD, Ozinsky A, Hawn TR, Yi EC, Goodlett DR, Eng JK, Akira S, Underhill DM, Aderem A. 2001. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410:1099–1103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 16.Miao EA, Mao DP, Yudkovsky N, Bonneau R, Lorang CG, Warren SE, Leaf IA, Aderem A. 2010. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc Natl Acad Sci U S A 107:3076–3080. doi: 10.1073/pnas.0913087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, Xu J, Xie Y, Qiu Y, Fu S, Yuan X, Ke Y, Yu S, Du X, Cui M, Chen Y, Wang T, Wang Z, Yu Y, Huang K, Huang L, Peng G, Chen Z, Wang Y. 2012. Vaccination with recombinant flagellar proteins FlgJ and FliN induce protection against Brucella abortus 544 infection in BALB/c mice. Vet Microbiol 161:137–144. doi: 10.1016/j.vetmic.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 18.Lovewell RR, Collins RM, Acker JL, O'Toole GA, Wargo MJ, Berwin B. 2011. Step-wise loss of bacterial flagellar torsion confers progressive phagocytic evasion. PLoS Pathog 7:e1002253. doi: 10.1371/journal.ppat.1002253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang M, Wu H, Li X, Yang M, Chen T, Wang Q, Liu Q, Zhang Y. 2012. Edwardsiella tarda flagellar protein FlgD: a protective immunogen against edwardsiellosis. Vaccine 30:3849–3856. doi: 10.1016/j.vaccine.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 20.Mizel SB, Bates JT. 2010. Flagellin as an adjuvant: cellular mechanisms and potential. J Immunol 185:5677–5682. doi: 10.4049/jimmunol.1002156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vijay-Kumar M, Gewirtz AT. 2009. Flagellin: key target of mucosal innate immunity. Mucosal Immunol 2:197–205. doi: 10.1038/mi.2009.9. [DOI] [PubMed] [Google Scholar]
- 22.Ramos HC, Rumbo M, Sirard JC. 2004. Bacterial flagellins: mediators of pathogenicity and host immune responses in mucosa. Trends Microbiol 12:509–517. doi: 10.1016/j.tim.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 23.Andersen-Nissen E, Smith KD, Strobe KL, Barrett SL, Cookson BT, Logan SM, Aderem A. 2005. Evasion of Toll-like receptor 5 by flagellated bacteria. Proc Natl Acad Sci U S A 102:9247–9252. doi: 10.1073/pnas.0502040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guerry P, Szymanski CM. 2008. Campylobacter sugars sticking out. Trends Microbiol 16:428–435. doi: 10.1016/j.tim.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 25.Shanks KK, Guang W, Kim KC, Lillehoj EP. 2010. Interleukin-8 production by human airway epithelial cells in response to Pseudomonas aeruginosa clinical isolates expressing type a or type b flagellins. Clin Vaccine Immunol 17:1196–1202. doi: 10.1128/CVI.00167-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hogardt M, Heesemann J. 2013. Microevolution of Pseudomonas aeruginosa to a chronic pathogen of the cystic fibrosis lung. Curr Topics Microbiol Immunol 358:91–118. [DOI] [PubMed] [Google Scholar]
- 27.Jyot J, Sonawane A, Wu W, Ramphal R. 2007. Genetic mechanisms involved in the repression of flagellar assembly by Pseudomonas aeruginosa in human mucus. Mol Microbiol 63:1026–1038. doi: 10.1111/j.1365-2958.2006.05573.x. [DOI] [PubMed] [Google Scholar]
- 28.Wolfgang MC, Jyot J, Goodman AL, Ramphal R, Lory S. 2004. Pseudomonas aeruginosa regulates flagellin expression as part of a global response to airway fluid from cystic fibrosis patients. Proc Natl Acad Sci U S A 101:6664–6668. doi: 10.1073/pnas.0307553101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rutherford ST, Bassler BL. 2012. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med 2:a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi M, Wu L, Xia Y, Chen H, Luo Q, Sun L, Gao H. 2013. Exoprotein production correlates with morphotype changes of nonmotile Shewanella oneidensis mutants. J Bacteriol 195:1463–1474. doi: 10.1128/JB.02187-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 32.Haid S, Windisch MP, Bartenschlager R, Pietschmann T. 2010. Mouse-specific residues of claudin-1 limit hepatitis C virus genotype 2a infection in a human hepatocyte cell line. J Virol 84:964–975. doi: 10.1128/JVI.01504-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gentzsch J, Hinkelmann B, Kaderali L, Irschik H, Jansen R, Sasse F, Frank R, Pietschmann T. 2011. Hepatitis C virus complete life cycle screen for identification of small molecules with pro- or antiviral activity. Antiviral Res 89:136–148. doi: 10.1016/j.antiviral.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 34.Blanka A, Schulz S, Eckweiler D, Franke R, Bielecka A, Nicolai T, Casilag F, Duvel J, Abraham WR, Kaever V, Haussler S. 2014. Identification of the alternative sigma factor SigX regulon and its implications for Pseudomonas aeruginosa pathogenicity. J Bacteriol 196:345–356. doi: 10.1128/JB.01034-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Godeke J, Pustelny C, Haussler S. 2013. Recycling of peptidyl-tRNAs by peptidyl-tRNA hydrolase counteracts azithromycin-mediated effects on Pseudomonas aeruginosa. Antimicrob Agents Chemother 57:1617–1624. doi: 10.1128/AAC.02582-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haider SR, Sharp BL, Reid HJ. 2011. A comparison of Tris-glycine and Tris-tricine buffers for the electrophoretic separation of major serum proteins. J Sep Sci 34:2463–2467. doi: 10.1002/jssc.201100315. [DOI] [PubMed] [Google Scholar]
- 37.Olling A, Seehase S, Minton NP, Tatge H, Schroter S, Kohlscheen S, Pich A, Just I, Gerhard R. 2012. Release of TcdA and TcdB from Clostridium difficile cdi 630 is not affected by functional inactivation of the tcdE gene. Microb Pathog 52:92–100. doi: 10.1016/j.micpath.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 38.UniProt Consortium. 2014. Activities at the Universal Protein Resource (UniProt). Nucleic Acids Res 42:D191–D198. doi: 10.1093/nar/gkt1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, Villanueva J, Wei T, Ausubel FM. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A 103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inoue H, Massion PP, Ueki IF, Grattan KM, Hara M, Dohrman AF, Chan B, Lausier JA, Golden JA, Nadel JA. 1994. Pseudomonas stimulates interleukin-8 mRNA expression selectively in airway epithelium, in gland ducts, and in recruited neutrophils. Am J Respir Cell Mol Biol 11:651–663. doi: 10.1165/ajrcmb.11.6.7946394. [DOI] [PubMed] [Google Scholar]
- 41.Yoshino Y, Kitazawa T, Kamimura M, Tatsuno K, Ota Y, Yotsuyanagi H. 2011. Pseudomonas putida bacteremia in adult patients: five case reports and a review of the literature. J Infect Chemother 17:278–282. doi: 10.1007/s10156-010-0114-0. [DOI] [PubMed] [Google Scholar]
- 42.Thibodeaux BA, Caballero AR, Marquart ME, Tommassen J, O'Callaghan RJ. 2007. Corneal virulence of Pseudomonas aeruginosa elastase B and alkaline protease produced by Pseudomonas putida. Curr Eye Res 32:373–386. doi: 10.1080/02713680701244181. [DOI] [PubMed] [Google Scholar]
- 43.Harwood CS, Fosnaugh K, Dispensa M. 1989. Flagellation of Pseudomonas putida and analysis of its motile behavior. J Bacteriol 171:4063–4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dasgupta N, Arora SK, Ramphal R. 2000. fleN, a gene that regulates flagellar number in Pseudomonas aeruginosa. J Bacteriol 182:357–364. doi: 10.1128/JB.182.2.357-364.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soutourina OA, Bertin PN. 2003. Regulation cascade of flagellar expression in Gram-negative bacteria. FEMS Microbiol Rev 27:505–523. doi: 10.1016/S0168-6445(03)00064-0. [DOI] [PubMed] [Google Scholar]
- 46.Winzer K, Williams P. 2001. Quorum sensing and the regulation of virulence gene expression in pathogenic bacteria. Int J Med Microbiol 291:131–143. doi: 10.1078/1438-4221-00110. [DOI] [PubMed] [Google Scholar]
- 47.Bhaduri S, Turner-Jones C, Taylor MM, Lachica RV. 1990. Simple assay of calcium dependency for virulent plasmid-bearing clones of Yersinia enterocolitica. J Clin Microbiol 28:798–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Braun P, de Groot A, Bitter W, Tommassen J. 1998. Secretion of elastinolytic enzymes and their propeptides by Pseudomonas aeruginosa. J Bacteriol 180:3467–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olson JC, Ohman DE. 1992. Efficient production and processing of elastase and LasA by Pseudomonas aeruginosa require zinc and calcium ions. J Bacteriol 174:4140–4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dekimpe V, Deziel E. 2009. Revisiting the quorum-sensing hierarchy in Pseudomonas aeruginosa: the transcriptional regulator RhlR regulates LasR-specific factors. Microbiology 155:712–723. doi: 10.1099/mic.0.022764-0. [DOI] [PubMed] [Google Scholar]
- 51.Sarkisova S, Patrauchan MA, Berglund D, Nivens DE, Franklin MJ. 2005. Calcium-induced virulence factors associated with the extracellular matrix of mucoid Pseudomonas aeruginosa biofilms. J Bacteriol 187:4327–4337. doi: 10.1128/JB.187.13.4327-4337.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Braun P, Bitter W, Tommassen J. 2000. Activation of Pseudomonas aeruginosa elastase in Pseudomonas putida by triggering dissociation of the propeptide-enzyme complex. Microbiology 146(Part 10):2565–2572. doi: 10.1099/00221287-146-10-2565. [DOI] [PubMed] [Google Scholar]
- 53.Lewenza S. 2013. Extracellular DNA-induced antimicrobial peptide resistance mechanisms in Pseudomonas aeruginosa. Front Microbiol 4:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mulcahy H, Charron-Mazenod L, Lewenza S. 2008. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog 4:e1000213. doi: 10.1371/journal.ppat.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Woodcock DM, Crowther PJ, Doherty J, Jefferson S, DeCruz E, Noyer-Weidner M, Smith SS, Michael MZ, Graham MW. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res 17:3469–3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saltikov CW, Newman DK. 2003. Genetic identification of a respiratory arsenate reductase. Proc Natl Acad Sci U S A 100:10983–10988. doi: 10.1073/pnas.1834303100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nelson KE, Weinel C, Paulsen IT, Dodson RJ, Hilbert H, Martins dos Santos VA, Fouts DE, Gill SR, Pop M, Holmes M, Brinkac L, Beanan M, DeBoy RT, Daugherty S, Kolonay J, Madupu R, Nelson W, White O, Peterson J, Khouri H, Hance I, Chris Lee P, Holtzapple E, Scanlan D, Tran K, Moazzez A, Utterback T, Rizzo M, Lee K, Kosack D, Moestl D, Wedler H, Lauber J, Stjepandic D, Hoheisel J, Straetz M, Heim S, Kiewitz C, Eisen JA, Timmis KN, Dusterhoft A, Tummler B, Fraser CM. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ Microbiol 4:799–808. doi: 10.1046/j.1462-2920.2002.00366.x. [DOI] [PubMed] [Google Scholar]
- 58.Duque E, de la Torre J, Bernal P, Molina-Henares MA, Alaminos M, Espinosa-Urgel M, Roca A, Fernandez M, de Bentzmann S, Ramos JL. 2013. Identification of reciprocal adhesion genes in pathogenic and non-pathogenic Pseudomonas. Environ Microbiol 15:36–48. doi: 10.1111/j.1462-2920.2012.02732.x. [DOI] [PubMed] [Google Scholar]
- 59.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.