Abstract
Antimicrobial fatty acids (AFAs) protect the human epidermis against invasion by pathogenic bacteria. In this study, we questioned whether human skin fatty acids (FAs) can be incorporated into the lipid moiety of lipoproteins and whether such incorporation would have an impact on innate immune stimulation in the model organism Staphylococcus aureus USA300 JE2. This organism synthesized only saturated FAs. However, when feeding USA300 with unsaturated FAs present on human skin (C16:1, C18:1, or C18:2), those were taken up, elongated stepwise by two carbon units, and finally found in the bacterial (phospho)lipid fraction. They were also observed in the lipid moiety of lipoproteins. When USA300 JE2 was fed with the unsaturated FAs, the cells and cell lysates showed an increased innate immune activation with various immune cells and peripheral blood mononuclear cells (PBMCs). Immune activation was highest with linoleic acid (C18:2). There are several pieces of evidence that the enhanced immune stimulating effect was due to the incorporation of unsaturated FAs in lipoproteins. First, the enhanced stimulation was dependent on Toll-like receptor 2 (TLR2). Second, an lgt mutant, unable to carry out lipidation of prolipoproteins, was unable to carry out immune stimulation when fed with unsaturated FAs. Third, the supplied FAs did not significantly affect growth, protein release, or expression of the model lipoprotein Lpl1. Although S. aureus is unable to synthesize unsaturated FAs, it incorporates long-chain unsaturated FAs into its lipoproteins, with the effect that the cells are better recognized by the innate immune system. This is an additional mechanism how our skin controls bacterial colonization and infection.
INTRODUCTION
The skin is the largest organ at the interface between the environment and the host. The skin plays a major protective role not only as physical barrier but also as the site of first recognition of microbes and other exogenous substances and is an orchestrator of consecutive immune responses (1–3). Staphylococcus aureus is one of the most potent skin pathogens. About 30 to 50% of healthy adults are colonized; 10 to 20% are colonized persistently (4). Coming from the skin, S. aureus can infect any tissue of the body and cause life-threatening diseases.
Besides low pH and antimicrobial peptides, antimicrobial fatty acids (AFAs) also protect human skin against invasion by pathogenic bacteria (5, 6). Free fatty acids (FFAs) are ubiquitously found on the surface of human skin and are the most predominant components in human sebum (7). Particularly medium- to long-chain FFAs (C8 to C18) have antibacterial activity against a broad range of Gram-positive bacteria and are thought to be responsible for at least part of the direct antimicrobial activity of the skin surface against pathogen colonization and infection (8, 9). Lauric acid (C12:0), for example, had a high activity toward Clostridium perfringens (10); furthermore, it is also highly active against Propionibacterium acnes, Staphylococcus aureus, and Staphylococcus epidermidis but did not induce cytotoxicity to human sebocytes (9). For S. aureus, the seven most effective AFAs are lauric acid (C12:0), glycerol monolaurate, capric acid (C10:0), myristic acid (C14:0), and the polyunsaturated linoleic acid (C18:2) (11).
On skin, FFAs are produced by lipases that are secreted from the members of the commensal bacterial flora, such as P. acnes and staphylococci, by hydrolyzing sebum triacylglycerides secreted from sebaceous glands (12–15). The lipase SAL2 of S. aureus USA300 has been shown to hydrolyze triglycerides into FFAs that inhibit growth of the bacterium (16). However, FFAs can also be produced by sebocytes without the presence of bacteria (17, 18). In addition to having direct antimicrobial activity, some sebum FFAs (e.g., lauric acid [C12:0], palmitic acid [C16:0] and oleic acid [C18:1,Δ9]), enhance the skin innate immune defense by inducing human beta-defensin-2 (HBD-2) in the human sebocytes (19). Therefore, the sebaceous glands may play a significant role in skin's innate immunity by providing antimicrobial agents to the external skin surface. By their antimicrobial activity, AFAs may allow survival and growth of only compatible microorganisms. Indeed, it has been shown that some areas (abdomen, leg, face, and plantar site) of the human stratum corneum showed marked differences as to the concentration of FFAs (20). Interestingly, skin microbiota studies showed that staphylococcal colonization was lower in areas with large amounts of FFAs (21), suggesting an inverse correlation between levels of staphylococcal colonization and FFAs (22).
The mode of action of AFAs is manifold, but the primary target appears to be the cell membrane. High concentrations of long-chain unsaturated FFAs (UFFAs) increase membrane disorder (fluidity), and in S. aureus, inhibitory concentrations of linoleic acid (C18:2Δ9,Δ12) cause protein leakage and interference with metabolic pathways such as the electron transport chain and nutrient uptake (23, 24). The human skin AFA cis-6-hexadecenoic acid (C6H) swiftly kills S. aureus by causing a loss of membrane integrity (25, 26). However, staphylococci, as classical skin colonizers, possess mechanisms for increased tolerance of AFAs, such as the surface protein IsdA and wall teichoic acids reducing bacterial cellular hydrophobicity and preventing AFA binding (27, 28).
Not all FAs are harmful, and certain FAs are even used as nutrients. In S. aureus, oleic acid (C18:1Δ9) is a substrate for phospholipid biosynthesis, while the AFA sapienic acid (C16:1Δ9) is elongated before it is incorporation into cellular pathways (29, 30).
While the antimicrobial activity of FAs is well documented, little is known about the impact of skin FAs on recognition of skin-colonizing bacteria by the innate immune system. In S. aureus, among the major players in innate immune activation are the lipoproteins (Lpp) (31, 32). Lpp are important microbe-associated molecular patterns (MAMPs) in Gram-positive bacteria. They are recognized by Toll-like receptor 2 (TLR2) in connection with the coreceptors TLR1 and TLR6 (33, 34). In S. aureus, there are between 55 and 70 Lpp, and many are involved in nutrient acquisition (31). The lipidation of the S. aureus Lpp is crucial for TLR2-MyD88 activation and is important in systemic infections (35).
Here we show that some of the predominant unsaturated FAs on human skin can be incorporated into the lipid moiety of S. aureus Lpp and that these cells have an increased TLR2-dependent immune stimulating activity rendering the pathogen more recognizable by the immune defense system.
MATERIALS AND METHODS
Chemicals.
The standard free fatty acids (FFAs)—caproic acid (C6:0), palmitic acid (C16:0), stearic acid (C18:0), palmitoleic acid (C16:1), oleic acid (C18:1), linoleic acid (C18:2), and cerotic acid (C26:0)—were from Sigma (Taufkirchen, Germany). Stocks (0.1 M) of these fatty acids were prepared by dissolving in dimethyl sulfoxide (DMSO).
Bacterial strains and growth condition.
Staphylococcus aureus USA300 JE2 is cured from the three plasmids present in the parent strain USA300 LAC, a highly characterized community-associated methicillin-resistant S. aureus (MRSA) strain isolated from the Los Angeles County jail (36). USA300 JE2 was obtained from Ken Bayles, Department of Pathology & Microbiology, University of Nebraska Medical Center. For fatty acid extraction, the staphylococcal strains were first precultivated aerobically in tryptic soy broth (TSB) at 37°C for 6 to 8 h; this culture was used to inoculate fresh TSB (optical density at 600 nm [OD600] of 0.1), which was cultivated at 37°C for 16 to 17 h under constant shaking at 150 rpm. For feeding the cells with standard FFAs, the TSB medium was supplied with 50 μM FFAs from 0.1 M stock solutions or with DMSO as a control. The OD600 of overnight cultures was monitored over time and measured every hour. Bacterial culture supernatants were obtained by centrifugation and filtered through 0.2-μm-pore-size filters. The Bradford assay (Quickstart from Bio-Rad) was used to evaluate membrane permeability by measuring protein amounts of culture filtrates. This assay was performed according to manufacturer's instructions. Bacteria were harvested by centrifugation at 5,000 × g for 10 min and washed at least twice with phosphate-buffered saline (PBS) or 0.9% NaCl prior to further use.
Deletion of lgt in S. aureus USA300 JE2.
lgt is the diacylglyceryl transferase enzyme-encoding gene that is involved in lipidation of lipoproteins (31). For deletion of lgt in USA300 JE2, knockout plasmid pBASE6 containing the upstream and the downstream flanking regions of lgt was constructed. The flanking regions were amplified by PCR from the genome of USA300 JE2, the upstream one using primers Fr_up (EcoRI) (GATGAATTCGCTGGTGAAGAAGGAC) and Re_up (SacI) (CGTGAGCTCAGTGGTCCTAAGTTAAATGCC) and the downstram one using primers Fr_down (SacI) (CGTGAGCTCAGTGGTCCTAAGTTAAATGCC) and Re_down (BgIII) (TTGAGATCTAAGATATTGGAATAGTATTTG) (restriction sites are underlined). The amplified upstream fragment was cut with EcoRI and SacI, and the amplified downstream fragment was cut with SacI and BgIII. These fragments were then ligated with vector pBASE6 digested by EcoRI and BgIII, resulting in pBASE6Δlgt, which was transformed into Escherichia coli DH5α. The clones containing plasmid pBASE6Δlgt were isolated, purified, and checked for the correct sequence with primers Fr_pBASE6 (CCTCAAGCTAGAGAGTCATTACCCC) and Re_pBASE6 (CTACTTCTTTCAAACTCTCTCTACG). The correct plasmid pBASE6 Δlgt was subsequently transformed by electroporation into S. aureus RN4220 as an intermediary host and then transformed into S. aureus USA300 JE2. The procedure for deletion of lgt from USA300 JE2 was followed as described previously (37).
Construction of pCtuf-sitC18.
For expression of a truncated sitC gene, the plasmid pCtuf was used (38). Downstream of the tuf promoter, truncated sitC, composed of the lipo signal sequence followed by 10 codons of sitC and ending with 8 codons of the Strep tag and two stop codons, was cloned. Truncated sitC was amplified from the USA300 genome by using a forward primer with a PacI cleavage site (underlined sequence) (CGCTTAATTAATGAAAAAATTAGTACCTTTATTATTAG). The reversed primer comprised the StreptagII coding sequence (italic sequence), two stop codons (bold sequence), and a HindIII site (underlined sequence) (CCGAAGCTTTTATTATTTTTCAAATTGTGGATGTGACCAATCACTGCTTTGTTTACCAC). The amplified fragment was ligated into the pCtuf plasmid after digestion with PacI and HindIII to yield plasmid pCtuf-sitC18. This plasmid was transformed into USA300 Je2 by electroporation.
Purification of lipopeptide SitC18.
SitC18 was isolated from the membrane fraction of USA300 Je2(pCtuf-sitC18). The clone was cultivated aerobically at 37°C in TSB medium for 16 h. From 3 liters of culture, the bacterial cells were harvested by centrifugation at 4,000 × g at 4°C. The cell pellets were washed two times with Tris buffer (20 mM Tris, 100 mM HCl [pH 8.0]). Then the pellet was resuspended with Tris buffer containing a protease inhibitor (Merck, Darmstadt, Germany) and lysostaphin (30 μg/ml) and incubated at 37°C for 2 h to disrupt the cell wall. Membrane proteins were pelleted by ultracentrifugation (235,000 × g for 45 min at 4°C) and extracted according to a previous study (39). SitC18 was purified by affinity chromatography with Strep-Tactin Superflow beads (Qiagen, Germany) overnight in cold condition (6°C) under mild rotation (20 rpm). Then, the resin suspension was loaded into a 5-ml column and the flowthrough was collected. One volume of Strep-Tactin resin (5 ml) was washed 3 times with 4 volumes of washing buffer (100 mM Tris-Cl, 150 mM NaCl, 1 mM EDTA [pH 8]). SitC18 was eluted with 3 volumes of elution buffer (100 mM Tris-Cl, 150 mM NaCl, 2.5 mM desthiobiotin, 1 mM EDTA [pH 8]) for 3 h at 6°C. Finally, 0.5 ml of the eluate was concentrated by StrataClean Resin for running SDS-PAGE, and the remaining part was lyophilized overnight for fatty acid extraction.
Western blotting.
USA300 JE2 was overnight cultured in TSB and TSB supplemented with 50 μM C16:1, C18:1, and C18:2 UFFAs. Bacteria were harvested by centrifugation at 5,000 × g for 10 min at 4°C and washed 2 times with 20 mM Tris buffer (pH 8). The cell pellets were again dissolved in Tris buffer and incubated with lysostaphin (30 μg/ml) at 37°C for 30 min. The total proteins were subsequently extracted by the methanol-chloroform method, dissolved in SDS running buffer, and separated by SDS-PAGE. The amount of Lpl1 protein was detected by Western blotting using anti-Lpl1 antibody as described previously (39).
Lipid extraction.
The total lipids were extracted by the method of Bligh and Dyer. Briefly, the bacterial cells from the 50-ml overnight culture were harvested by centrifugation at 5,000 × g in 10 min at 4°C and washed twice with 0.9% NaCl. For the lipid extraction, the wet pellets were mixed with 15 ml of methanol-chloroform (2:1, vol/vol) and incubated on the rotary shaker for 2 h. The supernatant was collected by centrifugation at 5,000 × g in 10 min at 4°C. The lipids were repeatedly extracted by mixing the pellet with 15 ml of methanol-chloroform-water (2:1:0.8, vol/vol/vol) for 2 h. This extraction was repeated twice. The collected supernatants were mixed with 11 ml of chloroform and 11 ml of water in the separator funnel. After overnight incubation, two layers as an organic phase and an aqueous phase were formed. The lipid in the organic phase was collected. The chloroform was removed by rotary evaporator.
Fatty acid extraction and analysis by GC-MS.
Fatty acids were extracted from the total lipid or lipopeptides according to a previous study (40). The samples were suspended in 1 ml of reagent I (22.5 g of NaOH plus 75 ml of MeOH and 75 ml water) and transferred to 10-ml glass jars with screw-top lids with a Teflon seal for saponification. The suspension was well vortexed and incubated for 35 min at 100°C. After cooling, 2 ml of reagent II (162.5 ml of 6 N HCl plus 137.5 ml of MeOH) was added for esterification. The suspension was incubated for 12 min at 80°C and cooled down again. The FA methyl esters were extracted by adding 1.25 ml of hexane (C6H14). The suspension was then shaken for 5 min, until phase separation occurred. After a short centrifugation step, the upper phase was transferred to a fresh glass vial and used for gas chromatography-mass spectrometry (GC-MS).
FAs were analyzed by GC-MS with a nonpolar capillary column (Optima 5; methylsilicone phase; length, 15 m; inside diameter, 0.25 mm; layer thickness, 0.25 μm; Macherey-Nagel). The gradient temperature conditions were 5 min at 45°C, 45 to 200°C at 18°C min−1, 200 to 340°C at 84°C min−1 and 2 min at 340°C. GC parameters were as follows: injector temperature, 300°C; interface temperature, 300°C; column pressure, 18.7 kPa; spitless; injection volume, 1 μl; column flow rate, 1.2 ml min−1; and total flow rate, 11.4 ml min−1. The data were analyzed using the GC-17A program (Shimadzu). FAs were identified based on the retention times and by comparing the MS peaks with library spectra and the spectra from authentic standards.
Preparation of bacteria and bacterial lysates for stimulation.
A preculture of 20 ml of TSB was inoculated with bacteria from a fresh tryptic soy agar (TSA) or blood agar plate. After 6 h of growth, the main culture (20 ml of TSB) was inoculated to an OD600 of 0.1 and supplemented with 50 μM free fatty acids (FFAs) or DMSO as a control. After 17 h of growth at 37°C with agitation, bacteria were harvested by centrifugation at 5,000 × g for 10 min and washed 3 times with PBS. Bacteria were set to the distinct OD/CFU in the corresponding medium for stimulation of Mono Mac 6, HEK 293, or HEK293-TLR2 cells or peripheral blood mononuclear cells (PBMCs) isolated from fresh blood of healthy human donors.
Bacterial lysates were made using FastPrep-24 from MP Biomedicals. Bacteria were washed three times with PBS, resuspended in lysis buffer (20 mM Tris-HCl, 0.1 M NaCl [pH 8]) to an OD600 of 28, and heat inactivated at 70°C for 30 min. Then 500 μl of sterile glass beads was mixed with 500 μl of heat-inactivated bacteria. The FastPrep speed was set to 5, and bacteria were lysed by three runs for 30 s. The protein amounts in the lysates were determined using the Bradford assay (Quickstart from Bio-Rad). Lysates were used to stimulate PBMCs.
Cell culture and stimulation assays.
Mono Mac 6, a human monocytic leukemia cell line (41), was obtained from DSMZ (Braunschweig, Germany) and cultured in RPMI 1640 (Biochrom AG, Berlin, Germany) supplemented with 10% FBS Superior (BiochromAG, Berlin, Germany), and 1% OPI (O5003; Sigma, Taufkirchen, Germany) in nonessential amino acids (NEA; Biochrom AG, Berlin, Germany) and 1% Zell Shield (Minerva Biolabs GmbH, Berlin, Germany) at 37°C with 5% CO2. Prior to stimulation, 106 cells per 24-well microtiter plate were seeded out in 1 ml of culture medium and incubated for 1 h at 37°C with a 5% CO2 supplement. Mono Mac 6 cells were stimulated with an MOI of 30:1. In addition, UFAs (50 μM) were applied to 106 Mono Mac 6 cells and Pam3Cys (EMC, Tübingen, Germany) was used as a control. The period of stimulation was 4 h for tumor necrosis factor alpha (TNF-α) or 24 h for interleukin-6 (IL-6) and IL-10. The supernatants were collected and stored at −20°C until use.
Human embryonic kidney (HEK 293) cells, stably transfected with the human TLR2 gene, were purchased from Invivogen. HEK-TLR2 cells were cultivated in 75-cm2 culture flasks using 20 ml of growth medium (Dulbecco modified Eagle medium [DMEM], 10% fetal calf serum [FCS], 100 μg/ml of Normocin, and 10 μg/ml of blasticidin). HEK-TLR2 cells were seeded on 24-well cell culture plates and cultivated until confluence was reached. HEK-TLR2 cells were stimulated with bacteria using 1 × 106 bacteria per ml and well. In addition, HEK 293 cells without TLR2 expression (in DMEM, 10% FCS, 20 mM l-glutamine, and 1,000 U/ml of penicillin-streptomycin) were used as a control. P2C was used at 1 nM as a positive control. Bacteria were unable to grow due to antibiotic supplementation in stimulation medium. Stimulation was carried out for 18 h at 37°C and 5% CO2. The supernatants were collected and stored at −20°C until use.
Human PBMCs were isolated from fresh blood of healthy human donors by standard Ficoll/Histopaque gradient centrifugation. Culture medium consisting of very low endotoxin (VLE) RPMI 1640 was supplemented with 2 mM sodium pyruvate, 2 mM l-glutamine, 100 U/ml of penicillin-streptomycin (Pen-Strep), and 10 mM HEPES. PBMCs were stimulated with bacteria in a 96-well plate. Therefore, 5 × 105 PBMCs were stimulated with 0.2 × 105 bacteria per well for 5 h (multiplicity of infection [MOI] = 25:1). A total of 200 μg/ml of gentamicin was added to prevent bacterial growth. Growth and stimulation of cells were performed at 37°C and 5% CO2. Then, cultures were centrifuged for 10 min at 250 × g and 4°C, and supernatants were collected and stored at −20°C until use.
Detection of cytokines by ELISA.
Human IL-8, TNF-α, IL-6, and IL-10 secretion was measured in cellular supernatants using the BD OptEIA enzyme-linked immunosorbent assay (ELISA) kits according to the manufacturer's instructions.
Isolation and analysis of PGN.
Cells were centrifuged at 4,700 rpm for 10 min, and the pellets were washed twice with a 1:1 ethanol-water mixture to get rid of remaining fatty acids. The isolation of ultrapure peptidoglycan (PGN; free of DNA, RNA, proteins, wall teichoic acids, lipoteichoic acids, and salts) was done by following the method described for previous studies (42, 43).
Lyophilized PGN (1 mg/ml) was digested with 500 U of mutanolysin of Streptomyces globisporus ATCC 21553 (Sigma, Steinheim, Germany) in a 12.5 mM phosphate buffer (pH 5.5) at 37°C for 16 h. The sample was boiled for 3 min and centrifuged for 5 min at 10,000 × g. The FAs were reduced with sodium borohydride solved in 0.5 M borate buffer (pH 9) for 20 min at room temperature. The pH was subsequently adjusted to 2 with orthophosphoric acid.
Analysis of the peptidoglycan pattern was performed by ultraperformance liquid chromatography (UPLC) using a Waters Acquity H class UPLC system.
A 100- by 2.1-mm reversed-phase column (Acquity CSH C18; 1.7 μm; Waters, Eschborn, Germany), guarded by a Vanguard 2.1- by 5-mm precolumn (CSH C18; 1.7 μm; Waters), was used. The samples were eluted at a flow rate of 0.176 ml/min using a linear gradient starting from 100% buffer A (0.1% trifluoroacetic acid [TFA], 5% [vol/vol] methanol) to 100% buffer B (0.1% TFA, 30% [vol/vol] methanol) within 60 min. The column temperature was set to 52°C. The eluted muropeptides were detected by UV absorption at 205 nm.
Ethics statement.
Human PBMCs were isolated from venous blood of healthy volunteers in accordance with protocols approved by the Institutional Review Board for Human Subjects at the University of Tübingen. Informed written consent was obtained from all volunteers.
Statistical analysis.
Unpaired two-tailed Student's t test or one-way analysis of variance (ANOVA) was used to determine the significance of the differences between means. Statistical analysis was performed using GraphPad Prism 5.0 or SPSS v.19. The significance level was set as follows: a P value of >0.05 was considered not significant. In figures, significant differences are depicted as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
RESULTS
FA composition in the lipid fraction of S. aureus USA300 JE2.
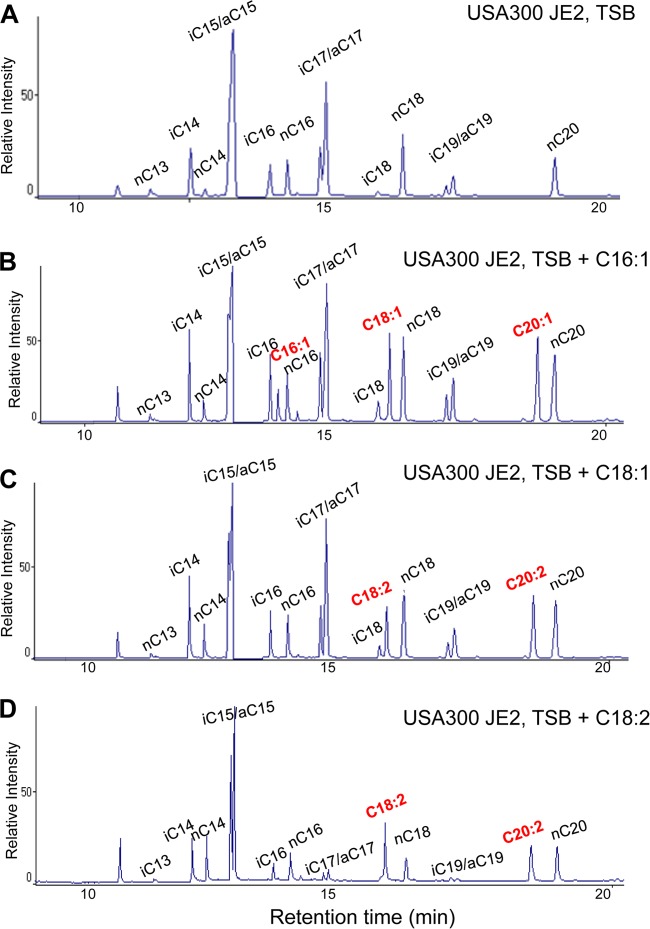
When cultivated in TSB, the lipid fraction of USA300 JE2 contained only saturated FAs with a chain length ranging from 13 to 20 carbon atoms (Fig. 1A). The normally saturated (n) and iso-saturated (i) FAs were mostly detected in even carbon numbers (iC14 and nC14, iC16 and nC16, iC18 and nC18, and nC20); nC13 was an exception, but this FA was detectable only in tiny amounts. The iso- and anteiso-saturated (a) FAs were mainly found in odd carbon numbers (iC15 and aC15, iC17 and aC17, and iC19 and aC19). Interestingly, there were no unsaturated FAs (UFAs) detectable in the lipid fraction.
FIG 1.
GC analysis of the fatty acids isolated by total lipid extraction from S. aureus USA300 JE2 in TSB medium (A), TSB medium with a supplement of 50 μM C16:1 FA (B), TSB medium with 50 μM C18:1 FA (C), and TSB medium with 50 μM C18:2 FA (D).
The question therefore was whether USA300 JE2 is able to take up UFAs from the medium. As some FAs are reported to inhibit growth of S. aureus at higher concentrations, it was necessary to find the right concentration that will not inhibit growth. In this study, we have chosen 50 μM C16:1, C18:1, and C18:2 UFAs, as these are the predominant UFAs on human skin, sebaceous glands, and nasal sebum, where S. aureus frequently has its first contact with the healthy human host (7, 20, 44). When testing the effect of C16:1, C18:1, and C18:2 UFAs, it turned out that a concentration of 50 μM UFAs in TSB hardly affected growth (see Fig. S1 in the supplemental material). For our feeding studies, we therefore used always this concentration.
UFAs were taken up and elongated by S. aureus USA300 JE2.
When we fed USA300 JE2 with 50 μM UFAs, we not only detected these unsaturated FAs in the lipid fraction but also observed elongated forms. The elongation occurs apparently stepwise by the addition of each two-carbon unit. For example, when we fed bacteria with C16:1 UFA, it was elongated to C18:1 and C20:1 (Fig. 1B). When the medium was supplied with C18:1 UFA, we detected also C20:1 UFA (Fig. 1C). When we fed them double unsaturated FAs like C18:2 FA, we detected also C20:2 UFA (Fig. 1D). The composition of other saturated FAs was not changed when bacteria were fed these UFAs. In addition, we verified whether also short- or long-chain FAs (C6:0 and C26:0) can be taken up. However, none of these FAs could be detected in the lipid fraction (see Fig. S2 in the supplemental material), suggesting that S. aureus is only able to take up FAs with a certain chain length. Here we showed that exogenously supplied UFAs were not only taken up by the cells but also elongated. However, whether the supplied UFAs were also incorporated into the lipid moiety of lipoproteins is unknown.
Exogenously supplied UFAs were incorporated into the lipid moiety of lipopeptide SitC18.
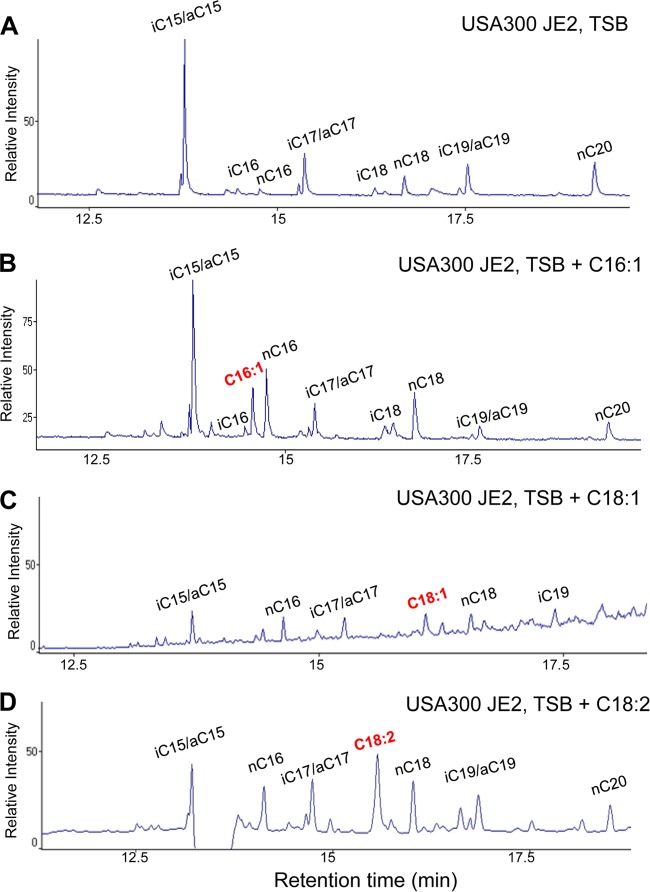
To better investigate the incorporation of exogenously supplied UFAs into the lipid moiety of lipoproteins, we constructed a plasmid that expresses a SitC-derived lipopeptide provided with a C-terminal Strep tag (Fig. 2A). SitC is involved in Fe acquisition and is one of the predominant lipoproteins in S. aureus (31). Plasmid pCtuf-sitC18 expresses the lipopeptide SitC-18 which is composed of the N-terminal lipid moiety, the first 10 amino acids of SitC, and a Strep tag for affinity purification (Fig. 2B). SitC18 was purified from the membrane fraction of USA300 JE2 (pCtuf-sitC18) by Strep-Tactin resin. By this method, we could purify sufficient amounts of lipidated SitC18, as shown by SDS-PAGE (Fig. 2C). Of the purified lipopeptide SitC18, the fatty acids were extracted and analyzed by GC-MS. If SitC18 was isolated from the clone cultivated only in TSB, the normally saturated and iso- and anteiso-saturated FAs with chain lengths of C15 to C20 were detectable (Fig. 3A). SitC18, however, was isolated from the clone cultivated in TSB supplemented with UFAs like C16:1, C18:1, or C18:2. We found that they were all incorporated into the lipid moiety of SitC18 (Fig. 3B to D). As lipoproteins are the predominant innate immune activators in staphylococci, we wondered whether the incorporation of the UFAs has an effect on innate immune stimulation. However, first of all, we investigated whether UFAs alone showed innate immune stimulation.
FIG 2.
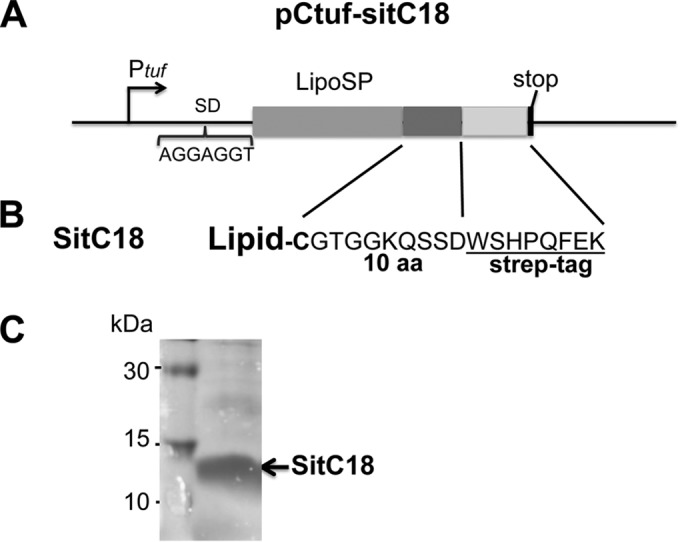
Construction of and purification of lipopeptide SitC18. (A) pCtuf-SitC18 is the elongation expression plasmid for the truncated SitC protein gene consisting of a signal peptide and 10 amino acids. The 3′ end is extended by a Strep tag sequence and a stop codon. (B) Amino acid sequence of SitC18. (C) SDS-PAGE with purified SitC-18.
FIG 3.
GC analysis of the fatty acids isolated from the lipid moiety of the purified lipopeptide SitC18 from S. aureus USA300 JE2 in TSB medium (A), TSB medium with a supplement of 50 μM C16:1 FA (B), TSB medium with 50 μM C18:1 FA (C), and TSB medium with 50 μM C18:2 FA (D).
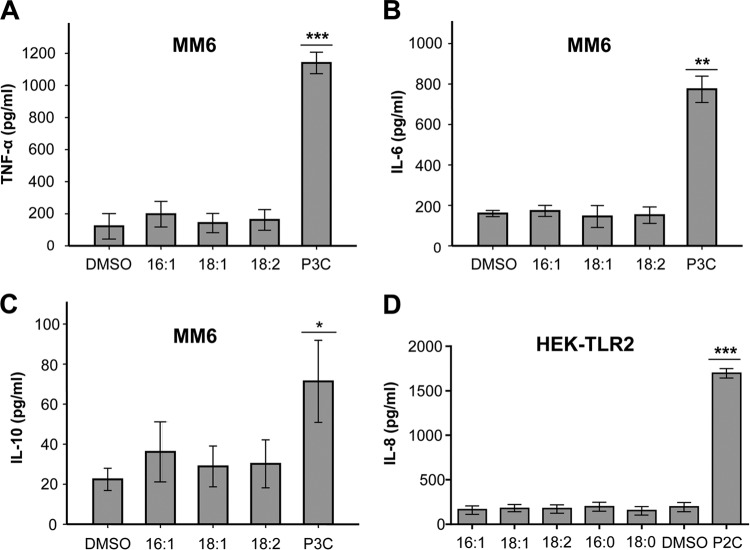
UFAs do not induce cytokines in human Mono Mac 6 or HEK-TLR2 cells.
We evaluated the function of three UFAs (C16:1, C18:1, and C18:2) on inducing cytokines in human Mono Mac 6 cells. Applied in a concentration of 50 μM, they do not induce TNF-α, IL-6, or IL-10, in contrast to the positive control, 200 nM P3C (Fig. 4A to C). In HEK-TLR2 cells, saturated and unsaturated fatty acids have no effect on inducing IL-8 cytokine, except P2C (1 nM) as a control (Fig. 4D).
FIG 4.
Free fatty acids (FFAs) do not stimulate Mono Mac 6 or HEK-TLR2 cytokines. A total of 106 Mono Mac 6 cells were infected with 50 μM UFFA. (A) TNF-α was determined after 4 h of stimulation. (B and C) IL-6 and IL-10 were determined after 24 h. (D) HEK-TLR2 cells were stimulated with 50 μM FFAs, and IL-8 release was measured after 18 h. The negative control was buffer with 10% DMSO; positive controls were 200 nM P3C and 1 nM P2C. The experiments in duplicate were conducted at least 3 times. Error bars indicate SEMs. Statistical significance was calculated to compare test groups and the positive control (P3C or P2C) with the negative control (DMSO) by using one-way ANOVA or Student's t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
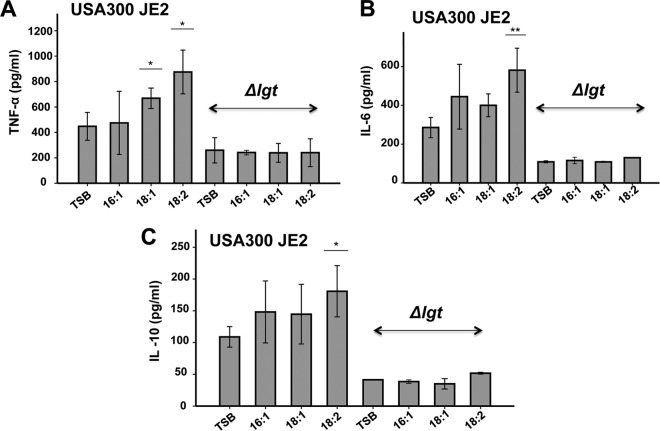
UFAs fed S. aureus induced cytokine production with human Mono Mac 6 and HEK-TLR2 cells and PBMCs.
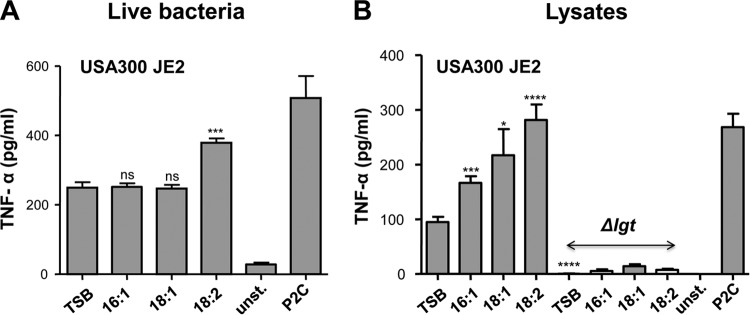
We have shown that UFAs not only are found in the lipid fraction of S. aureus but also are incorporated into the lipid moiety of lipopeptide SitC18. Lipoproteins and lipopeptides are classical TLR2 ligands. To see whether UFAs have an effect on host signaling, we cultured USA300 JE2 in TSB with and without supplemented 50 μM UFAs. As a control, we also generated a Δlgt mutant in USA300 JE2 which is unable to lipidate prolipoproteins and is thus severely affected in TLR2-mediated activation of the innate immune system (31). The staphylococcal strains were cultivated overnight in TSB with and without supplemented 50 μM UFAs (C16:1, C18:1, or C18:2), and live bacteria were subsequently used to infected Mono Mac 6 cells with an MOI of 30. The production of proinflammatory cytokines, such as TNF-α, was determined after 4 h and in the case of IL-6 and IL-10 after 24 h of stimulation.
The result showed that S. aureus fed with UFAs significantly increased production of TNF-α, IL-6, and IL-10 (Fig. 5). The most pronounced effect was observed with the doubly unsaturated C18:2 FA. Compared to TSB alone, the addition of C18:2 FA caused almost a 2-fold increase in cytokine production. On the other hand, the Δlgt mutant was severely affected in cytokine induction and completely uninfluenced by UFAs used to supplement the medium. These results suggest that cytokine induction is mainly triggered by the lipoproteins and that it is TLR2 dependent.
FIG 5.
Cytokine expression by Mono Mac 6 cells upon infection with USA300 JE2 and its Δlgt mutant cultured in TSB with and without supplemented 50 μM UFFAs. USA300 JE2 and its Δlgt mutant were cultured in TSB medium supplemented with 50 μM UFFAs (C16:1, C18:1, or C18:2) for 16 h. For immune stimulation, Mono Mac 6 cells were infected with bacterial cells at an MOI of 30:1. TNF-α was determined after 4 h of stimulation (A), while IL-6 and IL-10 were determined after 24 h of stimulation (B and C). The experiments in duplicate were conducted at least 3 times. Error bars indicate standard errors. Statistical significances were calculated to compare test groups with the control (TSB) by using Student's t test. *, P < 0.05; **, P < 0.01.
To further assess TLR2-dependent signaling, we carried out the same experiments with and without TLR2-expressing HEK cells. IL-8 production was only observed with HEK-TLR2 cells and not with HEK cells alone upon stimulation with USA300 JE2 (Fig. 6). Again, when USA300 JE2 was fed with C18:2 FA, significantly greater stimulation was observed than with TSB alone. The Δlgt mutant was completely silent regarding IL-8 stimulation even when cells were cultivated in the presence of UFAs. It exerted no effect with either HEK or HEK-TLR2 cells, indicating that the signaling is mediated essentially by the lipoproteins and that it is TLR2 dependent.
FIG 6.
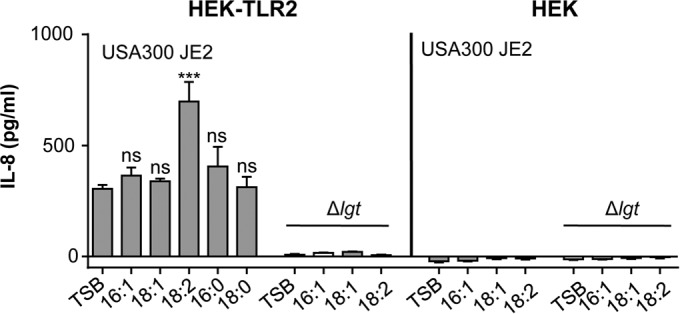
Stimulation of HEK293 and HEK293-TLR2 cells by USA300 JE2 and its Δlgt mutant cultured in TSB with and without supplemented 50 μM FFAs. HEK cells were stimulated with 106 bacteria. USA300 JE2 and the Δlgt mutant were cultured in TSB medium alone or supplemented with 50 μM UFFAs (C16:1, C18:1, C18:2, C16:0, and C18:0) for 17 h. IL-8 was measured after 18 h of stimulation. The experiments in duplicate were conducted at least 3 times. Error bars indicate SEMs. Statistical significances were calculated to compare test groups with the control (TSB) by using unpaired Student's t test. ns, not significant (P > 0.05); ***, P < 0.001.
For more relevance to the situation in vivo, we carried out signaling studies with PBMCs isolated from fresh blood of healthy volunteers. When we applied live USA300 JE2 to PBMCs, again the highest induction of TNF-α was achieved when the bacteria were cultivated in TSB supplied with C18:2 UFA, while cells fed with C16:1 or C18:1 UFA showed no difference (Fig. 7A). Unstimulated and P2C (1 nM)-stimulated PBMCs were used as controls. When stimulation experiments were carried out with cell lysates (FastPrep), a continuous increase was seen when USA300 JE2 was cultivated in the presence of C16:1, C18:1, or C18:2 UFA; feeding with the latter UFA led to an almost 3-fold increase in TNF-α production (Fig. 7B). On the other hand, lysates from the Δlgt mutant showed no induction, corroborating the above-described finding that signaling is essentially due to lipoproteins.
FIG 7.
Stimulation of PBMCs by USA300 JE2 and its Δlgt mutant cultured in TSB with and without supplemented 50 μM UFFAs. A total of 5 × 105 PBMCs per well (n = 8) were stimulated with S. aureus strains (USA300 JE2 and Δlgt mutant) cultured in TSB medium alone or supplemented with 50 μM UFFAs (C16:1, C18:1, or C18:2); TNF-α production was the readout after 5 h of stimulation. Stimulation was carried out with live bacteria (A) and with 20 ng of crude cell lysates prepared by FastPrep (B). Controls were unstimulated (unst.) and P2C (1 nM) stimulated PMBCs. The experiments in duplicate were conducted at least 3 times. Error bars indicate mean+/−SEM. Statistical significances were calculated to compare test groups with control (TSB) by using unpaired Student's t test. ns, not significant (P > 0.05); *, P < 0.05; ***, P < 0.001; ****, P < 0.0001.
Supplied UFAs did not affect S. aureus peptidoglycan composition.
Previously it has been reported that long-chain UFAs affect peptidoglycan synthesis in S. aureus (45). As peptidoglycan also contributes to immune stimulation, particularly via NOD2, we wanted to see whether the exogenously supplied UFAs (50 μM) had an effect on peptidoglycan composition in USA300 JE2 cultivated in the presence of UFAs. However, the muropeptide pattern of peptidoglycan after mutanolysin digestion was almost superimposable after separation by high-performance liquid chromatography (HPLC) (see Fig. S3 in the supplemental material). There is no evidence that the UFAs applied at the concentrations used had an effect on peptidoglycan cross-linking or composition. Thus, the increased cytokine production is due to the incorporation of UFAs into the lipid moiety of lipoproteins.
Supplied FAs did not significantly affect growth, protein release, or Lpl1 expression.
We also verified whether the addition of FAs to the culture medium TSB had an influence on growth or protein release into the supernatant. Growth analysis showed that they had no apparent influence on growth in USA300 JE2 (see Fig. S4A in the supplemental material). The increased immune response could also not be due to an increased release of proteins into the supernatant, as in the presence of FAs, there was no increased protein content in the supernatant observed (see Fig. S4B); in contrast, with some FAs such as C16:1 and C18:2 FAs, the protein amount was even slightly decreased. To check whether lipoprotein expression was influenced by the FAs, we determined Lpl1 expression by Western blotting (see Fig. S4C); there was no apparent difference in Lpl1 amount in total protein extraction. Lpl1 is encoded by the first gene of the tandem lipoprotein cluster and can be regarded as a prototype lipoprotein (39). Altogether, the enhanced innate immune stimulation due to feeding with unsaturated FAs is indeed caused by the incorporation of the UFAs in lipoproteins; it is not due to altered growth, increased release of proteins into the supernatant due to increased membrane leakiness, or enhanced expression of lipoproteins.
DISCUSSION
The lipid fraction of S. aureus mainly contains iC14:0, iC15:0, aC15:0, C16:0, iC17:0, aC17:0, C18:0, and C20:0 FAs; no FAs below a chain length of C10:0 or above C22:0 were found (46). Most of the FAs were associated with the complex lipids lysyl phosphatidyl glycerol, phosphatidyl glycerol, cardiolipin, monoglycosyl diglyceride, and diglycosyl diglyceride. When we investigated the FA composition of the lipid fraction of S. aureus USA300, we obtained similar results. In the lipid fraction, we found in measurable amounts only saturated FAs with chain lengths of C13:0 to C20:0. Feeding with C6:0, C16:0, or C26:0 FAs did not influence the other FA composition. As only saturated FAs were found, S. aureus is apparently unable to synthesize UFAs. However, when the cells were fed with C16:1, C18:1, or C18:2 UFFAs, they were not only incorporated into lipids but also elongated by two carbon units: C16:1 was elongated to C18:1 and C20:1, C18:1 was elongated to C20:1, and C18:2 was elongated to C20:2. The results are summarized in Table 1.
TABLE 1.
FA composition in the lipid fraction of aerobically grown S. aureus USA300 JE2 as well as incorporation and elongation of fed UFFAs
| Chain length of FAMEa | Presence of FAME in S. aureus |
||||
|---|---|---|---|---|---|
| In TSB control | Fed with C6:0, C16:0, and C26:0 FAs | Fed with C16:1 FA | Fed with C18:1 FA | Fed with C18:2 FA | |
| C6:0 | − | − | − | − | − |
| C13:0 | + | + | + | + | + |
| iC14:0 | + | + | + | + | + |
| nC14:0 | + | + | + | + | + |
| iC15:0 | + | + | + | + | + |
| aC15:0 | + | + | + | + | + |
| iC16:0 | + | + | + | + | + |
| C16:1 | − | − | + | − | − |
| nC16:0 | + | + | + | + | + |
| iC17:0 | + | + | + | + | + |
| aC17:0 | + | + | + | + | + |
| nC17:0 | + | + | + | + | + |
| iC18:0 | + | + | + | + | + |
| C18:2 | − | − | − | − | + |
| C18:1 | − | − | + | + | − |
| nC18:0 | + | + | + | + | + |
| iC19:0 | + | + | + | + | + |
| aC19:0 | + | + | + | + | + |
| C20:2 | − | − | − | − | + |
| C20:1 | − | − | + | + | − |
| C20:0 | + | + | + | + | + |
| C26:0 | − | − | − | − | − |
FAMEs, fatty acid methyl esters. The FAMEs shown in boldface type are not found in S. aureus cultured in TSB medium as a control.
So far, FAs have been studied mainly with respect to their antimicrobial activity (8, 9, 25, 47). In our feeding studies, we therefore took care to apply a subinhibitory concentration of UFFAs. As shown in Fig. S1 in the supplemental material, a concentration of 50 μM hardly affected the growth of USA300. This concentration was close to the in vivo situation. Nasal fluid, for example, was described to have a concentration of FFAs of about 35 μM, which was sufficient to inhibit growth of Pseudomonas aeruginosa but not of S. aureus (44).
New insight into how exogenous FAs are activated and incorporated into sn-glycerol-3-phosphates to produce phospholipids came from the group of Charles O. Rock (48). They identified an FA kinase that phosphorylates incoming FAs, which is most likely the first step in FA activation. The FA kinase is part of a multiple-enzyme complex consisting of FakAB1B2: FakA represents the FA kinase, FakB1 selectively binds saturated FAs, and FakB2 binds unsaturated FAs. The stepwise diacylation of sn-glycerol-3-phosphates is then carried out by two enzymes: PlsY transfers the FA from acyl-phosphate to the 1 position, while PlsC uses acyl acyl carrier protein (acyl-ACP) as the substrate for acylation at the 2 position (29). Acyl-ACP is produced by PlsX, which catalyzes the interconversion of acyl-ACP and acyl-phosphate. The observed elongation of the exogenous FAs is carried out via acyl-ACP by the FASII system. The previously described elongation of oleic acid (C18:1Δ9) to C20:1Δ11 in S. aureus suggests that unlike in the case of E. coli, exogenous FAs have direct access to the FASII system (49).
While the steps in the activation and incorporation of externally supplied FAs into sn-glycerol-3-phosphates are comparatively well studied, little is known about whether unsaturated FAs are incorporated into the lipid moiety of lipoproteins and whether such incorporation has an effect on innate immune activation. Feeding of USA300 JE2 with three predominant skin UFFAs—palmitoleic acid (C16:1), oleic acid (C18:1), and linoleic acid (C18:2)—led to their incorporation into the lipid moiety of the lipopeptide SitC18 (Fig. 3). In bacteria, the transfer of an sn-1,2-diacylglyceryl group from phosphatidylglycerol to prolipoproteins is catalyzed by the membrane-bound diacylglyceryl transferase encoded by the lgt gene (31, 50). An lgt mutant of S. aureus is unable to carry out the lipidation of prelipoproteins, and such mutants are severely affected in innate immune stimulation (31, 35).
The most surprising result was, however, that S. aureus cells fed with UFFAs enhanced significantly the activation of human immune cells, like Mono Mac 6 cells or PBMCs, in a TLR2-dependent way (Fig. 5 to 7). It is unlikely that the increased TLR2 stimulation is caused by increased phagocytosis. This could play a role in Mono Mac 6 cells, but we see the effect also in the nonprofessional HEK-TLR2 cells. Therefore, phagocytosis alone could not account for the increased immune stimulation. There are several pieces of evidence that this activation is due to the presence of UFFAs in the lipid moiety of lipoproteins: (i) UFFAs alone showed no signaling activity; (ii) signaling was dependent on TLR2, the receptor for lipoproteins; (iii) signaling was Lgt dependent; and (iv) the supplied FAs did not significantly affect growth, protein release, or expression of the model lipoprotein Lpl1. As in S. aureus the lipid moiety of lipoproteins is crucial for TLR2-mediated activation of the innate immunity, it was not astonishing that the lgt mutant was uninfluenced by feeding with UFFAs (31, 51). The highest signaling activity was always observed with linoleic acid (C18:2), which caused a 2- to 3-fold increase in cytokine production.
Such an increase in innate immune response could play a role in vivo. The epidermis consists of 90% keratinocytes, which represent the immune sentinels in the skin and consequently provide a first line of defense against microbial pathogens. They express almost all TLRs, which is crucial for promoting skin immune responses to adherent bacteria (52). Activation of these receptors on human keratinocytes leads to a predominant TH1-type immune response and to the production of type I interferons (IFNs) (53). Moreover, keratinocytes also secrete, or are induced to release, numerous cytokines, including IL-1, IL-6, IL-10, IL-18, and TNF-α. For example, the S. aureus lipoprotein SitC colocalizes with TLR2 in murine keratinocytes and elicits both intracellular TLR2 accumulation and release of cytokines (54). Other lipoproteins, such as those encoded by the “lipoprotein-like” gene cluster (lpl) present in epidemic S. aureus strains, like USA300, trigger invasion into human keratinocytes (39). Recently, it has been shown that cutaneous S. aureus can negatively regulate skin-driven immune responses by inducing myeloid-derived suppressor cells (MDSCs) via TLR2 to -6 activation by diacylated lipopeptides (55).
We think that the incorporation of unsaturated fatty acids in lipoproteins increases the affinity for TLR2. Particularly synthetic lipopeptides have been extensively studied, as they have strong adjuvant activity and are also potential vaccine candidates (56–58). The self-adjuvanting effect of lipopeptides was demonstrated to act through stimulation of TLR2 and dendritic cell (DC) activation (59, 60). By screening a combinatorial lipohexapeptide collection based on synthetic Pam3Cys-lipopeptides in an in vitro IL-8 induction assay, it has been demonstrated that both the peptide sequence and the lipid moiety influence signaling (56). Indeed, it has already been shown that the ester-bound oleic acid and/or linoleic acid gave best the results in the IL-8 release assay. However, host cells may react differently on lipopeptides by heterodimerization of TLR2 with TLR1 or TLR6, which expands the ligand spectrum (61). It is therefore easily conceivable that incorporation of different FAs into lipoproteins influences the immune response.
An increased immune response is not beneficial for the pathogen but is beneficial for the host. We think that the uptake of unsaturated FAs, particularly the uptake of linoleic acid (C18:2) and its incorporation into lipoproteins, is an Achilles' heel of this pathogen and may explain species-specific preferences of skin colonization; S. aureus and Staphylococcus epidermidis, for example, are predominant and persistent in the nares, whereas S. epidermidis and Staphylococcus hominis were the most predominant and persistent staphylococci isolated from the axillae, head, legs, and arms (62). In this respect, it would be interesting to also study the effect of UFAs in other staphylococcal species.
Conclusion.
UFFAs do not only act as antimicrobial FAs affecting the bacterial membrane. Here we show that they are incorporated into the bacterial phospholipids and lipoproteins. The presence of UFA in lipoproteins, particularly the presence of linoleic acid, significantly increased the innate immune-stimulating activity of the bacterial cells, as indicated by enhanced production of proinflammatory cytokines. As the stimulation is TLR2 dependent, we assume that the presence of UFA esters in the lipid moiety of lipoproteins increases the affinity of these lipoproteins for the TLR2 receptor domain. We describe here a further function of the UFFAs on mammalian skin in defending against and controlling bacterial colonization and infection. As previously shown, keratinocytes, which are the predominant cells in the epidermis, are very responsive to Lpp (39, 54). Thus, we assume that they respond first upon exposure to UFA-Lpp. Figure 8 illustrates the interaction of skin UFFAs and enhanced immune stimulation by S. aureus.
FIG 8.
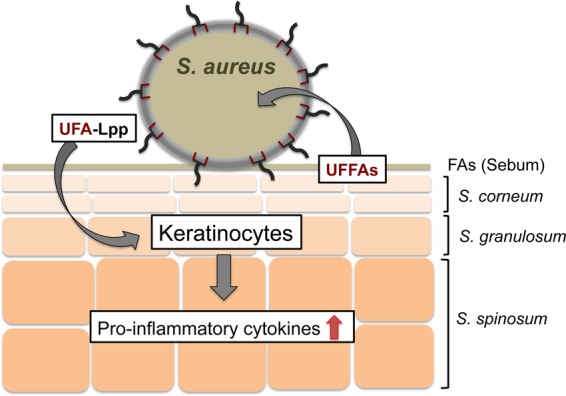
Schematic representation of the interplay of skin's UFFAs and enhanced immune stimulation by S. aureus. UFFAs produced by the sebum glands are incorporated into the S. aureus phospholipids and lipoproteins. Particularly linoleic acid in lipoproteins increased TLR2 dependently on the immune response of skin cells, leading to an increased production of proinflammatory cytokines. The UFFAs on the skin act as bait for bacteria; as soon as bacteria incorporate the bait into their lipoproteins, they become more visible to the host's innate immune system.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dogan Doruk Demircioglu and Daniel Kühner for help with the peptidoglycan analysis.
Funding Statement
This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG; GO 371/9-1, PE805/5-1, SFB766, SFB685, TRR34, and TRR156), the Baden-Württemberg Biosynthesis netWork (BW2), and the Fortüne Program of the Medical Faculty, University of Tübingen.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00822-15.
REFERENCES
- 1.Swamy M, Jamora C, Havran W, Hayday A. 2010. Epithelial decision makers: in search of the ‘epimmunome.’ Nat Immunol 11:656–665. doi: 10.1038/ni.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Volz T, Kaesler S, Biedermann T. 2012. Innate immune sensing 2.0—from linear activation pathways to fine tuned and regulated innate immune networks. Exp Dermatol 21:61–69. doi: 10.1111/j.1600-0625.2011.01393.x. [DOI] [PubMed] [Google Scholar]
- 3.Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, Deming C, Quinones M, Koo L, Conlan S, Spencer S, Hall JA, Dzutsev A, Kong H, Campbell DJ, Trinchieri G, Segre JA, Belkaid Y. 2012. Compartmentalized control of skin immunity by resident commensals. Science 337:1115–1119. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lowy FD. 1998. Staphylococcus aureus infections. N Engl J Med 339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 5.Nicolaides N. 1974. Skin lipids: their biochemical uniqueness. Science 186:19–26. doi: 10.1126/science.186.4158.19. [DOI] [PubMed] [Google Scholar]
- 6.Ricketts CR, Squire JR, Topley E. 1951. Human skin lipids with particular reference to the self-sterilising power of the skin. Clin Sci 10:80–110. [PubMed] [Google Scholar]
- 7.Wille JJ, Kydonieus A. 2003. Palmitoleic acid isomer (C16:1delta6) in human skin sebum is effective against gram-positive bacteria. Skin Pharmacol Appl Skin Physiol 16:176–187. doi: 10.1159/000069757. [DOI] [PubMed] [Google Scholar]
- 8.Drake DR, Brogden KA, Dawson DV, Wertz PW. 2008. Thematic review series: skin lipids. Antimicrobial lipids at the skin surface. J Lipid Res 49:4–11. [DOI] [PubMed] [Google Scholar]
- 9.Nakatsuji T, Kao MC, Fang JY, Zouboulis CC, Zhang L, Gallo RL, Huang CM. 2009. Antimicrobial property of lauric acid against Propionibacterium acnes: its therapeutic potential for inflammatory acne vulgaris. J Invest Dermatol 129:2480–2488. doi: 10.1038/jid.2009.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skrivanová E, Marounek M, Dlouha G, Kanka J. 2005. Susceptibility of Clostridium perfringens to C-C fatty acids. Lett Appl Microbiol 41:77–81. doi: 10.1111/j.1472-765X.2005.01709.x. [DOI] [PubMed] [Google Scholar]
- 11.Kelsey JA, Bayles KW, Shafii B, McGuire MA. 2006. Fatty acids and monoacylglycerols inhibit growth of Staphylococcus aureus. Lipids 41:951–961. doi: 10.1007/s11745-006-5048-z. [DOI] [PubMed] [Google Scholar]
- 12.Holland KT, Ingham E, Cunliffe WJ. 1981. A review, the microbiology of acne. J Appl Bacteriol 51:195–215. doi: 10.1111/j.1365-2672.1981.tb01234.x. [DOI] [PubMed] [Google Scholar]
- 13.Götz F, Verheij HM, Rosenstein R. 1998. Staphylococcal lipases: molecular characterisation, secretion, and processing. Chem Phys Lipids 93:15–25. doi: 10.1016/S0009-3084(98)00025-5. [DOI] [PubMed] [Google Scholar]
- 14.Simons JW, Götz F, Egmond MR, Verheij HM. 1998. Biochemical properties of staphylococcal (phospho)lipases. Chem Phys Lipids 93:27–37. doi: 10.1016/S0009-3084(98)00026-7. [DOI] [PubMed] [Google Scholar]
- 15.Simons JW, Adams H, Cox RC, Dekker N, Götz F, Slotboom AJ, Verheij HM. 1996. The lipase from Staphylococcus aureus. Expression in Escherichia coli, large-scale purification and comparison of substrate specificity to Staphylococcus hyicus lipase. Eur J Biochem 242:760–769. [DOI] [PubMed] [Google Scholar]
- 16.Cadieux B, Vijayakumaran V, Bernards MA, McGavin MJ, Heinrichs DE. 2014. Role of lipase from community-associated methicillin-resistant Staphylococcus aureus strain USA300 in hydrolyzing triglycerides into growth-inhibitory free fatty acids. J Bacteriol 196:4044–4056. doi: 10.1128/JB.02044-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujie T, Shikiji T, Uchida N, Urano Y, Nagae H, Arase S. 1996. Culture of cells derived from the human sebaceous gland under serum-free conditions without a biological feeder layer or specific matrices. Arch Dermatol Res 288:703–708. doi: 10.1007/BF02505281. [DOI] [PubMed] [Google Scholar]
- 18.Zouboulis CC, Seltmann H, Neitzel H, Orfanos CE. 1999. Establishment and characterization of an immortalized human sebaceous gland cell line (SZ95). J Invest Dermatol 113:1011–1020. doi: 10.1046/j.1523-1747.1999.00771.x. [DOI] [PubMed] [Google Scholar]
- 19.Nakatsuji T, Kao MC, Zhang L, Zouboulis CC, Gallo RL, Huang CM. 2010. Sebum free fatty acids enhance the innate immune defense of human sebocytes by upregulating beta-defensin-2 expression. J Invest Dermatol 130:985–994. doi: 10.1038/jid.2009.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lampe MA, Burlingame AL, Whitney J, Williams ML, Brown BE, Roitman E, Elias PM. 1983. Human stratum corneum lipids: characterization and regional variations. J Lipid Res 24:120–130. [PubMed] [Google Scholar]
- 21.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. 2009. Bacterial community variation in human body habitats across space and time. Science 326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coates R, Moran J, Horsburgh MJ. 2014. Staphylococci: colonizers and pathogens of human skin. Future Microbiol 9:75–91. doi: 10.2217/fmb.13.145. [DOI] [PubMed] [Google Scholar]
- 23.Galbraith H, Miller TB. 1973. Effect of long chain fatty acids on bacterial respiration and amino acid uptake. J Appl Bacteriol 36:659–675. doi: 10.1111/j.1365-2672.1973.tb04151.x. [DOI] [PubMed] [Google Scholar]
- 24.Greenway DL, Dyke KG. 1979. Mechanism of the inhibitory action of linoleic acid on the growth of Staphylococcus aureus. J Gen Microbiol 115:233–245. doi: 10.1099/00221287-115-1-233. [DOI] [PubMed] [Google Scholar]
- 25.Cartron ML, England SR, Chiriac AI, Josten M, Turner R, Rauter Y, Hurd A, Sahl HG, Jones S, Foster SJ. 2014. Bactericidal activity of the human skin fatty acid cis-6-hexadecanoic acid on Staphylococcus aureus. Antimicrob Agents Chemother 58:3599–3609. doi: 10.1128/AAC.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neumann Y, Ohlsen K, Donat S, Engelmann S, Kusch H, Albrecht D, Cartron M, Hurd A, Foster SJ. 2015. The effect of skin fatty acids on Staphylococcus aureus. Arch Microbiol 197:245–267. doi: 10.1007/s00203-014-1048-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clarke SR, Mohamed R, Bian L, Routh AF, Kokai-Kun JF, Mond JJ, Tarkowski A, Foster SJ. 2007. The Staphylococcus aureus surface protein IsdA mediates resistance to innate defenses of human skin. Cell Host Microbe 1:199–212. doi: 10.1016/j.chom.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 28.Köhler T, Weidenmaier C, Peschel A. 2009. Wall teichoic acid protects Staphylococcus aureus against antimicrobial fatty acids from human skin. J Bacteriol 191:4482–4484. doi: 10.1128/JB.00221-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parsons JB, Frank MW, Jackson P, Subramanian C, Rock CO. 2014. Incorporation of extracellular fatty acids by a fatty acid kinase-dependent pathway in Staphylococcus aureus. Mol Microbiol 92:234–245. doi: 10.1111/mmi.12556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parsons JB, Yao J, Frank MW, Jackson P, Rock CO. 2012. Membrane disruption by antimicrobial fatty acids releases low-molecular-weight proteins from Staphylococcus aureus. J Bacteriol 194:5294–5304. doi: 10.1128/JB.00743-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stoll H, Dengjel J, Nerz C, Götz F. 2005. Staphylococcus aureus deficient in lipidation of prelipoproteins is attenuated in growth and immune activation. Infect Immun 73:2411–2423. doi: 10.1128/IAI.73.4.2411-2423.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hashimoto M, Tawaratsumida K, Kariya H, Kiyohara A, Suda Y, Krikae F, Kirikae T, Götz F. 2006. Not lipoteichoic acid but lipoproteins appear to be the dominant immunobiologically active compounds in Staphylococcus aureus. J Immunol 177:3162–3169. doi: 10.4049/jimmunol.177.5.3162. [DOI] [PubMed] [Google Scholar]
- 33.Takeda K, Takeuchi O, Akira S. 2002. Recognition of lipopeptides by Toll-like receptors. J Endotoxin Res 8:459–463. doi: 10.1177/09680519020080060101. [DOI] [PubMed] [Google Scholar]
- 34.Takeuchi O, Kawai T, Muhlradt PF, Morr M, Radolf JD, Zychlinsky A, Takeda K, Akira S. 2001. Discrimination of bacterial lipoproteins by Toll-like receptor 6. Int Immunol 13:933–940. doi: 10.1093/intimm/13.7.933. [DOI] [PubMed] [Google Scholar]
- 35.Schmaler M, Jann NJ, Ferracin F, Landolt LZ, Biswas L, Götz F, Landmann R. 2009. Lipoproteins in Staphylococcus aureus mediate inflammation by TLR2 and iron-dependent growth in vivo. J Immunol 182:7110–7118. doi: 10.4049/jimmunol.0804292. [DOI] [PubMed] [Google Scholar]
- 36.Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 37.Geiger T, Francois P, Liebeke M, Fraunholz M, Goerke C, Krismer B, Schrenzel J, Lalk M, Wolz C. 2012. The stringent response of Staphylococcus aureus and its impact on survival after phagocytosis through the induction of intracellular PSMs expression. PLoS Pathog 8:e1003016. doi: 10.1371/journal.ppat.1003016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mauthe M, Yu W, Krut O, Kronke M, Götz F, Robenek H, Proikas-Cezanne T. 2012. WIPI-1 positive autophagosome-like vesicles entrap pathogenic Staphylococcus aureus for lysosomal degradation. Int J Cell Biol 2012:179207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen MT, Kraft B, Yu W, Demicrioglu DD, Hertlein T, Burian M, Schmaler M, Boller K, Bekeredjian-Ding I, Ohlsen K, Schittek B, Götz F. 2015. The νSaα specific lipoprotein like cluster (lpl) of S. aureus USA300 contributes to immune stimulation and invasion in human cells. PLoS Pathog 11:e1004984. doi: 10.1371/journal.ppat.1004984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heinzelmann E, Berger S, Müller C, Hartner T, Poralla K, Wohlleben W, Schwartz D. 2005. An acyl-CoA dehydrogenase is involved in the formation of the Delta cis3 double bond in the acyl residue of the lipopeptide antibiotic friulimicin in Actinoplanes friuliensis. Microbiology 151:1963–1974. doi: 10.1099/mic.0.27844-0. [DOI] [PubMed] [Google Scholar]
- 41.Ziegler-Heitbrock HW, Schraut W, Wendelgass P, Strobel M, Sternsdorf T, Weber C, Aepfelbacher M, Ehlers M, Schutt C, Haas JG. 1994. Distinct patterns of differentiation induced in the monocytic cell line Mono Mac 6. J Leukoc Biol 55:73–80. [DOI] [PubMed] [Google Scholar]
- 42.Kühner D, Stahl M, Demircioglu DD, Bertsche U. 2014. From cells to muropeptide structures in 24 h: peptidoglycan mapping by UPLC-MS. Sci Rep 4:7494. doi: 10.1038/srep07494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nega M, Dube L, Kull M, Ziebandt AK, Ebner P, Albrecht D, Krismer B, Rosenstein R, Hecker M, Götz F. 2015. Secretome analysis revealed adaptive and non-adaptive responses of the Staphylococcus carnosus femB mutant. Proteomics 15:1268–1279. doi: 10.1002/pmic.201400343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Do TQ, Moshkani S, Castillo P, Anunta S, Pogosyan A, Cheung A, Marbois B, Faull KF, Ernst W, Chiang SM, Fujii G, Clarke CF, Foster K, Porter E. 2008. Lipids including cholesteryl linoleate and cholesteryl arachidonate contribute to the inherent antibacterial activity of human nasal fluid. J Immunol 181:4177–4187. doi: 10.4049/jimmunol.181.6.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kenny JG, Ward D, Josefsson E, Jonsson IM, Hinds J, Rees HH, Lindsay JA, Tarkowski A, Horsburgh MJ. 2009. The Staphylococcus aureus response to unsaturated long chain free fatty acids: survival mechanisms and virulence implications. PLoS One 4:e4344. doi: 10.1371/journal.pone.0004344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.White DC, Frerman FE. 1968. Fatty acid composition of the complex lipids of Staphylococcus aureus during the formation of the membrane-bound electron transport system. J Bacteriol 95:2198–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arsic B, Zhu Y, Heinrichs DE, McGavin MJ. 2012. Induction of the staphylococcal proteolytic cascade by antimicrobial fatty acids in community acquired methicillin resistant Staphylococcus aureus. PLoS One 7:e45952. doi: 10.1371/journal.pone.0045952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parsons JB, Broussard TC, Bose JL, Rosch JW, Jackson P, Subramanian C, Rock CO. 2014. Identification of a two-component fatty acid kinase responsible for host fatty acid incorporation by Staphylococcus aureus. Proc Natl Acad Sci U S A 111:10532–10537. doi: 10.1073/pnas.1408797111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parsons JB, Frank MW, Subramanian C, Saenkham P, Rock CO. 2011. Metabolic basis for the differential susceptibility of Gram-positive pathogens to fatty acid synthesis inhibitors. Proc Natl Acad Sci U S A 108:15378–15383. doi: 10.1073/pnas.1109208108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pailler J, Aucher W, Pires M, Buddelmeijer N. 2012. Phosphatidylglycerol::prolipoprotein diacylglyceryl transferase (Lgt) of Escherichia coli has seven transmembrane segments, and its essential residues are embedded in the membrane. J Bacteriol 194:2142–2151. doi: 10.1128/JB.06641-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmaler M, Jann NJ, Götz F, Landmann R. 2010. Staphylococcal lipoproteins and their role in bacterial survival in mice. Int J Med Microbiol 300:155–160. doi: 10.1016/j.ijmm.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 52.Kobayashi M, Yoshiki R, Sakabe J, Kabashima K, Nakamura M, Tokura Y. 2009. Expression of Toll-like receptor 2, NOD2 and dectin-1 and stimulatory effects of their ligands and histamine in normal human keratinocytes. Br J Dermatol 160:297–304. doi: 10.1111/j.1365-2133.2008.08897.x. [DOI] [PubMed] [Google Scholar]
- 53.Nestle FO, Di Meglio P, Qin JZ, Nickoloff BJ. 2009. Skin immune sentinels in health and disease. Nat Rev Immunol 9:679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Müller P, Müller-Anstett M, Wagener J, Gao Q, Kaesler S, Schaller M, Biedermann T, Götz F. 2010. The Staphylococcus aureus lipoprotein SitC colocalizes with Toll-like receptor 2 (TLR2) in murine keratinocytes and elicits intracellular TLR2 accumulation. Infect Immun 78:4243–4250. doi: 10.1128/IAI.00538-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Skabytska Y, Wolbing F, Gunther C, Koberle M, Kaesler S, Chen KM, Guenova E, Demircioglu D, Kempf WE, Volz T, Rammensee HG, Schaller M, Rocken M, Götz F, Biedermann T. 2014. Cutaneous innate immune sensing of Toll-like receptor 2-6 ligands suppresses T cell immunity by inducing myeloid-derived suppressor cells. Immunity 41:762–775. doi: 10.1016/j.immuni.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 56.Spohn R, Buwitt-Beckmann U, Brock R, Jung G, Ulmer AJ, Wiesmüller KH. 2004. Synthetic lipopeptide adjuvants and Toll-like receptor 2—structure-activity relationships. Vaccine 22:2494–2499. doi: 10.1016/j.vaccine.2003.11.074. [DOI] [PubMed] [Google Scholar]
- 57.Schild H, Deres K, Wiesmüller KH, Jung G, Rammensee HG. 1991. Efficiency of peptides and lipopeptides for in vivo priming of virus-specific cytotoxic T cells. Eur J Immunol 21:2649–2654. doi: 10.1002/eji.1830211102. [DOI] [PubMed] [Google Scholar]
- 58.Deres K, Schild H, Wiesmüller KH, Jung G, Rammensee HG. 1989. In vivo priming of virus-specific cytotoxic T lymphocytes with synthetic lipopeptide vaccine. Nature 342:561–564. doi: 10.1038/342561a0. [DOI] [PubMed] [Google Scholar]
- 59.Zaman M, Toth I. 2013. Immunostimulation by synthetic lipopeptide-based vaccine candidates: structure-activity relationships. Front Immunol 4:318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zaman M, Abdel-Aal AB, Fujita Y, Phillipps KS, Batzloff MR, Good MF, Toth I. 2012. Immunological evaluation of lipopeptide group A streptococcus (GAS) vaccine: structure-activity relationship. PLoS One 7:e30146. doi: 10.1371/journal.pone.0030146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Farhat K, Riekenberg S, Heine H, Debarry J, Lang R, Mages J, Buwitt-Beckmann U, Roschmann K, Jung G, Wiesmüller KH, Ulmer AJ. 2008. Heterodimerization of TLR2 with TLR1 or TLR6 expands the ligand spectrum but does not lead to differential signaling. J Leukoc Biol 83:692–701. [DOI] [PubMed] [Google Scholar]
- 62.Kloos WE, Musselwhite MS. 1975. Distribution and persistence of Staphylococcus and Micrococcus species and other aerobic bacteria on human skin. Appl Microbiol 30:381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.