Abstract
The type III secretion system (T3SS) of Edwardsiella tarda is crucial for its intracellular survival and pathogenesis in fish. The orf13 gene (escE) of E. tarda is located 84 nucleotides (nt) upstream of esrC in the T3SS gene cluster. We found that EscE is secreted and translocated in a T3SS-dependent manner and that amino acids 2 to 15 in the N terminus were required for a completely functional T3SS in E. tarda. Deletion of escE abolished the secretion of T3SS translocators, as well as the secretion and translocation of T3SS effectors, but did not influence their intracellular protein levels in E. tarda. Complementation of the escE mutant with a secretion-incompetent EscE derivative restored the secretion of translocators and effectors. Interestingly, the effectors that were secreted and translocated were positively correlated with the EscE protein level in E. tarda. The escE mutant was attenuated in the blue gourami fish infection model, as its 50% lethal dose (LD50) increased to 4 times that of the wild type. The survival rate of the escE mutant-strain-infected fish was 69%, which was much higher than that of the fish infected with the wild-type bacteria (6%). Overall, EscE represents a secreted T3SS regulator that controls effector injection and translocator secretion, thus contributing to E. tarda pathogenesis in fish. The homology of EscE within the T3SSs of other bacterial species suggests that the mechanism of secretion and translocation control used by E. tarda may be commonly used by other bacterial pathogens.
INTRODUCTION
Type III secretion systems (T3SSs) are crucial structures used by many Gram-negative bacterial pathogens for the delivery of virulence factors (1). Three classes of proteins are secreted from the T3SS: (i) subunits of a surface-exposed needle-like structure, (ii) translocon proteins that form pores in host cell membranes, and (iii) effectors that pass through the entire T3SS to host cells, where they suppress innate immunity or remodel the host cells to benefit bacterial survival (2).
Bacteria use multiple strategies to ensure a temporal and hierarchical release of proteins through their T3SSs (3–8). Within Edwardsiella tarda, a number of regulatory systems are used, including the two-component EsrA-EsrB system, which controls the expression and secretion of T3SS proteins in conjunction with a third regulator in the T3SS gene cluster called EsrC (9); PhoP-PhoQ, which detects changes in the environmental temperature and Mg2+ concentration to coordinate the regulation of virulence through EsrB (10); and the PhoB-PhoR system, which senses phosphate. Finally, the ferric uptake regulator (Fur) senses the iron concentration in E. tarda to regulate the expression of proteins to be secreted from the T3SS through EsrC (11).
E. tarda T3SS genes are clustered within seven transcriptional units (11). Thirty-four potential open reading frames included in the T3SS gene cluster are directly involved in the assembly and regulation of the T3SS (12, 13). Included among their products are the Esa proteins, which are primarily involved in the assembly of the syringe-like type III secretion (T3S) injectisome. Additionally, the effectors EseG (14) and EseJ (13), as well as the translocators EseB, EseC, and EseD, all use the T3S machinery (12). Stable expression of the effectors and translocators requires the chaperones EscA for EseC (15), EscC for EseB and EseD (16), and EscB for the effector EseG (14).
One gene carried within the E. tarda T3SS gene cluster encodes a small (74-amino-acid [aa]) hypothetical protein called EscE. Using Delta-BLAST for a domain search, the EscE protein was found to belong to the type III secretion system YscE family. Here, we show that, unlike the YscE family proteins, which function as chaperones for the T3SS needle proteins, EscE itself could be secreted and translocated into host cells. EscE expression also triggers the secretion of other T3SS translocators and effectors, and the effectors that are translocated are positively correlated with the intracellular EscE protein level.
MATERIALS AND METHODS
Cells and cell culture.
J774A.1 murine macrophages and HeLa cells were cultured in Dulbecco minimal essential medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Gibco) and 10 mM l-glutamine in a 5% CO2 atmosphere.
Bacterial strains, media, and culture conditions.
The bacterial strains and plasmids used in this study are described in Table 1. E. tarda PPD130/91 (17) and its derived strains were grown at 25°C in tryptic soy broth (TSB) (BD Biosciences) or DMEM (Gibco), while Escherichia coli strains were cultured at 37°C in Luria-Bertani broth (LB) (BD Biosciences). To induce T3SS proteins, E. tarda strains were grown in DMEM at 25°C in a 5% (vol/vol) CO2 atmosphere. When required, the medium was supplemented with appropriate antibiotics at the following concentrations: 100 μg/ml ampicillin (Amp) (Sigma), 12.5 μg/ml colistin (Col) (Sigma), 100 μg/ml gentamicin (Gem) (Sigma), 15 μg/ml tetracycline (Tet) (Amresco), and 34 μg/ml chloramphenicol (Cm) (Amresco).
TABLE 1.
Strains and plasmid vectors used in this study
| Strain or plasmid | Description or relevant characteristicsa | Reference or source |
|---|---|---|
| Strains | ||
| E. tarda | ||
| PPD130/91 | Wild type; Kms Colr Amps; LD50 = 105.0 | 17 |
| ΔescE | PPD130/91 with in-frame deletion of escE | This study |
| ΔesaN | PPD130/91 with in-frame deletion of esaN | 14 |
| wt/pACYC-escE-2HA | PPD130/91 with pACYC-escE-2HA | This study |
| ΔesaN/pACYC-escE-2HA | ΔesaN with pACYC-escE-2HA | This study |
| ΔescE/pACYC-escE-2HA | ΔescE with pACYC-escE-2HA | This study |
| ΔescE/pACYC-escEΔ2–15-2HA | ΔescE with pACYC-escEΔ2–15-2HA | This study |
| ΔescE/pACYC-gst-2HA | ΔescE with pACYC-gst-2HA | This study |
| ΔescE/pACYC-gst::escE-2HA | ΔescE with pACYC-gst::escE-2HA | This study |
| wt/pACYC-eseG::cyaA | PPD130/91 with pACYC-eseG::cyaA | 13 |
| ΔesaN/pACYC-eseG::cyaA | ΔesaN with pACYC-eseG::cyaA | 13 |
| wt/pACYC-escE::cyaA | PPD130/91 with pACYC-escE::cyaA | This study |
| ΔesaN/pACYC-escE::cyaA | ΔesaN with pACYC-escE::cyaA | This study |
| ΔescE/pJN105-gst::escE-2HA | ΔescE with pJN105-gst::escE-2HA | This study |
| ΔescE/pJN105-gst::escE-2HA/pACYC-eseG::cya | ΔescE with pJN105-gst::escE-2HA and pACYC-eseG::cyaA | This study |
| E. coli | ||
| DH5α | α complementation; Amps | Stratagene |
| S17-1 λpir | RK2 tra regulon; pir; host for pir-dependent plasmid | 19 |
| Plasmids | ||
| pMD18-T | Cloning vector; Ampr | TaKaRa |
| pRE112 | pGP704 suicide plasmid; pir dependent; Cmr oriT oriV sacB | 18 |
| pACYC-184 | Tetr Cmr | Amersham |
| pJN105 | Arabinose-inducible gene expression vector; araC-PBAD; Gmr | 20 |
| pRE112-escE | pRE112 containing escE flanking fragments with aa 2 to 74 deleted | This study |
| pACYC-escE-2HA | pACYC184 with escE-2HA; Tetr | This study |
| pACYC-escEΔ2–15-2HA | pACYC184 with escE lacking 2nd to 15th amino acid; Tetr | This study |
| pACYC-escE::cyaA | pACYC184 with cyaA fused to C terminus of escE; Cmr | This study |
| pACYC-eseG::cyaA | pACYC184 with cyaA fused to C terminus of full-length eseG gene; Cmr | 13 |
| pJN105-gst::escE-2HA | pJN105 with gst::escE-2HA; Gmr | This study |
| pACYC-gst-2HA | pACYC184 with gst-2HA; Tetr | This study |
| pACYC-gst::escE-2HA | pACYC184 with gst fused to N terminus of escE-2HA; Tetr | This study |
Km, kanamycin; Col, colistin; Amp, ampicillin; Cm, chloramphenicol; Gm, gentamicin; Tet, tetracycline.
Construction of mutants and plasmids.
Nonpolar deletion mutants of escE were generated by sacB-based allelic exchange (18), as described previously (9). In brief, two PCR fragments were generated from PPD130/91 genomic DNA with the primer pairs escE-for plus escE-int-rev and escE-int-for plus escE-rev. The resulting products were a 743-bp fragment containing the upstream region of escE and a 725-bp fragment containing the downstream region of escE. A 16-bp overlap in the sequences permitted their fusion by a second round of PCR using the escE-for and escE-rev primers. The resulting PCR product, with the deletion of the whole coding sequence of escE, was digested and ligated into the KpnI restriction site of the pRE112 suicide vector (18) to create pRE-ΔescE and subsequently transferred to E. coli strain S17-1 λpir (19) for conjugation with E. tarda PPD130/91. Deletion mutant strains were screened on 10% sucrose-tryptic soy agar (TSA) plates and were verified by PCR, SDS-PAGE, and Western blotting analysis. None of the mutant strains tested showed growth defects when cultured in TSB or DMEM. All the primers used in this work are listed in Table 2.
TABLE 2.
Oligonucleotides used in this study
The DNA sequence (including the escE gene and its ribosome binding site) was amplified with the escE-com-for and escE-com-rev primers and ligated into the EcoRI and ScaI restriction sites of pACYC184 (Amersham) to create pACYC-escE-2HA (2HA indicates two repeats of the hemagglutinin tag), which was introduced into the ΔescE mutant for complementing in trans. The influenza virus hemagglutinin (HA) epitope amino acid sequence, YDYDVPDYASL, was used because the expression and secretion of the tagged protein could be easily examined using an anti-HA antibody. By overlapping PCR with the primer pairs escE-com-for plus pACYC-escE-int-rev and pACYC-escE-int-for plus escE-com-rev, the upstream flanking sequence, including the ribosome binding site and the first codon of escE, was fused to the downstream flanking sequence escE16–74-2HA. The resulting PCR product was ligated into the EcoRI and ScaI sites of pACYC-184 to obtain pACYC-escEΔ2–15-2HA. This plasmid was transformed into wild-type (wt) E. tarda and the ΔescE mutant to learn whether the N terminus was necessary for EscE secretion.
To produce EscE that could not pass through the T3SS secretion machine, gst::escE-2HA (synthesized by Genscript, China) was ligated into the EcoRV restriction site of pUC57-simple to obtain pUC-gst::escE-2HA, from which gst::escE-2HA was digested and ligated into the EcoRI and NcoI sites of pACYC-184 to produce pACYC-gst::escE-2HA. Next, using pUC57-gst::escE-2HA as the template, gst-2HA was amplified with the primer pair pACYC-gst-for plus pACYC-gst-rev and ligated into the EcoRI and NcoI sites of pACYC-184 to produce pACYC-gst-2HA. pACYC-gst-2HA was used as the control for pACYC-gst::escE-2HA. The two plasmids were transformed into wild-type E. tarda and the ΔescE mutant to learn whether EscE secretion triggered secretion of the T3SS translocon and effectors.
Using pUC57-gst::escE-2HA as the template and the pJN105-gst::escE-for and pJN105-gst::escE-rev primers, gst::escE-2HA was amplified and ligated into the EcoRI and XbaI sites of pJN105 (20) to produce pJN-gst::escE-2HA. This plasmid was transformed into the ΔescE mutant, and GST::EscE expression was induced by including l-arabinose in the culture media.
All of the plasmids constructed as described above were verified by DNA sequencing.
Expression and secretion assays.
E. tarda strains cultured overnight were subcultured at 1:200 into DMEM and grown without shaking at 25°C for 24 h. The extracellular proteins (ECP) and the total bacterial proteins (TBP) were prepared as described by Zheng and Leung (21). Five percent of TBP and 10% of ECP were separated by Tricine–SDS-PAGE and transferred onto polyvinylidene difluoride (PVDF) membranes (0.22 μm). After fixation in 2.5% glutaraldehyde, the membrane was subjected to immunoblotting to detect EscE. TBP and ECP were also separated by 12% SDS-PAGE and subjected to Coomassie blue staining or were transferred onto PVDF membranes (0.45 μm) for immunoblotting. The membranes were probed with rabbit anti-EscE antibody at a dilution of 1:1,000, rabbit anti-EseC at 1:1,000, rabbit anti-EseD at 1:1,000, rabbit anti-EseG at 1:2,000 (14), rabbit anti-EseJ at 1:2,000 (13), and rabbit anti-EvpC at 1: 5,000 (21). The membranes were then probed using a horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG at 1:2,500 (Millipore). The EscE antibody was raised against amino acids 9 to 22 (DSLRHDPRGHQRQR), the EseC antibody was raised against amino acids 67 to 80 (PLRPPPQRQQDHDP), and the EseD antibody was raised against amino acids 16 to 29 (PPDGHRYVSQDRGG); all of them were produced by Genscript, China, and purified using the specific peptide as the ligand. Some membranes were also probed with the mouse anti-EseB at 1:1,000 (22), mouse anti-DnaK at 1:2,000 (Abcam), or mouse anti-HA at 1:3,000 (Sigma), followed by HRP-conjugated goat anti-mouse IgG (1:2,500) (Millipore). Antigen-antibody complexes were visualized with Super Signal West Pico chemiluminescent substrate (Thermo), followed by exposure using a ChemiDoc MP imaging system (Bio-Rad). SDS-PAGE gel or immune-blotting experiments were repeated at least three times on independently collected samples. Densitometric calculations of immune-blotting bands were conducted on the images of blots using Image Lab 4.1 software.
CyaA-based translocation assay.
CyaA translocation assays were performed following the general procedures of Urbanowski et al. (23). J774A.1 cells were seeded into 24-well plates and infected at a multiplicity of infection (MOI) of 5 with E. tarda strains expressing the EscE::CyaA or the EseG::CyaA fusion protein. J774A.1 cells were also infected with E. tarda strains harboring both pACYC-eseG::cyaA and pJN105-gst::escE-2HA, and the cell culture media were supplemented with l-arabinose at 0 mM, 1 mM, 10 mM, or 50 mM during the infections. At 5 h postinfection, the media were removed, and the cells were washed twice with PBS and then lysed with sample diluents (supplied with the cyclic AMP [cAMP] immunoassay kit) supplemented with 0.2% Triton X-100. After lysis at room temperature for 15 min, the protein concentrations of the supernatants of the cell lysates were examined using a Bradford assay before the evaluation of cAMP levels using a cAMP enzyme immunoassay (EIA) system (Arbor Assays). The translocation efficiency was calculated from the mean of four wells per infection, and the experiment was repeated three times independently.
Single-strain infection in blue gourami fish.
Healthy blue gourami (Trichogaster trichopterus Pallas; 11.38 ± 0.62 g) were infected with E. tarda strains using the procedures of Ling et al. (17). Wild-type E. tarda PPD130/91 and the ΔescE strain were cultured and subcultured separately at 25°C before injection. Doses of 106, 105, and 104 bacteria were injected intramuscularly near the dorsal fins using 10 fish at a time. The 50% lethal dose (LD50) was calculated using the method of Reed and Muench (24). For each strain, the LD50 estimation was performed at least three times. To determine the survival rates of fish infected with the wild-type and ΔescE strains, 16 blue gourami fish per strain were injected at a dosage of 5 × 105 CFU per fish. The mortality was recorded over a period of 7 days postinjection.
The experiments with fish were performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the Chinese Academy of Sciences. The protocol was approved by the Committee on the Ethics of Animal Experiments of the Institute of Hydrobiology (permit number Y213201301). Dissection was performed under 3-aminobenzoic acid ethyl ester methanesulfonate (MS-222) anesthesia.
Statistical analysis.
All data are representative of the results of at least three independent experiments and are presented as means and standard deviations (SD). Comparisons between groups were performed using one-way analysis of variance (ANOVA) with Tukey's multiple-comparison test.
RESULTS
Sequence analysis of EscE.
The orf13 gene, located upstream of esrC and downstream of esaD in the E. tarda T3SS gene cluster (12), encodes an 8.7-kDa protein with a pI of 7.9. The Orf13 protein was predicted to contain three α-helices by PSIPRED (25) and shares 86% identity and 91% similarity with a hypothetical protein (GenBank accession no. WP_015870338.1) from the related bacterium Edwardsiella ictaluri 93-146 (26) and only 36% identity and 45.3% similarity to the CV2595 hypothetical protein from Chromobacterium violaceum (12). When domains were searched using Delta-Blast, the Orf13 protein was found to belong to the type III secretion system YscE family. However, within the family, Orf13 shares only 17% identity with YscE of Yersinia enterocolitica, 27% identity with VseE of Vibrio alginolyticus, 26% identity with SsaE of Salmonella enterica, and 20% identity with PscE and AscE of Pseudomonas aeruginosa and Aeromonas salmonicida, respectively. Orf13 was thus named EscE (Edwardsiella secretion chaperone E).
EscE is secreted in a T3SS-dependent manner.
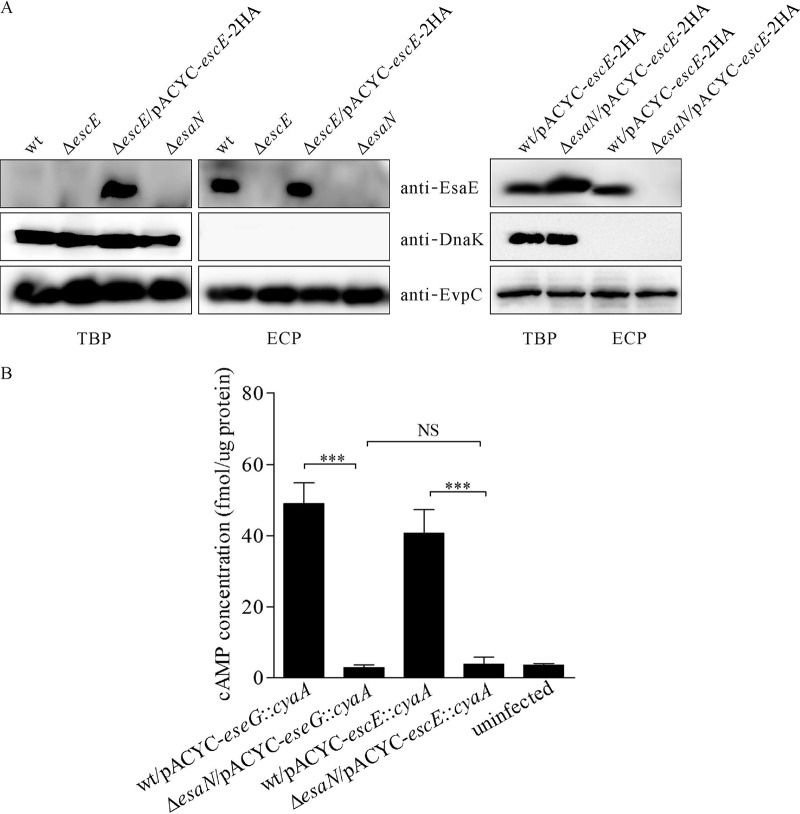
YscE family proteins function as chaperones for T3SS needle proteins. To study this function with EscE, equal amounts of ECP or TBP from wild-type E. tarda, the ΔescE strain, the ΔescE/pACYC-escE-2HA complemented mutant strain, and the ΔesaN strain were probed against EscE, DnaK, and EvpC. EsaN is an ATPase that energizes the transportation of T3SS effectors and translocon components, and mutation of esaN abolishes the secretion or translocation of T3SS substrates (13, 14). As shown in Fig. 1A, EscE was detected from ECP of wild-type E. tarda and the ΔescE/pACYC-escE-2HA strain, but not from that of the ΔescE or ΔesaN strain. EscE was not detected from TBP of the wild-type or ΔesaN strain but was detected readily from TBP of the ΔescE/pACYC-escE-2HA strain (Fig. 1A). There is one copy of escE in the genome, and pACYC184 is a medium-copy-number plasmid. This could explain why EscE was detected in the bacterial pellet of the ΔescE/pACYC-escE-2HA strain, but not from TBP of the wild-type or ΔesaN strain. It is also possible that in T3SS-inducing medium, most of the native EscE was secreted, making the trace EscE that remained hard to detect. To validate the T3S-dependent release of EscE, pACYC-escE-2HA was introduced into wild-type E. tarda and a ΔesaN mutant. Expression of EscE-2HA was readily detected from both wild-type E. tarda and the ΔesaN mutant, while EscE-2HA secretion was detected only from wild-type E. tarda (Fig. 1A). The cytoplasmic protein DnaK was not detected in any ECP, suggesting that the EscE that was detected from the ECP was not due to leakage from the bacterial pellets. EvpC is one of the major type VI secretion system (T6SS)-secreted proteins (21) and was used as a loading control in this study. Overall, our results indicate that EscE could be secreted and that the secretion of EscE is T3SS dependent.
FIG 1.
EscE is secreted and translocated in a T3SS-dependent manner. (A) EscE is secreted into the culture supernatant in a T3SS-dependent manner. Five percent of the TBP and 10% of the ECP from similar amounts of bacteria grown in DMEM were separated using Tricine–SDS-PAGE and transferred onto PVDF membranes for immunoblotting. (B) EscE is translocated into J774A.1 cells. J774A.1 cells were infected with E. tarda strains carrying the plasmid pACYC-eseG::cyaA or pACYC-escE::cyaA, and intracellular cAMP levels were determined at 5 h postinfection. Means and SD from the results of one representative experiment are shown. ***, P < 0.001; NS, not significant.
EscE is translocated into host cells in a T3SS-dependent manner.
To examine whether EscE could be translocated into host cells, we constructed a reporter plasmid, pACYC-escE::cyaA, from which a chimeric protein, EscE::CyaA, was expressed by fusing EscE to amino acids 2 to 406 of the adenylate cyclase toxin (CyaA) from Bordetella pertussis. In this system, if the CyaA fusion is translocated into the host cell cytoplasm, CyaA will convert ATP into cAMP in the presence of the eukaryotic-cell cytoplasmic protein calmodulin. The resulting increase in cAMP can then be measured by immunoassay (27). CyaA fusions have been used in many bacteria, including E. tarda, to successfully monitor the translocation of bacterial effectors (13, 28–30). J774A.1 cells were infected with E. tarda strains expressing the CyaA hybrid proteins to study their translocation, and EseG::CyaA was used as a positive control. As shown in Fig. 1B, the cAMP level in wt-infected cells was 40.75 ± 6.64 fmol/μg of host protein for EscE::CyaA and 49.02 ± 5.82 fmol/μg of host protein for EseG::CyaA, whereas the cAMP level of EseG::CyaA or EscE::CyaA was less than 3 fmol/μg of host protein in cells infected with the ΔesaN strain, which is similar to the uninfected control. These results indicate that EscE is translocated into J774A.1 cells in a T3SS-dependent manner.
EscE is required for the secretion of T3SS translocators and effectors.
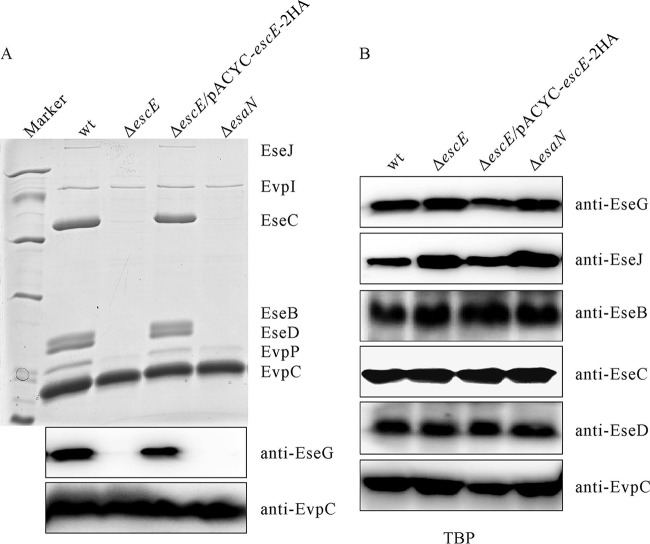
The T3SS translocator component EseB is involved in E. tarda autoaggregation (14, 22). However, the ΔescE mutant strain did not autoaggregate when cultured in DMEM (data not shown). To examine whether deletion of escE influences the secretion profile of E. tarda, the ECP of wild-type E. tarda and the ΔescE, ΔescE/pACYC-escE-2HA, and ΔesaN strains were compared. On SDS-PAGE gels, EseB, as well as two other T3SS translocator components (EseC and EseD), was absent from the ΔescE mutant strain. Unexpectedly, deletion of escE also abolished the secretion of the T3SS effector EseJ (Fig. 2A, top) and the T3SS effector EseG (Fig. 2A, bottom). The secretion levels of EvpI and EvpC did not change in the ΔescE mutant strain (Fig. 2A, top). The protein levels of T3SS translocators and effectors inside E. tarda did not change with the deletion of escE (Fig. 2B). Equivalent loading levels were evident when EvpC abundances among the lanes were examined. Together, these results indicate that EscE is necessary for the secretion of T3SS translocators and effectors in E. tarda.
FIG 2.
EscE controls the secretion of T3SS translocators and effectors. (A) (Top) Secretion profiles of E. tarda strains. ECP from equal amounts of bacteria grown in DMEM were separated using SDS-PAGE and stained with Coomassie blue. T3SS proteins (EseJ, EseC, EseB, and EseD) and T6SS proteins (EvpI, EvpP, and EvpC) were labeled. (Bottom) ECP separated using SDS-PAGE was transferred onto a PVDF membrane for immunoblotting against EseG. EvpC, a T6SS-secreted protein, was used to indicate the protein-loading level. (B) EscE does not interfere with the protein abundance of T3SS translocators or effectors. Bacteria grown in DMEM were washed twice with PBS, and proteins from equal amounts of E. tarda wild-type PPD130/91, ΔescE, ΔescE/pACYC-escE-2HA, and ΔesaN strains were separated by 12% SDS-PAGE and transferred onto PVDF membranes for immunoblotting.
The N terminus of EscE is crucial for a fully functional E. tarda T3SS.
The N terminus is commonly the region of effector proteins involved in directing their secretion (27, 29). To investigate whether the N terminus of EscE is engaged in its own secretion, pACYC-escEΔ2–15-2HA was introduced into wild-type and ΔescE E. tarda, and their secretion profiles were compared through immunoblotting. As shown in Fig. 3, expression of EscEΔ2–15-2HA in the ΔescE strain failed to restore the secretion of T3SS translocators, effectors, and EscEΔ2–15-2HA. EscEΔ2–15-2HA could be secreted by the wild type, albeit at a decreased level. This suggests that full-length EscE is required for an active T3SS and that the N terminus of EscE is not indispensable for its own secretion in wild-type E. tarda.
FIG 3.
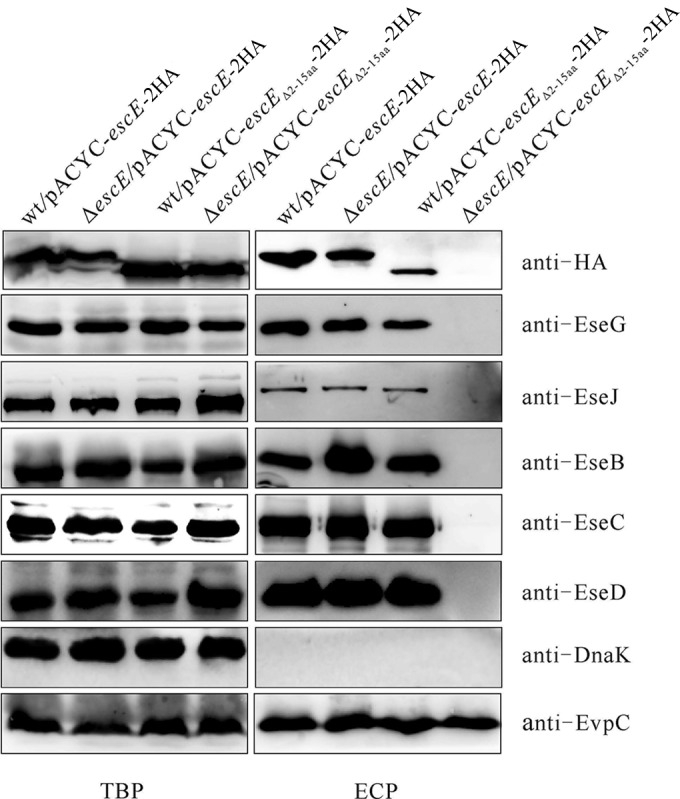
The N terminus of EscE is required for a fully functional T3SS. pACYC-escE-2HA and pACYC-escEΔ2–15-2HA were transformed into the E. tarda wild-type and ΔescE mutant strains, and their TBP and ECP from similar amounts of bacteria cultured in DMEM were compared by immunoblotting against HA, EseG, EseJ, EseB, EseC, EseD, EvpC, and DnaK.
The cell-bound form of EscE restores translocon and effector secretion from the escE mutant.
EscEΔ2–15 could not be secreted by the ΔescE mutant, whereas it could be released by wild-type E. tarda (Fig. 3). This suggested to us that expression or secretion of full-length EscE may trigger T3SS protein secretion. To examine this, we fused glutathione S-transferase (GST) to the N termini of type III-secreted substrates to block them from being released by the T3SS (31). pACYC-gst::escE-2HA was introduced into the wild-type and ΔescE strains to produce GST::EscE-2HA. As expected, this EscE derivative was not secreted by the escE mutant; however, secretion of the translocons and effectors was restored to wild-type levels (Fig. 4). These results indicate that the intracellular level of EscE ensures the secretion of T3SS proteins and that this secretion is independent of the secretion ability of EscE itself.
FIG 4.
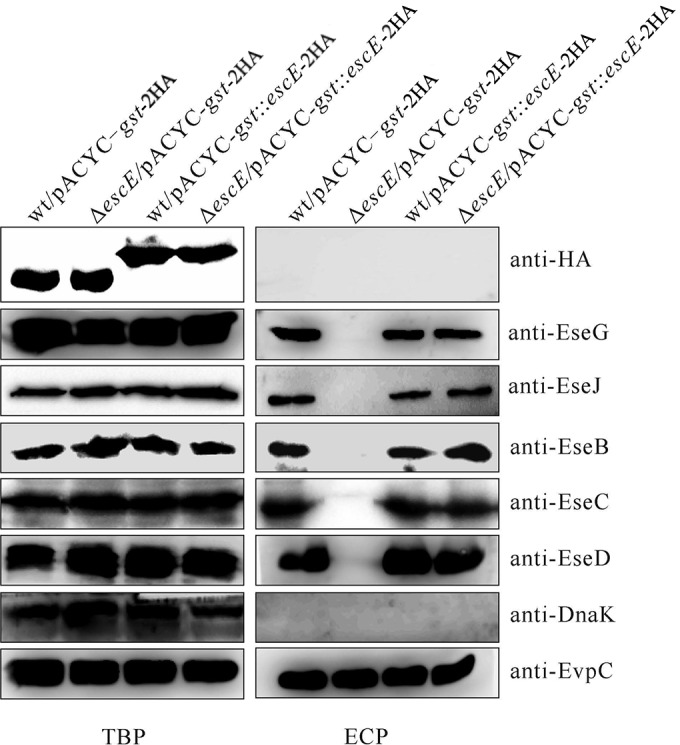
EscE controls T3SS protein secretion from within E. tarda. pACYC-gst-2HA and pACYC-gst::escE-2HA were introduced into the E. tarda wild-type and escE mutant strains, and their TBP and ECP from similar amounts of bacteria cultured in DMEM were compared by immunoblotting against HA, EseG, EseJ, EseB, EseC, EseD, EvpC, and DnaK.
The secretion and translocation of T3SS effectors are triggered by EscE expression, and effector translocation is positively correlated with bacterial EscE expression.
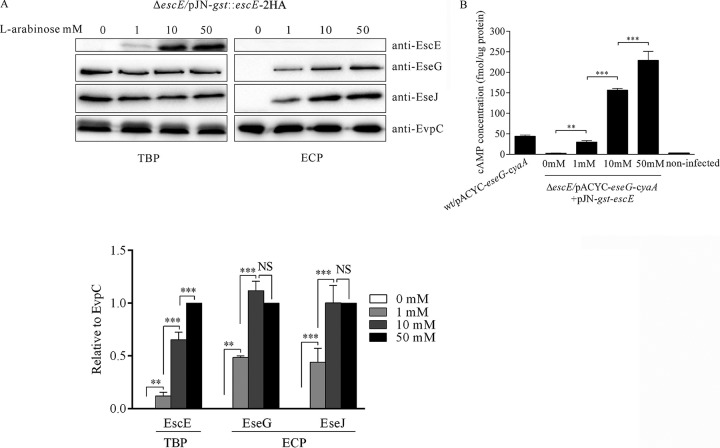
To examine whether the secretion of E. tarda effectors was influenced by EscE expression, we introduced the expression-inducible plasmid pJN105-gst::escE-2HA into E. tarda ΔescE and induced GST::EscE-2HA expression with l-arabinose. As shown in Fig. 5A, no GST::EscE-2HA was detected in the absence of l-arabinose, and no EseG or EseJ was secreted. When induced with 1 mM to 50 mM l-arabinose, the GST::EscE-2HA protein level increased accordingly, as did EseG and EseJ secretion (Fig. 5A). Protein levels of intracellular EscE, as well as the secreted EseG and EseJ, were analyzed by densitometry using EvpC as a loading control. We found that EseG and EseJ secretion increased proportionally to the EscE protein level when induced with l-arabinose at 0 mM to 10 mM. However, when l-arabinose increased from 10 mM to 50 mM, an obvious increase in EseG and EseJ secretion was not observed, suggesting a saturated level of effector secretion (Fig. 5A).
FIG 5.
EscE expression triggers the secretion and translocation of T3SS effectors. (A) The T3SS effectors secreted are proportional to the EscE level accumulated within bacteria. Five percent of the bacterial pellets (TBP) and 10% of the culture supernatants (ECP) from similar amounts of the ΔescE/pJN105-gst::escE-2HA strain were grown in DMEM; induced with 0 mM, 1 mM, 10 mM, or 50 mM l-arabinose; and then separated using SDS-PAGE and transferred onto PVDF membranes for immunoblotting against EscE, EseG, EseJ, and EvpC. The relative protein levels were normalized by EvpC densitometry. The graphs show the relative ratios (plus SD) of intracellular EscE, secreted EseG, and secreted EseJ, which are averages of the results of three independent experiments. (B) EseG translocation is proportional to EscE accumulation within E. tarda. J774A.1 cells were infected with the indicated E. tarda strains carrying the plasmids pACYC-eseG::cyaA and pACYC-gst::escE-2HA. During the infection, the DMEM was supplemented with 0 mM, 1 mM, 10 mM, or 50 mM l-arabinose. Intracellular cAMP levels were determined at 5 h postinfection. Means and SD from the results of one representative experiment are presented. ***, P < 0.001; **, P < 0.01; NS, not significant.
Next, we examined whether T3SS effector translocation would also be proportional to the protein level of EscE accumulated within E. tarda. pACYC-eseG::cyaA was introduced into the ΔescE/pJN105-gst::escE-2HA strain to infect J774A.1 cells, and the cell culture medium DMEM was supplemented with l-arabinose at 0 mM, 1 mM, 10 mM, and 50 mM. At 5 h postinfection, the intracellular cAMP level was measured as a readout of the translocation of EseG::CyaA. As shown in Fig. 5B, when l-arabinose was absent, the translocation of EseG::CyaA was almost identical to that of the uninfected control, which was less than 3.0 fmol/μg host protein. When l-arabinose was added, the cAMP level increased comparably (Fig. 5B). These data demonstrate that EseG translocation is positively correlated with the EscE accumulated inside E. tarda.
EscE contributes to E. tarda pathogenesis in vivo.
To determine the contribution of EscE to E. tarda pathogenesis, the survival rates of blue gourami fish were assessed following bacterial infections. To do this, fish were injected with 5 × 105 CFU of E. tarda per fish. Seven days postinfection, 6% of the fish infected with the wild type survived, whereas 69% of the fish infected with E. tarda ΔescE survived (Fig. 6A). To extend our results described above, we determined the LD50 of the ΔescE strain in blue gourami fish. Here, the LD50 refers to the minimum lethal dose of E. tarda required to kill half of the blue gourami fish tested within 7 days. We found that the LD50 of the E. tarda ΔescE strain was 105.43±0.12, which was 3 times higher than that of the wild-type strain (104.83±0.12). From the LD50 curve, when 104 CFU was injected per fish, 80% of the fish infected with the wild type survived, whereas 95% of those infected with the ΔescE strain remained alive. When 105 CFU was injected per fish, only 3.33% of the fish survived the wild-type infection compared to 35% of those infected with the ΔescE strain. No fish survived when 106 CFU of wild-type E. tarda was used, whereas 22.5% of those with ΔescE mutant strain infection remained alive (Fig. 6B). Together, these in vivo experiments demonstrate that EscE contributes to E. tarda pathogenesis.
FIG 6.
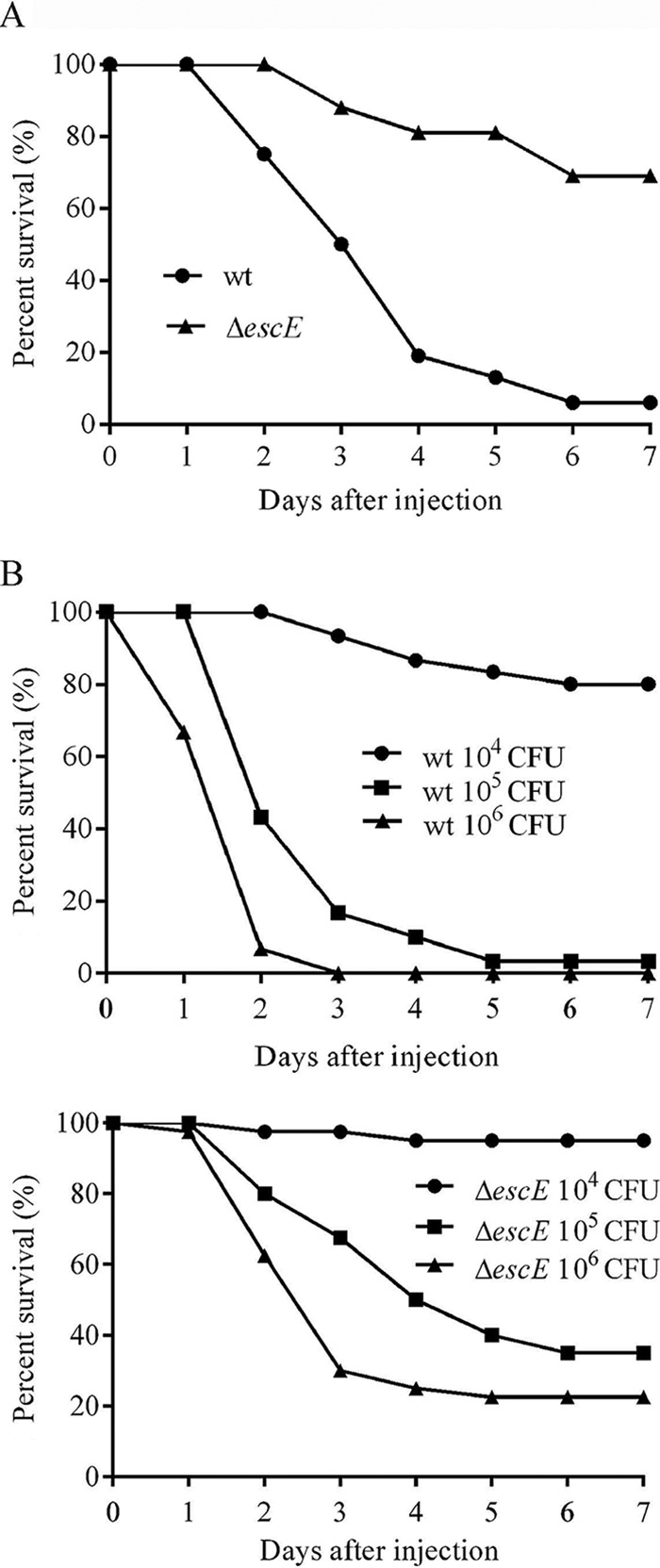
Survival curve of blue gourami fish infected with wild-type and ΔescE E. tarda. Fish survival was determined for a period of 7 days. (A) Groups of 16 fish were injected at a dosage of 5 × 105 CFU. (B) LD50 curves of wild-type and ΔescE mutant strains.
DISCUSSION
Bacterial type III secretion machines are used to translocate effector proteins directly from the bacterial cytosol into eukaryotic cells (2), and effector secretion/translocation are tightly controlled (32–34). In this study, we demonstrated that EscE expression triggers the secretion of T3SS translocators and effectors, and we also demonstrated that the secretion and translocation of T3SS effectors are positively correlated with the EscE protein level inside E. tarda.
When domains were examined, EscE was found to share similarities with T3SS YscE family proteins. YscE, together with YscG, functions as the chaperone of the T3SS needle protein, YscF, and deletion of yscE blocks the secretion of Yersinia outer proteins (Yops) (35). During infection, needle proteins are secreted first to complete the assembly of a fully functional protein export apparatus. This is required for the secretion of the T3SS intermediate (translocators) and late (effectors) substrates (3, 36, 37). Deletion of escE may abolish the secretion of the T3SS needle protein, which leads to the defect in the secretion of translocators and effectors. It is possible that EscE functions as a transient T3SS needle chaperone.
The requirement for EscE for an active T3SS in E. tarda is similar to that for HrpJ in Pseudomonas syringae. Like EscE, a hrpJ mutant cannot secrete translocators or inject effectors into plant cells (8, 38, 39). In addition, HrpJ itself is also a T3SS substrate that can be translocated, and secretion-incompetent HrpJ derivatives retain their regulatory roles (8). Another protein, HpaA from Xanthomonas campestris pathovar vesicatoria, is the T3SS control protein that promotes the secretion of many unrelated T3S substrates, including pilus, translocon, and effector proteins, and acts as a secreted regulator (40). Like the above-mentioned two proteins, EscE is also a control protein that titrates effectors to be secreted or translocated from within E. tarda.
In P. aeruginosa, the translocation of ExsE into Chinese hamster ovary cells is required for transcriptional induction of its T3SS (23). In the present study, EscE was not detected in cell lysates of wild-type E. tarda. Is EscE also required for the transcriptional induction of the T3SS through translocation or secretion? The expression of an EscE construct that was unable to pass through the T3SS in a ΔescE strain restored T3S protein secretion to the level in the wild type (Fig. 4), indicating that secretion and translocation of EscE were not required for transcriptional activation of T3SS in E. tarda. On the other hand, when expression of secretion-incompetent GST::EscE-2HA was not induced in the ΔescE mutant strain, we found a high level of effectors inside the bacteria, but no effector translocation was detected. When the medium was supplemented with 1 mM l-arabinose, GST::EscE-2HA expression was switched on and an increase in EseG translocation was detected. These observations indicate that the expression of EscE turned on T3SS effector translocation in E. tarda. Thus, it will be interesting to investigate how T3SS protein secretion or translocation is triggered by EscE expression and if another protein(s), together with EscE, ensures a defined order of T3SS secretion in E. tarda.
EscE expression triggers the secretion of T3SS translocators and switches on effector translocation. This led us to speculate that the intracellular EscE protein level might be fine-tuned through EscE secretion/translocation to accurately control the timing and the amount of proteins being secreted or translocated. EscE may control type III secretion near the top of the secretion hierarchy in E. tarda. It will be interesting to learn in the future what signals switch on EscE expression.
REFERENCES
- 1.Cornelis GR. 2006. The type III secretion injectisome. Nat Rev Microbiol 4:811–825. doi: 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- 2.Galán JE, Wolf-Watz H. 2006. Protein delivery into eukaryotic cells by type III secretion machines. Nature 444:567–573. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- 3.Deane JE, Abrusci P, Johnson S, Lea SM. 2010. Timing is everything: the regulation of type III secretion. Cell Mol Life Sci 67:1065–1075. doi: 10.1007/s00018-009-0230-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osborne SE, Coombes BK. 2011. Expression and secretion hierarchy in the nonflagellar type III secretion system. Future Microbiol 6:193–202. doi: 10.2217/fmb.10.172. [DOI] [PubMed] [Google Scholar]
- 5.Pallen MJ, Beatson SA, Bailey CM. 2005. Bioinformatics analysis of the locus for enterocyte effacement provides novel insights into type-III secretion. BMC Microbiol 5:9. doi: 10.1186/1471-2180-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Botteaux A, Sory MP, Biskri L, Parsot C, Allaoui A. 2009. MxiC is secreted by and controls the substrate specificity of the Shigella flexneri type III secretion apparatus. Mol Microbiol 71:449–460. doi: 10.1111/j.1365-2958.2008.06537.x. [DOI] [PubMed] [Google Scholar]
- 7.Ferracci F, Schubot FD, Waugh DS, Plano GV. 2005. Selection and characterization of Yersinia pestis YopN mutants that constitutively block Yop secretion. Mol Microbiol 57:970–987. doi: 10.1111/j.1365-2958.2005.04738.x. [DOI] [PubMed] [Google Scholar]
- 8.Crabill E, Karpisek A, Alfano JR. 2012. The Pseudomonas syringae HrpJ protein controls the secretion of type III translocator proteins and has a virulence role inside plant cells. Mol Microbiol 85:225–238. doi: 10.1111/j.1365-2958.2012.08097.x. [DOI] [PubMed] [Google Scholar]
- 9.Zheng J, Tung SL, Leung KY. 2005. Regulation of a type III and a putative secretion system in Edwardsiella tarda by EsrC is under the control of a two-component system, EsrA-EsrB. Infect Immun 73:4127–4137. doi: 10.1128/IAI.73.7.4127-4137.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chakraborty S, Li M, Chatterjee C, Sivaraman J, Leung KY, Mok Y-K. 2010. Temperature and Mg2+ sensing by a novel PhoP-PhoQ two-component system for regulation of virulence in Edwardsiella tarda. J Biol Chem 285:38876–38888. doi: 10.1074/jbc.M110.179150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chakraborty S, Sivaraman J, Leung KY, Mok Y-K. 2011. Two-component PhoB-PhoR regulatory system and ferric uptake regulator sense phosphate and iron to control virulence genes in type III and VI secretion systems of Edwardsiella tarda. J Biol Chem 286:39417–39430. doi: 10.1074/jbc.M111.295188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan YP, Zheng J, Tung SL, Rosenshine I, Leung KY. 2005. Role of type III secretion in Edwardsiella tarda virulence. Microbiology 151:2301–2313. doi: 10.1099/mic.0.28005-0. [DOI] [PubMed] [Google Scholar]
- 13.Xie HX, Lu JF, Zhou Y, Yi J, Yu XJ, Leung KY, Nie P. 2015. Identification and functional characterization of a novel Edwardsiella tarda effector EseJ. Infect Immun 83:1650–1660. doi: 10.1128/IAI.02566-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie HX, Yu HB, Zheng J, Nie P, Foster LJ, Mok YK, Leung KY. 2010. EseG, an effector of the type III secretion system of Edwardsiella tarda, triggers microtubule destabilization. Infect Immun 78:5011–5021. doi: 10.1128/IAI.00152-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang B, Mo ZL, Mao YX, Zou YX, Xiao P, Li J, Yang JY, Ye XH, Leung KY, Zhang PJ. 2009. Investigation of EscA as a chaperone for the Edwardsiella tarda type III secretion system putative translocon component EseC. Microbiology 155:1260–1271. doi: 10.1099/mic.0.021865-0. [DOI] [PubMed] [Google Scholar]
- 16.Zheng J, Li N, Tan YP, Sivaraman J, Mok Y-K, Mo ZL, Leung KY. 2007. EscC is a chaperone for the Edwardsiella tarda type III secretion system putative translocon components EseB and EseD. Microbiology 153:1953–1962. doi: 10.1099/mic.0.2006/004952-0. [DOI] [PubMed] [Google Scholar]
- 17.Ling SHM, Wang XH, Xie L, Lim TM, Leung KY. 2000. Use of green fluorescent protein (GFP) to study the invasion pathways of Edwardsiella tarda in in vivo and in vitro fish models. Microbiology 146:7–19. doi: 10.1099/00221287-146-1-7. [DOI] [PubMed] [Google Scholar]
- 18.Edwards RA, Keller LH, Schifferli DM. 1998. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207:149–157. doi: 10.1016/S0378-1119(97)00619-7. [DOI] [PubMed] [Google Scholar]
- 19.Simon R, Priefer U, Pühler A. 1983. A broad-host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. BioTechnology 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 20.Khlebnikov A, Skaug T, Keasling JD. 2002. Modulation of gene expression from the arabinose-inducible araBAD promoter. J Ind Microbiol Biotechnol 29:34–37. doi: 10.1038/sj.jim.7000259. [DOI] [PubMed] [Google Scholar]
- 21.Zheng J, Leung KY. 2007. Dissection of a type VI secretion system in Edwardsiella tarda. Mol Microbiol 66:1192–1206. doi: 10.1111/j.1365-2958.2007.05993.x. [DOI] [PubMed] [Google Scholar]
- 22.Gao ZP, Nie P, Lu JF, Liu LY, Xiao TY, Liu W, Liu JS, Xie HX. 2015. Type III secretion system translocon component EseB forms filaments on and mediates autoaggregation of and biofilm formation by Edwardsiella tarda. Appl Environ Microbiol 81:6078–6087. doi: 10.1128/AEM.01254-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Urbanowski ML, Brutinel ED, Yahr TL. 2007. Translocation of ExsE into Chinese hamster ovary cells is required for transcriptional induction of the Pseudomonas aeruginosa type III secretion system. Infect Immun 75:4432–4439. doi: 10.1128/IAI.00664-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reed LJ, Muench H. 1938. A simple method of estimating fifty percent end points. Am J Hyg 27:493–497. [Google Scholar]
- 25.McGuffin LJ, Bryson K, Jones DT. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 26.Williams ML, Gillaspy AF, Dyer DW, Thune RL, Waldbieser GC, Schuster SC, Gipson J, Zaitshik J, Landry C, Banes MM, Lawrence ML. 2012. Genome sequence of Edwardsiella ictaluri 93-146, a strain associated with a natural channel catfish outbreak of enteric septicemia of catfish. J Bacteriol 194:740–741. doi: 10.1128/JB.06522-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sory MP, Boland A, Lambermont I, Cornelis GR. 1995. Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc Natl Acad Sci U S A 92:11998–12002. doi: 10.1073/pnas.92.26.11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sory MP, Cornelis GR. 1994. Translocation of a hybrid YopE adenylatecyclase from Yersinia enterocolitica into HeLa cells. Mol Microbiol 14:583–594. doi: 10.1111/j.1365-2958.1994.tb02191.x. [DOI] [PubMed] [Google Scholar]
- 29.Schesser K, Frithz-Lindsten E, Wolf-Watz H. 1996. Delineation and mutational analysis of the Yersinia pseudotuberculosis YopE domains which mediate translocation across bacterial and eukaryotic cellular membranes. J Bacteriol 178:7227–7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Casper-Lindley C, Dahlbeck D, Clark ET, Staskawicz BJ. 2002. Direct biochemical evidence for type III secretion-dependent translocation of the AvrBs2 effector protein into plant cells. Proc Natl Acad Sci U S A 99:8336–8341. doi: 10.1073/pnas.122220299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riordan KE, Sorg JA, Berube BJ, Schneewind O. 2008. Impassable YscP substrates and their impact on the Yersinia enterocolitica type III secretion pathway. J Bacteriol 190:6204–6216. doi: 10.1128/JB.00467-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu XJ, Liu M, Holden DW. 2004. SsaM and SpiC interact and regulate secretion of Salmonella Pathogenicity Island 2 type III secretion system effectors and translocators. Mol Microbiol 54:604–619. doi: 10.1111/j.1365-2958.2004.04297.x. [DOI] [PubMed] [Google Scholar]
- 33.Wang D, Roe AJ, McAteer S, Shipston MJ, Gally DL. 2008. Hierarchal type III secretion of translocators and effectors from Escherichia coli O157:H7 requires the carboxy terminus of SepL that binds to Tir. Mol Microbiol 69:1499–1512. doi: 10.1111/j.1365-2958.2008.06377.x. [DOI] [PubMed] [Google Scholar]
- 34.Yu XJ, McGourty K, Liu M, Unsworth KE, Holden DW. 2010. pH sensing by intracellular Salmonella induces effector translocation. Science 328:1040–1043. doi: 10.1126/science.1189000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun P, Tropea JE, Austin BP, Cherry S, Waugh DS. 2008. Structural characterization of the Yersinia pestis type III secretion system needle protein YscF in complex with its heterodimeric chaperone YscE/YscG. J Mol Biol 377:819–830. doi: 10.1016/j.jmb.2007.12.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lara-Tejero M, Kato J, Wagner S, Liu X, Galán JE. 2011. A sorting platform determines the order of protein secretion in bacterial type III systems. Science 331:1188–1191. doi: 10.1126/science.1201476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Archuleta TL, Spiller BW. 2014. A gatekeeper chaperone complex directs translocation secretion during type three secretion. PLoS Pathog 10:e1004498. doi: 10.1371/journal.ppat.1004498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu ZQ, Guo M, Alfano JR. 2006. Pseudomonas syringae HrpJ is a type III secreted protein that is required for plant pathogenesis, injection of effectors, and secretion of the HrpZ1 harpin. J Bacteriol 188:6060–6069. doi: 10.1128/JB.00718-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei HL, Collmer A. 2012. Multiple lesions from the multiple functions of a regulator of type III secretion system assembly in the plant pathogen Pseudomonas syringae. Mol Microbiol 85:195–200. doi: 10.1111/j.1365-2958.2012.08119.x. [DOI] [PubMed] [Google Scholar]
- 40.Lorenz C, Kirchner O, Egler M, Stuttmann J, Bonas U, Büttner D. 2008. HpaA from Xanthomonas is a regulator of type III secretion. Mol Microbiol 69:344–360. doi: 10.1111/j.1365-2958.2008.06280.x. [DOI] [PubMed] [Google Scholar]